Improving the Efficiency of CRISPR/Cas9-Mediated Non-Homologous End Joining Gene Knockout Using Small Molecules in Porcine Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
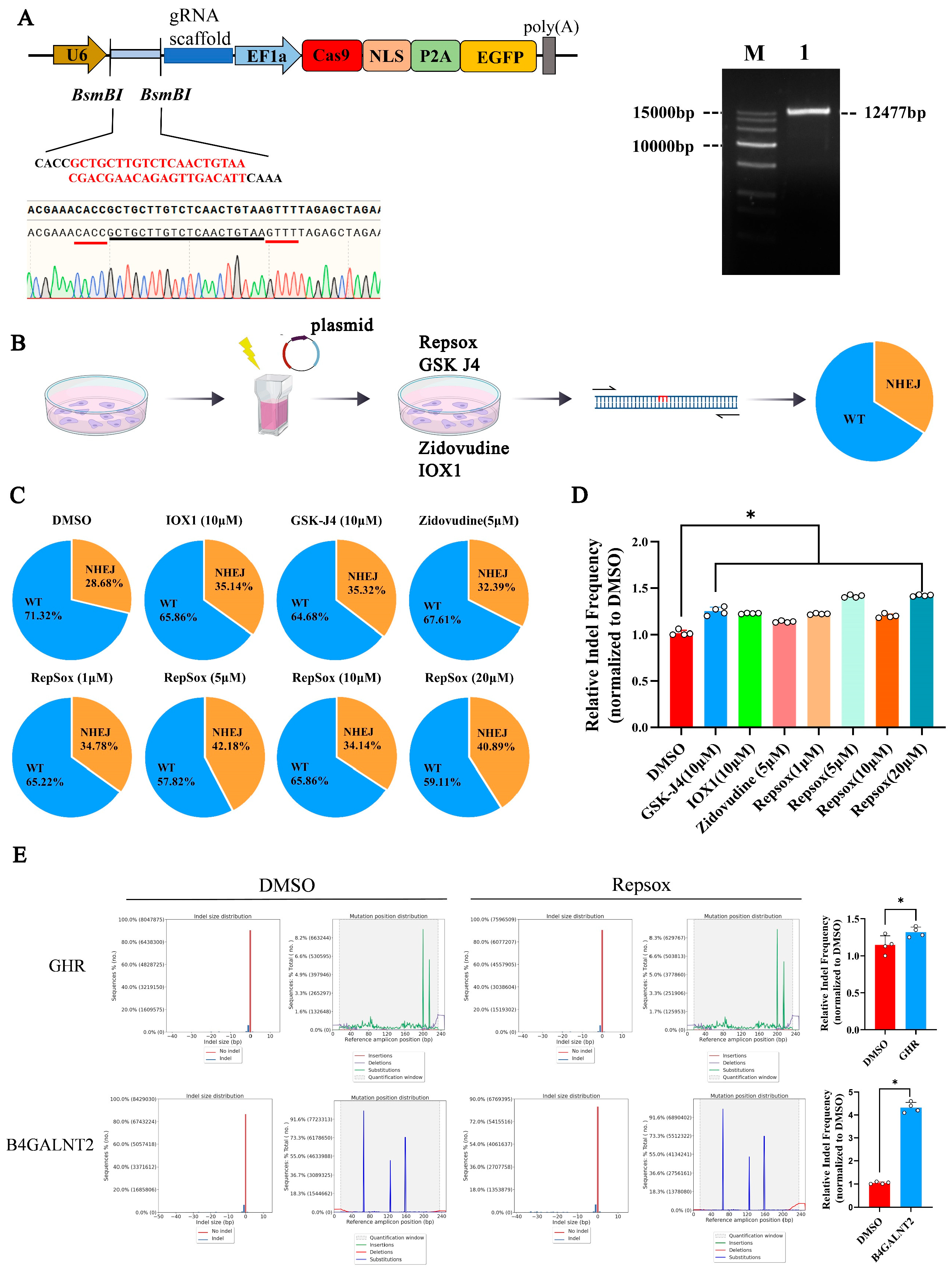
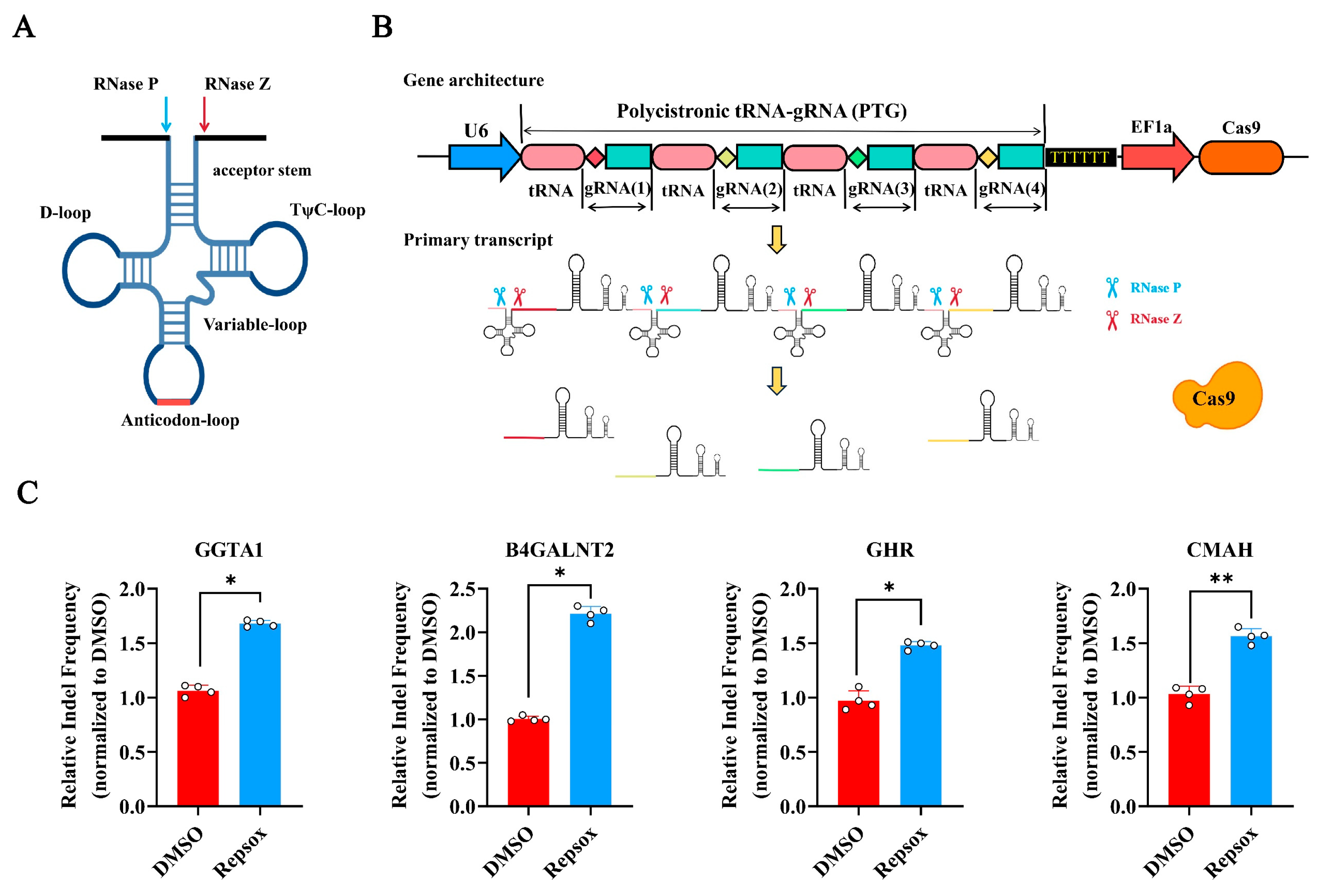
2.2. Design of gRNAs and Plasmid Construction
2.3. Cell Electroporation
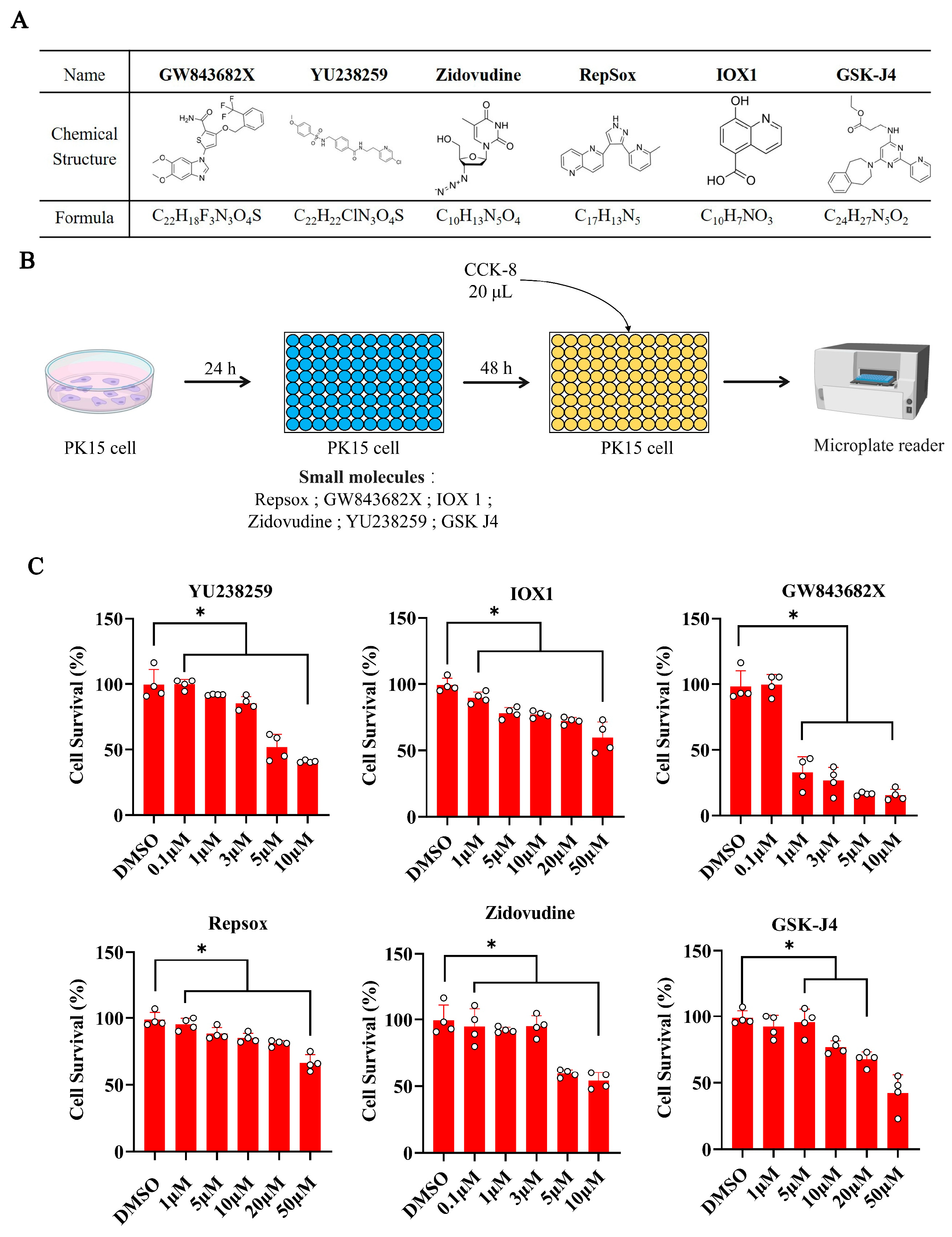
2.4. Cell Activity Assay
2.5. DNA Extraction and PCR
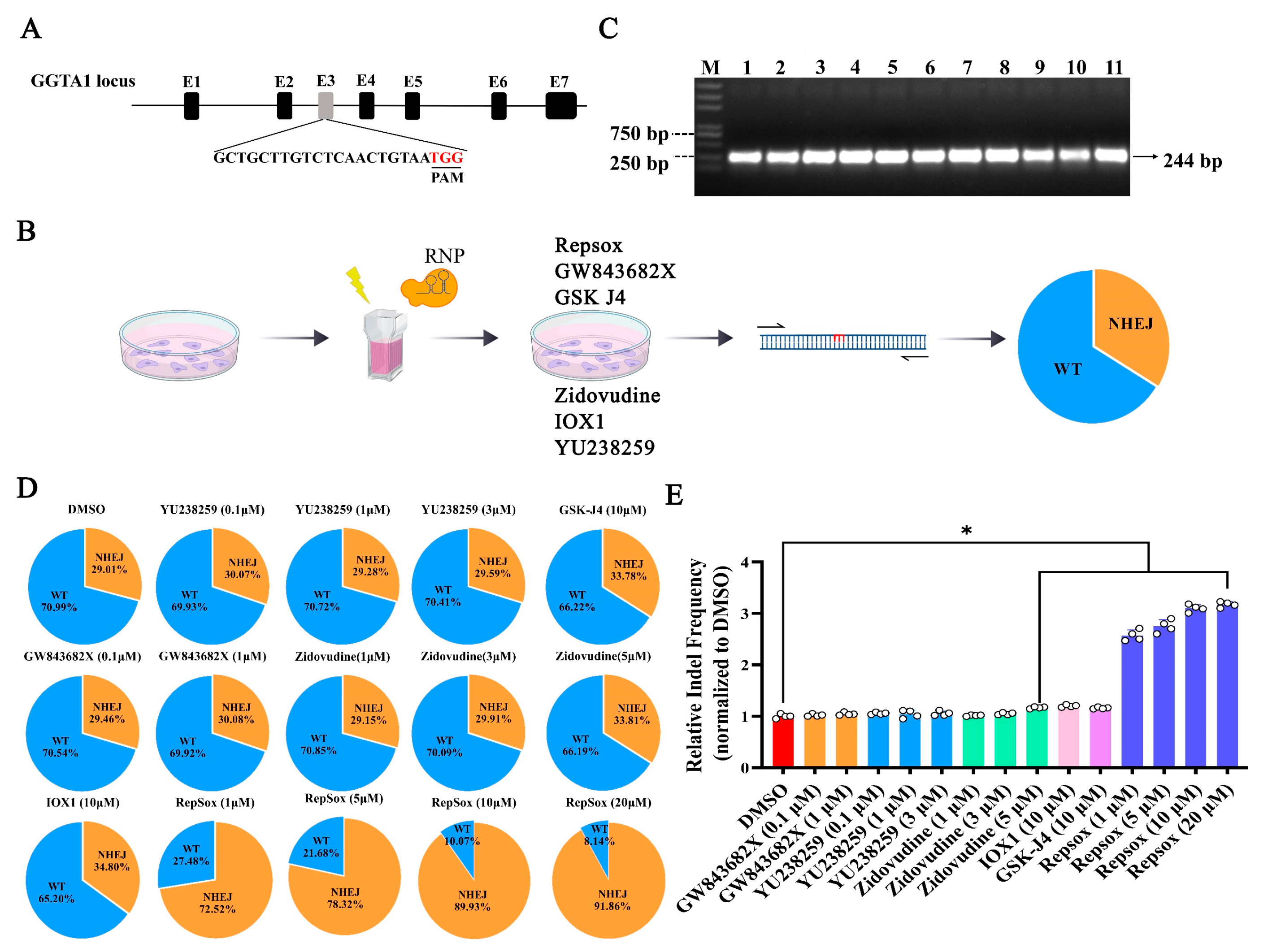
2.6. Deep Sequencing (Deep-Seq) and Analysis
2.7. Real-Time Quantitative PCR
2.8. Statistical Analysis
3. Results
3.1. The Effects of Different Concentrations of Small Molecules on Cell Viability
3.2. The Effects of Different Small Molecules on the Efficiency of CRISPR RNP-Mediated NHEJ Genome Editing
3.3. The Effects of Different Small Molecules on the Efficiency of CRISPR/Cas9 Plasmid Vector-Mediated NHEJ Genome Editing
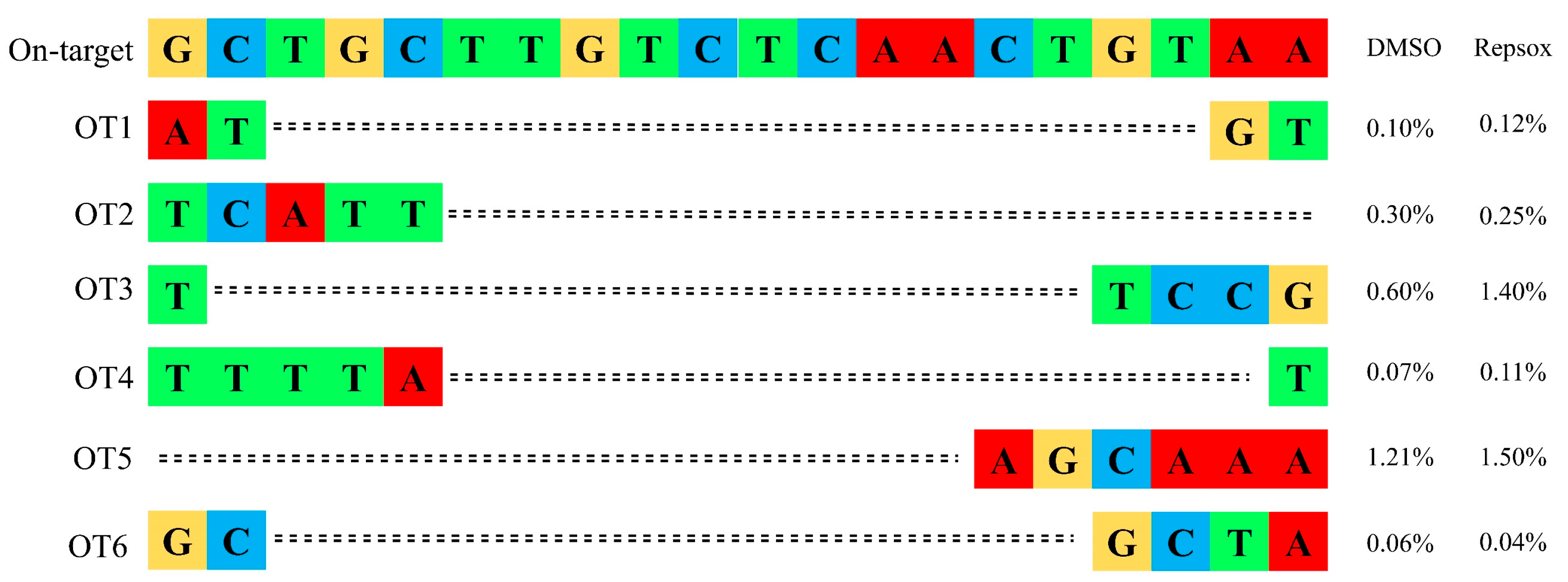
3.4. The Effect of Small Molecule Repsox on the Loading and Off-Target Efficiency of CRISPR/Cas9
3.5. The Effect of Small Molecule Repsox on the Efficiency of CRISPR/Cas9-Mediated Multi-Gene Editing
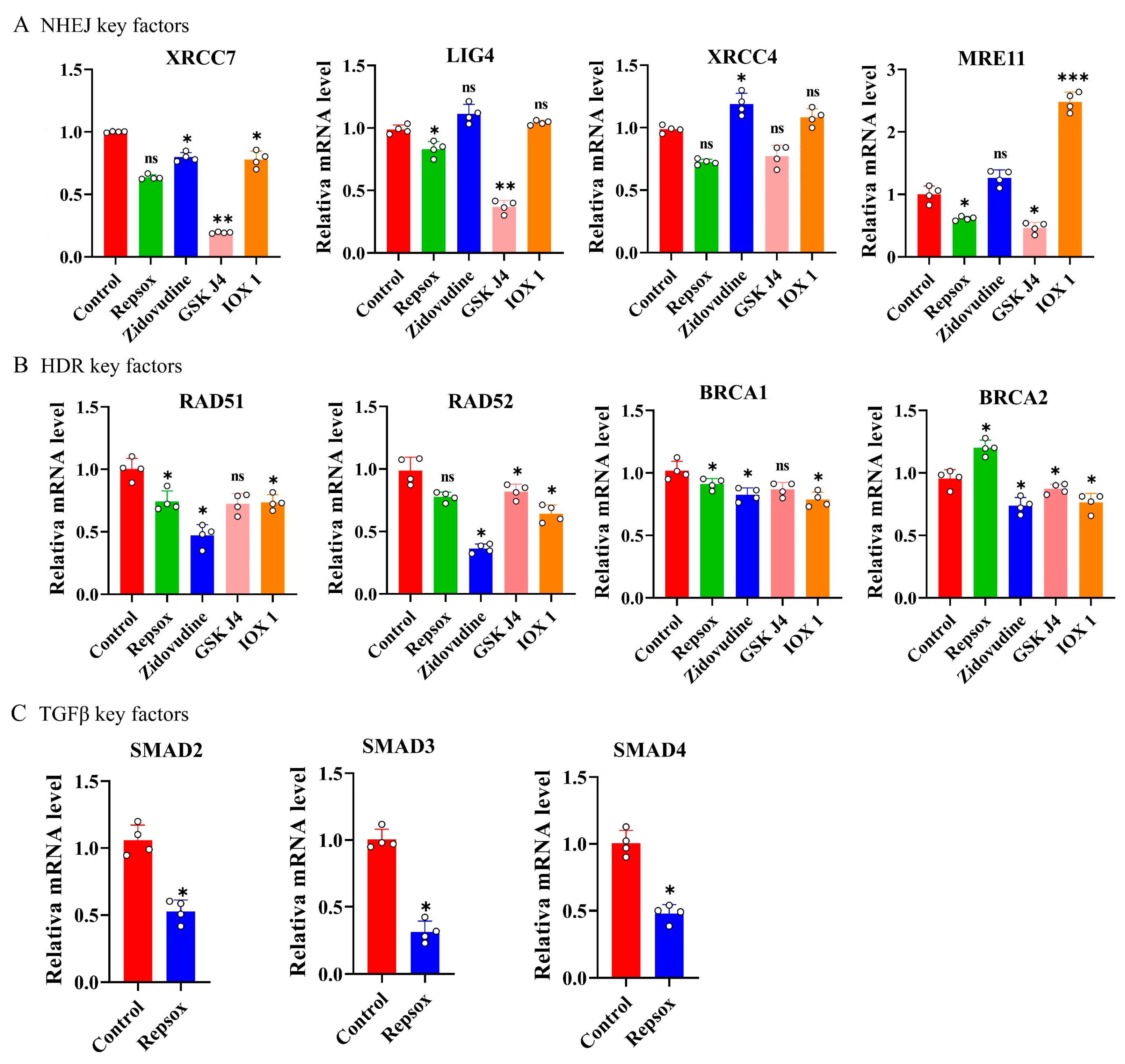
3.6. Expression Levels of Key Factors After Small-Molecule Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Doudna, J.A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Doench, J.G.; Hartenian, E.; Graham, D.B.; Tothova, Z.; Hegde, M.; Smith, I.; Sullender, M.; Ebert, B.L.; Xavier, R.J.; Root, D.E. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014, 32, 1262–1267. [Google Scholar] [CrossRef]
- Iyer, S.; Suresh, S.; Guo, D.; Daman, K.; Chen, J.C.J.; Liu, P.; Zieger, M.; Luk, K.; Roscoe, B.P.; Mueller, C.; et al. Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature 2019, 568, 561–565. [Google Scholar] [CrossRef]
- Zaboikin, M.; Zaboikina, T.; Freter, C.; Srinivasakumar, N. Non-Homologous End Joining and Homology Directed DNA Repair Frequency of Double-Stranded Breaks Introduced by Genome Editing Reagents. PLoS ONE 2017, 12, e0169931. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Yeh, C.D.; Richardson, C.D.; Corn, J.E. Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 2019, 21, 1468–1478. [Google Scholar] [CrossRef]
- Wu, W.Y.; Lebbink, J.H.G.; Kanaar, R.; Geijsen, N.; van der Oost, J. Genome editing by natural and engineered CRISPR-associated nucleases. Nat. Chem. Biol. 2018, 14, 642–651. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Doudna, J.A. The promise and challenge of therapeutic genome editing. Nature 2020, 578, 229–236. [Google Scholar] [CrossRef]
- Maresca, M.; Lin, V.G.; Guo, N.; Yang, Y. Obligate ligation-gated recombination (ObLiGaRe): Custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013, 23, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Tsunekawa, Y.; Hernandez-Benitez, R.; Wu, J.; Zhu, J.; Kim, E.J.; Hatanaka, F.; Yamamoto, M.; Araoka, T.; Li, Z.; et al. In Vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 2016, 540, 144–149. [Google Scholar] [CrossRef]
- Song, R.; Wang, Y.; Zheng, Q.; Yao, J.; Cao, C.; Wang, Y.; Zhao, J. One-step base editing in multiple genes by direct embryo injection for pig trait improvement. Sci. China Life Sci. 2022, 65, 739–752. [Google Scholar] [CrossRef]
- Xu, K.; Zhou, Y.; Mu, Y.; Liu, Z.; Hou, S.; Xiong, Y.; Fang, L.; Ge, C.; Wei, Y.; Zhang, X.; et al. CD163 and pAPN double-knockout pigs are resistant to PRRSV and TGEV and exhibit decreased susceptibility to PDCoV while maintaining normal production performance. Elife 2020, 9, e57132. [Google Scholar] [CrossRef]
- Anand, R.P.; Layer, J.V.; Heja, D.; Hirose, T.; Lassiter, G.; Firl, D.J.; Paragas, V.B.; Akkad, A.; Chhangawala, S.; Colvin, R.B.; et al. Design and testing of a humanized porcine donor for xenotransplantation. Nature 2023, 622, 393–401. [Google Scholar] [CrossRef]
- Stachelek, G.C.; Peterson-Roth, E.; Liu, Y.; Fernandez, R.J., 3rd; Pike, L.R.; Qian, J.M.; Abriola, L.; Hoyer, D.; Hungerford, W.; Merkel, J.; et al. YU238259 Is a Novel Inhibitor of Homology-Dependent DNA Repair That Exhibits Synthetic Lethality and Radiosensitization in Repair-Deficient Tumors. Mol. Cancer Res. 2015, 13, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Chen, S.; Rose, A.; Chen, D.; Cao, F.; Zwinderman, M.; Kiemel, D.; Aissi, M.; Dekker, F.J.; Haisma, H.J. Inhibition of histone deacetylase 1 (HDAC1) and HDAC2 enhances CRISPR/Cas9 genome editing. Nucleic Acids Res. 2020, 48, 517–532. [Google Scholar] [CrossRef]
- Yu, C.; Liu, Y.; Ma, T.; Liu, K.; Xu, S.; Zhang, Y.; Liu, H.; La Russa, M.; Xie, M.; Ding, S.; et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 2015, 16, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Mishra, T.; Bhardwaj, V.; Ahuja, N.; Gadgil, P.; Ramdas, P.; Shukla, S.; Chande, A. Improved loss-of-function CRISPR-Cas9 genome editing in human cells concomitant with inhibition of TGF-beta signaling. Mol. Ther.-Nucleic Acids 2022, 28, 202–218. [Google Scholar] [CrossRef] [PubMed]
- Clement, K.; Rees, H.; Canver, M.C.; Gehrke, J.M.; Farouni, R.; Hsu, J.Y.; Cole, M.A.; Liu, D.R.; Joung, J.K.; Bauer, D.E.; et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 2019, 37, 224–226. [Google Scholar] [CrossRef]
- Bento, A.P.; Gaulton, A.; Hersey, A.; Bellis, L.J.; Chambers, J.; Davies, M.; Kruger, F.A.; Light, Y.; Mak, L.; McGlinchey, S.; et al. The ChEMBL bioactivity database: An update. Nucleic Acids Res. 2014, 42, D1083–D1090. [Google Scholar] [CrossRef]
- Dong, F.; Xie, K.; Chen, Y.; Yang, Y.; Mao, Y. Polycistronic tRNA and CRISPR guide-RNA enables highly efficient multiplexed genome engineering in human cells. Biochem. Biophys. Res. Commun. 2017, 482, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Yang, Y. A Multiplexed CRISPR/Cas9 Editing System Based on the Endogenous tRNA Processing. Methods Mol. Biol. 2019, 1917, 63–73. [Google Scholar] [CrossRef]
- Li, G.; Zhang, X.; Zhong, C.; Mo, J.; Quan, R.; Yang, J.; Liu, D.; Li, Z.; Yang, H.; Wu, Z. Small molecules enhance CRISPR/Cas9-mediated homology-directed genome editing in primary cells. Sci. Rep. 2017, 7, 8943. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef]
- Ryan, D.E.; Taussig, D.; Steinfeld, I.; Phadnis, S.M.; Lunstad, B.D.; Singh, M.; Vuong, X.; Okochi, K.D.; McCaffrey, R.; Olesiak, M.; et al. Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2018, 46, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Sander, J.D.; Reyon, D.; Cascio, V.M.; Joung, J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014, 32, 279–284. [Google Scholar] [CrossRef]
- Perez-Pinera, P.; Kocak, D.D.; Vockley, C.M.; Adler, A.F.; Kabadi, A.M.; Polstein, L.R.; Thakore, P.I.; Glass, K.A.; Ousterout, D.G.; Leong, K.W.; et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 2013, 10, 973–976. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Amrani, N.; Gao, X.D.; Liu, P.; Edraki, A.; Mir, A.; Ibraheim, R.; Gupta, A.; Sasaki, K.E.; Wu, T.; Donohoue, P.D.; et al. NmeCas9 is an intrinsically high-fidelity genome-editing platform. Genome Biol. 2018, 19, 214. [Google Scholar] [CrossRef]
- Edraki, A.; Mir, A.; Ibraheim, R.; Gainetdinov, I.; Yoon, Y.; Song, C.Q.; Cao, Y.; Gallant, J.; Xue, W.; Rivera-Perez, J.A.; et al. A Compact, High-Accuracy Cas9 with a Dinucleotide PAM for In Vivo Genome Editing. Mol. Cell. 2019, 73, 714–726.e4. [Google Scholar] [CrossRef]
- Yang, H.; Ren, S.; Yu, S.; Pan, H.; Li, T.; Ge, S.; Zhang, J.; Xia, N. Methods Favoring Homology-Directed Repair Choice in Response to CRISPR/Cas9 Induced-Double Strand Breaks. Int. J. Mol. Sci. 2020, 21, 6461. [Google Scholar] [CrossRef] [PubMed]
- Kruidenier, L.; Chung, C.W.; Cheng, Z.; Liddle, J.; Che, K.; Joberty, G.; Bantscheff, M.; Bountra, C.; Bridges, A.; Diallo, H.; et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 2012, 488, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Schiller, R.; Scozzafava, G.; Tumber, A.; Wickens, J.R.; Bush, J.T.; Rai, G.; Lejeune, C.; Choi, H.; Yeh, T.L.; Chan, M.C.; et al. A cell-permeable ester derivative of the JmjC histone demethylase inhibitor IOX1. ChemMedChem 2014, 9, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Di Stazio, M.; Foschi, N.; Athanasakis, E.; Gasparini, P.; d’Adamo, A.P. Systematic analysis of factors that improve homologous direct repair (HDR) efficiency in CRISPR/Cas9 technique. PLoS ONE 2021, 16, e0247603. [Google Scholar] [CrossRef] [PubMed]
- Kagita, A.; Lung, M.S.Y.; Xu, H.; Kita, Y.; Sasakawa, N.; Iguchi, T.; Ono, M.; Wang, X.H.; Gee, P.; Hotta, A. Efficient ssODN-Mediated Targeting by Avoiding Cellular Inhibitory RNAs through Precomplexed CRISPR-Cas9/sgRNA Ribonucleoprotein. Stem Cell Rep. 2021, 16, 985–996. [Google Scholar] [CrossRef]
- Hirakawa, M.P.; Krishnakumar, R.; Timlin, J.A.; Carney, J.P.; Butler, K.S. Gene editing and CRISPR in the clinic: Current and future perspectives. Biosci. Rep. 2020, 40, BSR20200127. [Google Scholar] [CrossRef]
- Hu, Q.; Lu, H.; Wang, H.; Li, S.; Truong, L.; Li, J.; Liu, S.; Xiang, R.; Wu, X. Break-induced replication plays a prominent role in long-range repeat-mediated deletion. EMBO J. 2019, 38, e101751. [Google Scholar] [CrossRef]
- Le, B.V.; Podszywalow-Bartnicka, P.; Maifrede, S.; Sullivan-Reed, K.; Nieborowska-Skorska, M.; Golovine, K.; Yao, J.C.; Nejati, R.; Cai, K.Q.; Caruso, L.B.; et al. TGFbetaR-SMAD3 Signaling Induces Resistance to PARP Inhibitors in the Bone Marrow Microenvironment. Cell Rep. 2020, 33, 108221. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Khan, A.R.; Yang, X.; Dong, B.; Ji, J.; Zhai, G. The reversal of chemotherapy-induced multidrug resistance by nanomedicine for cancer therapy. J. Control. Release 2021, 335, 1–20. [Google Scholar] [CrossRef]
- Okamoto, S.; Amaishi, Y.; Maki, I.; Enoki, T.; Mineno, J. Highly efficient genome editing for single-base substitutions using optimized ssODNs with Cas9-RNPs. Sci. Rep. 2019, 9, 4811. [Google Scholar] [CrossRef]
- Ma, X.; Chen, X.; Jin, Y.; Ge, W.; Wang, W.; Kong, L.; Ji, J.; Guo, X.; Huang, J.; Feng, X.H.; et al. Small molecules promote CRISPR-Cpf1-mediated genome editing in human pluripotent stem cells. Nat. Commun. 2018, 9, 1303. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, S.; Xu, X.; Yang, S.; Feng, M.; Yuan, Z.; Liu, X.; Jiang, C.; Song, J.; Mu, Y. Improving the Efficiency of CRISPR/Cas9-Mediated Non-Homologous End Joining Gene Knockout Using Small Molecules in Porcine Cells. Biomolecules 2025, 15, 1132. https://doi.org/10.3390/biom15081132
Lv S, Xu X, Yang S, Feng M, Yuan Z, Liu X, Jiang C, Song J, Mu Y. Improving the Efficiency of CRISPR/Cas9-Mediated Non-Homologous End Joining Gene Knockout Using Small Molecules in Porcine Cells. Biomolecules. 2025; 15(8):1132. https://doi.org/10.3390/biom15081132
Chicago/Turabian StyleLv, Shihao, Xiaokang Xu, Sijia Yang, Mingjie Feng, Zhongyu Yuan, Xueqing Liu, Chaoqian Jiang, Jun Song, and Yanshuang Mu. 2025. "Improving the Efficiency of CRISPR/Cas9-Mediated Non-Homologous End Joining Gene Knockout Using Small Molecules in Porcine Cells" Biomolecules 15, no. 8: 1132. https://doi.org/10.3390/biom15081132
APA StyleLv, S., Xu, X., Yang, S., Feng, M., Yuan, Z., Liu, X., Jiang, C., Song, J., & Mu, Y. (2025). Improving the Efficiency of CRISPR/Cas9-Mediated Non-Homologous End Joining Gene Knockout Using Small Molecules in Porcine Cells. Biomolecules, 15(8), 1132. https://doi.org/10.3390/biom15081132





