Crosstalk Between N6-Methyladenosine and Other Epigenetic Mechanisms in Central Nervous System Development and Disorders
Abstract
1. Introduction
2. Molecular Regulatory Mechanisms and Biological Functions of m6A
2.1. Molecular Regulatory Mechanisms of m6A
2.2. Biological Functions of m6A
3. The Role of m6A in the Development of the Central Nervous System
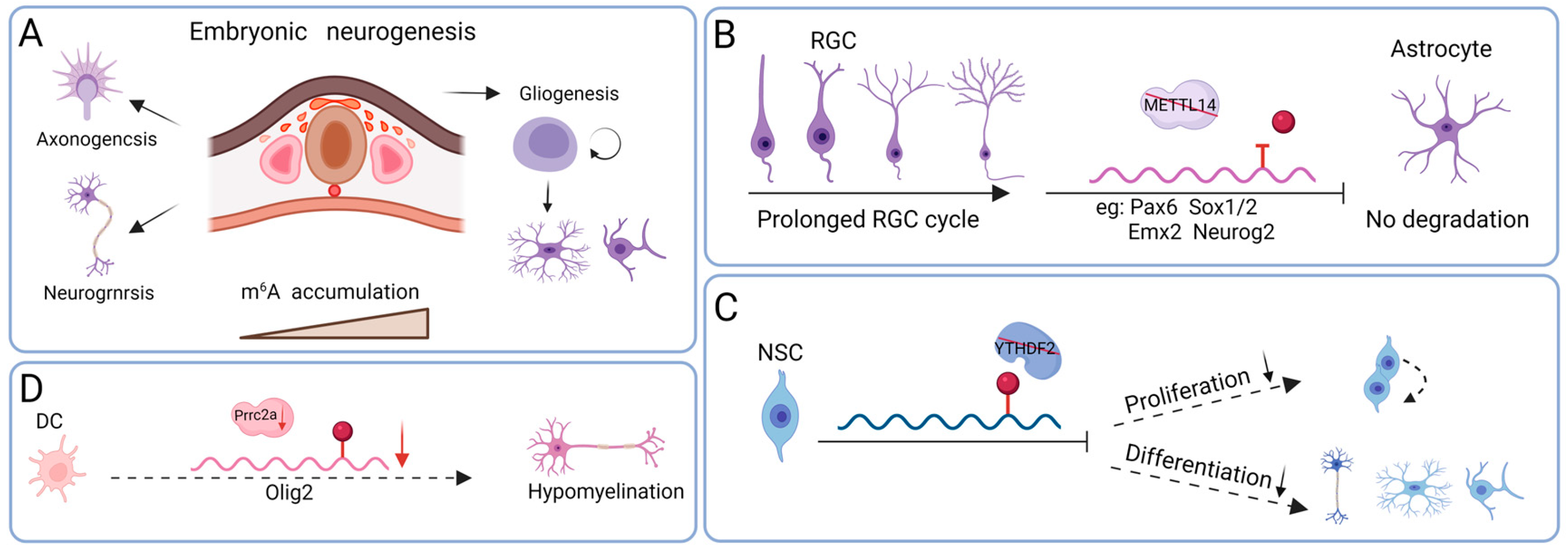
3.1. Neurogenesis
3.2. Gliogenesis
3.3. Axonal Growth
4. m6A Regulates Physiological Functions of the Central Nervous System
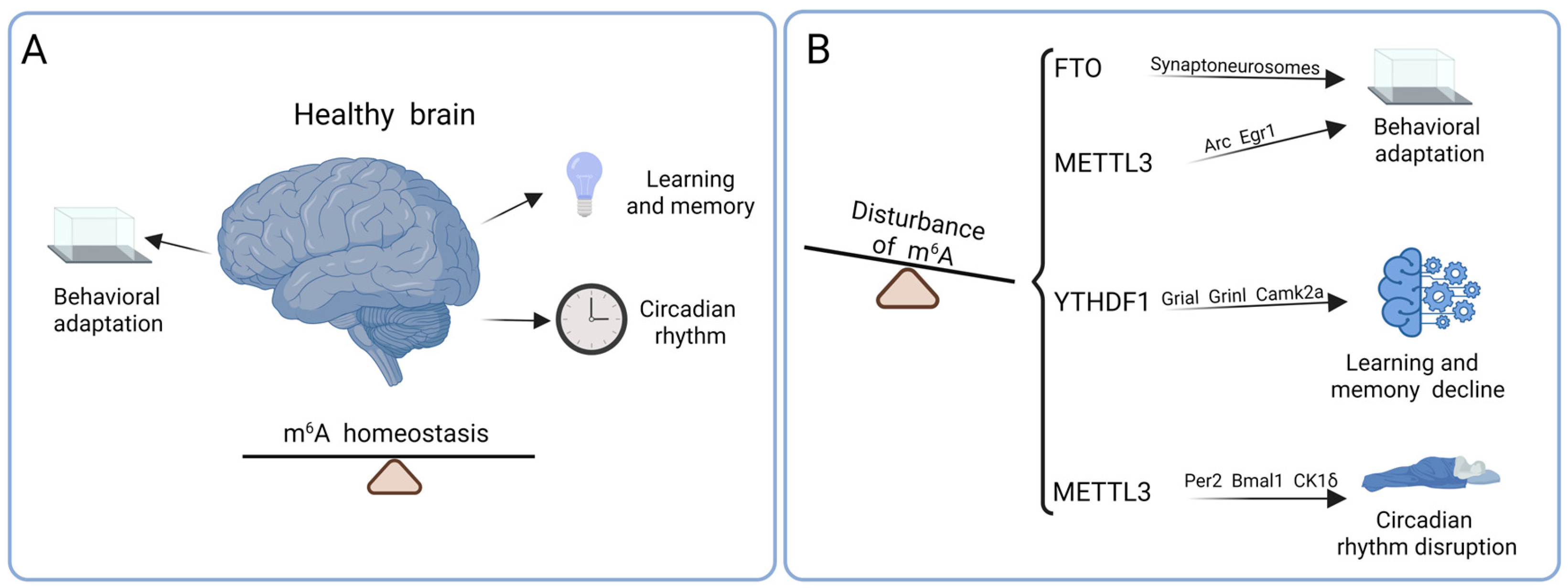
4.1. Behavioral Adaptation
4.2. Learning and Memory
4.3. Circadian Rhythm
5. The Regulation of Central Nervous System Development and Diseases by the Crosstalk Between m6A and Other Epigenetic Factors
5.1. Inter-Crosstalk Between m6A and Other Epigenetic Factors
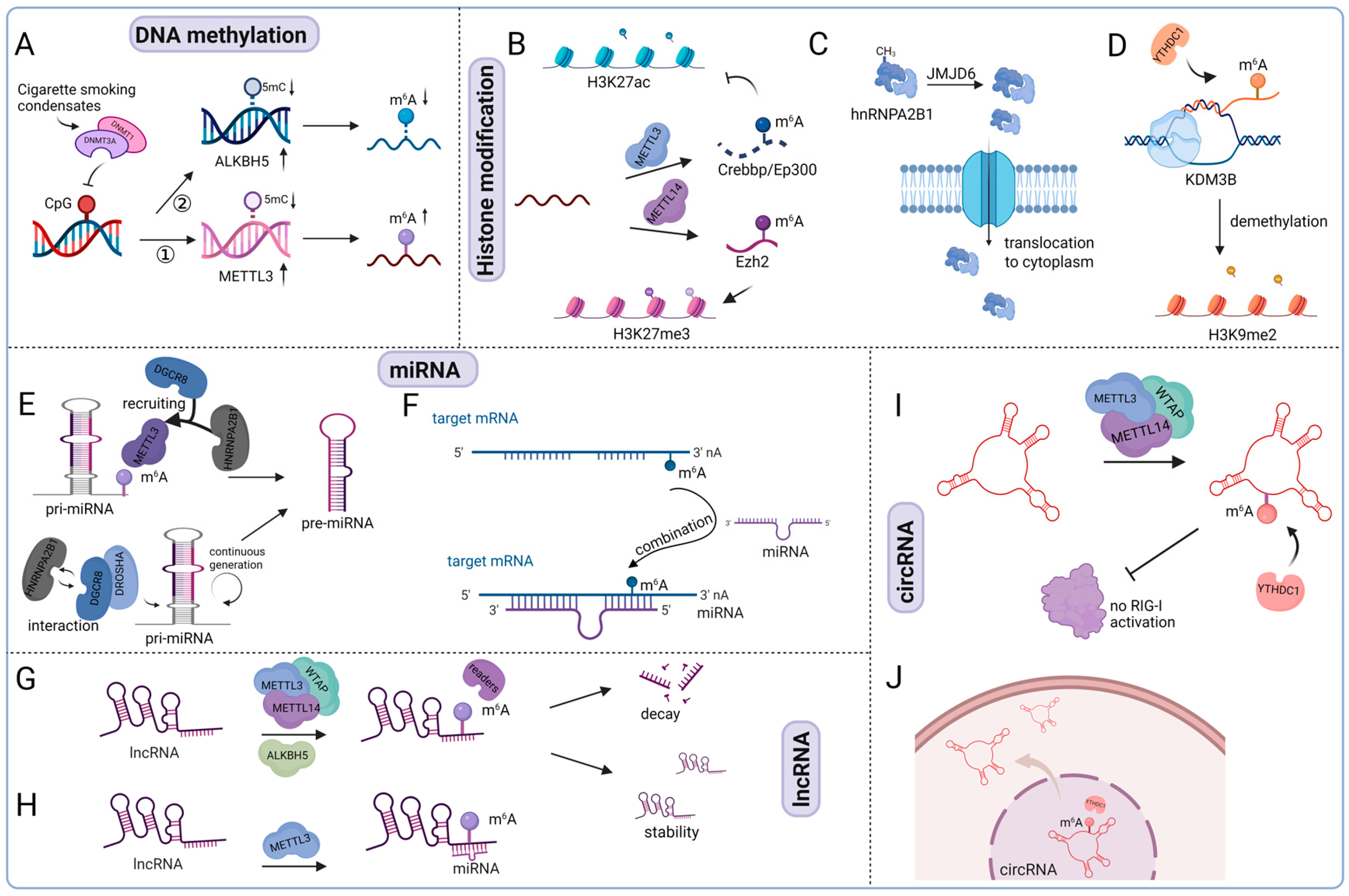
5.1.1. Inter-Crosstalk Between m6A and DNA Methylation
5.1.2. m6A and Histone Modification
5.1.3. m6A and ncRNAs (miRNAs, lncRNAs, and circRNAs)
5.2. The Regulatory Role of Crosstalk Between m6A and Other Epigenetic Factors in Central Nervous System Development and Diseases
5.2.1. Neurogenesis Disorder
5.2.2. Depression
5.2.3. Glioblastoma
5.2.4. Other Diseases
| Categories | m6A Related Enzymes | Categories of Epigenetics | Related Components | Mechanisms | Biological Influence | References |
|---|---|---|---|---|---|---|
| Neurodevelopmental disorders | METTL14 | Histone modification | Crebbp and Ep300 | Upregulated H3K27ac level, inhibited proliferation genes, and activated differentiation genes | Disordered NSC ground state | [98] |
| METTL3 | EZH2 | Inhibited H3K27me3 level; regulated P53 signaling pathway | Promoted ANSC to glial cell line; affected the development of newborn neurons | [13] | ||
| YTHDF1YTHDF3 | ncRNA | LncRNA Dubr | Accelerated degradation of YTHDF1 and YTHDF3; affected translation of Calmodulin and Tau | Inhibited axon growth; affected cortical neuron migration | [12] | |
| Depression | FTO | DNA methylation | Dnmt1and Dnmt3a | Downregulated FTO expression | Influenced plasticity-related gene expression | [58] |
| METTL3 | ncRNA | pri-miR-221 | Upregulated miR-221-3p; inhibited Gab1 expression | Induced cognitive impairment | [107] | |
| ALKBH5 | circRNA STAG1 | Reduced ALKBH5 level; promoted FAAH degradation | Induced depression-like behavior | [108] | ||
| Glioblastoma | METTL3 | Histone modification | EZH2 | Increased H3K27ac level; degraded nonsense-mediated mRNA | Increased drug resistance | [112] |
| JMJD1C | Inhibited SOCS2 expression | Inhibited GBM growth | [113] | |||
| IGF2BP2 | ncRNA | lncRNA CASC9 | Increased the stability of HK2; promoted aerobic sugar degradation | Promoted GBM proliferation | [114] | |
| — | lncRNA RP11-552D4.1 | — | Reflected immune infiltration disorders; predicted GBM risk | [116] | ||
| — | lncRNA AC005229.3/SOX21- AS1/AL133523.1/ AC004847.1 | — | Reflected immune response function; predicted GBM prognosis | [117] | ||
| Autism | METTL3 | ncRNA | lncRNA MALAT1 | Downregulated SFRP2 expression; inhibited Wnt/β-catenin signaling pathway | Reduced autism-like symptoms and hippocampal neuronal apoptosis | [118] |
| Parkinson’s disease | — | ncRNA | lncRNA CDC5L and lncRNA STAT3 | Increased ROS production; enhanced autophagy | Induced dopaminergic neuron damage and death | [119, 120] |
| Stroke | METTL3 | ncRNA | LncRNA D63785 | Increased the accumulation of miR-422a | Induced neuronal apoptosis | [123] |
6. Therapeutic Potential of Targeting Crosstalk Between m6A and Other Epigenetic Modulators
7. Summary and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Delaunay, S.; Helm, M.; Frye, M. RNA modifications in physiology and disease: Towards clinical applications. Nat. Rev. Genet. 2024, 25, 104–122. [Google Scholar] [CrossRef]
- Lu, Z.; Lyu, Z.; Dong, P.; Liu, Y.; Huang, L. N6-methyladenosine RNA modification in stomach carcinoma: Novel insights into mechanisms and implications for diagnosis and treatment. Biochim. Biophys. Acta–Mol. Basis Dis. 2025, 1871, 167793. [Google Scholar] [CrossRef]
- Xie, Y.-Y.; Zhong, Z.-D.; Chen, H.-X.; Ren, Z.-H.; Qiu, Y.-T.; Lan, Y.-L.; Wu, F.; Kong, J.-W.; Luo, R.-J.; Zhang, D.; et al. Single-molecule direct RNA sequencing reveals the shaping of epitranscriptome across multiple species. Nat. Commun. 2025, 16, 5119. [Google Scholar] [CrossRef]
- Yan, L.; Guo, L. The role and mechanism of m6A methylation in diabetic nephropathy. Life Sci. 2025, 363, 123355. [Google Scholar] [CrossRef]
- Luo, Z.; Lou, L.; Qiu, W.; Xu, Z.; Xiao, X. Predicting N6-Methyladenosine Sites in Multiple Tissues of Mammals through Ensemble Deep Learning. Int. J. Mol. Sci. 2022, 23, 15490. [Google Scholar] [CrossRef]
- Zaccara, S.; Jaffrey, S.R. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell 2020, 181, 1582–1595.e18. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef]
- Liu, J.; Eckert, M.A.; Harada, B.T.; Liu, S.-M.; Lu, Z.; Yu, K.; Tienda, S.M.; Chryplewicz, A.; Zhu, A.C.; Yang, Y.; et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018, 20, 1074–1083. [Google Scholar] [CrossRef]
- Livneh, I.; Moshitch-Moshkovitz, S.; Amariglio, N.; Rechavi, G.; Dominissini, D. The m6A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020, 21, 36–51. [Google Scholar] [CrossRef]
- Cheng, T.; Zhang, J.; Li, H.; Diao, J.; Zhang, W.; Niu, J.; Kawaguchi, T.; Nakayama, J.-I.; Kataoka, K.; Gao, S. Identification and characterization of the de novo methyltransferases for eukaryotic N6-methyladenine (6mA). Sci. Adv. 2025, 11, eadq4623. [Google Scholar] [CrossRef]
- Sun, Q.; Zhu, J.; Zhao, X.; Huang, X.; Qu, W.; Tang, X.; Ma, D.; Shu, Q.; Li, X. Mettl3-m6A-NPY axis governing neuron–microglia interaction regulates sleep amount of mice. Cell Discov. 2025, 11, 10. [Google Scholar] [CrossRef]
- Huang, J.; Jiang, B.; Li, G.W.; Zheng, D.; Li, M.; Xie, X.; Pan, Y.; Wei, M.; Liu, X.; Jiang, X.; et al. m6A-modified lincRNA Dubr is required for neuronal development by stabilizing YTHDF1/3 and facilitating mRNA translation. Cell Rep. 2022, 41, 111693. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, Y.-C.; Huang, C.; Shen, H.; Sun, B.; Cheng, X.; Zhang, Y.-J.; Yang, Y.-G.; Shu, Q.; Yang, Y.; et al. m6A Regulates Neurogenesis and Neuronal Development by Modulating Histone Methyltransferase Ezh2. Genom. Proteom. Bioinform. 2019, 17, 154–168. [Google Scholar] [CrossRef]
- Feng, S.; Tellaetxe-Abete, M.; Zhang, Y.; Peng, Y.; Zhou, H.; Dong, M.; Larrea, E.; Xue, L.; Zhang, L.; Koziol, M.J. Single-cell discovery of m6A RNA modifications in the hippocampus. Genome Res. 2024, 34, 822–836. [Google Scholar] [CrossRef]
- Eun, J.W.; Cheong, J.Y.; Jeong, J.-Y.; Kim, H.S. A New Understanding of Long Non-Coding RNA in Hepatocellular Carcinoma—From m6A Modification to Blood Biomarkers. Cells 2023, 12, 2272. [Google Scholar] [CrossRef] [PubMed]
- Chokkalla, A.K.; Mehta, S.L.; Vemuganti, R. Epitranscriptomic regulation by m6A RNA methylation in brain development and diseases. J. Cereb. Blood Flow Metab. 2020, 40, 2331–2349. [Google Scholar] [CrossRef]
- Dermentzaki, G.; Lotti, F. New Insights on the Role of N6-Methyladenosine RNA Methylation in the Physiology and Pathology of the Nervous System. Front. Mol. Biosci. 2020, 7, 555372. [Google Scholar] [CrossRef]
- Sacco, M.T.; Bland, K.M.; Horner, S.M.; Heise, M.T. WTAP Targets the METTL3 m6A-Methyltransferase Complex to Cytoplasmic Hepatitis C Virus RNA to Regulate Infection. J. Virol. 2022, 96, e0099722. [Google Scholar] [CrossRef]
- Gu, J.; Cao, H.; Chen, X.; Zhang, X.D.; Thorne, R.F.; Liu, X. RNA m6A modifications regulate crosstalk between tumor metabolism and immunity. Wiley Interdiscip. Rev. RNA 2024, 15, e1829. [Google Scholar] [CrossRef]
- Satterwhite, E.R.; Mansfield, K.D. RNA methyltransferase METTL16: Targets and function. Wiley Interdiscip. Rev. RNA 2022, 13, e1681. [Google Scholar] [CrossRef]
- Azzam, S.K.; Alsafar, H.; Sajini, A.A. FTO m6A Demethylase in Obesity and Cancer: Implications and Underlying Molecular Mechanisms. Int. J. Mol. Sci. 2022, 23, 3800. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Zha, X.; Li, M.; Xia, X.; Wang, S. Insights into the m6A demethylases FTO and ALKBH5: Structural, biological function, and inhibitor development. Cell Biosci. 2024, 14, 108. [Google Scholar] [CrossRef]
- Nabeel-Shah, S.; Pu, S.; Burke, G.L.; Ahmed, N.; Braunschweig, U.; Farhangmehr, S.; Lee, H.; Wu, M.; Ni, Z.; Tang, H.; et al. Recruitment of the m6A/m6Am demethylase FTO to target RNAs by the telomeric zinc finger protein ZBTB. Genome Biol. 2024, 25, 240. [Google Scholar] [CrossRef]
- Wei, J.; Liu, F.; Lu, Z.; Fei, Q.; Ai, Y.; He, P.C.; Shi, H.; Cui, X.; Su, R.; Klungland, A.; et al. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 2018, 71, 973–985.e5. [Google Scholar] [CrossRef] [PubMed]
- Relier, S.; Ripoll, J.; Guillorit, H.; Amalric, A.; Achour, C.; Boissière, F.; Vialaret, J.; Attina, A.; Debart, F.; Choquet, A.; et al. FTO-mediated cytoplasmic m6Am demethylation adjusts stem-like properties in colorectal cancer cell. Nat. Commun. 2021, 12, 1716. [Google Scholar] [CrossRef] [PubMed]
- De La Cruz, B.M.; Markus, R.; Malla, S.; Haig, M.I.; Gell, C.; Sang, F.; Bellows, E.; Sherif, M.A.; McLean, D.; Lourdusamy, A.; et al. Modifying the m6A brain methylome by ALKBH5-mediated demethylation: A new contender for synaptic tagging. Mol. Psychiatry 2021, 26, 7141–7153. [Google Scholar] [CrossRef]
- Yu, F.; Wei, J.; Cui, X.; Yu, C.; Ni, W.; Bungert, J.; Wu, L.; He, C.; Qian, Z. Post-translational modification of RNA m6A demethylase ALKBH5 regulates ROS-induced DNA damage response. Nucleic Acids Res. 2021, 49, 5779–5797. [Google Scholar] [CrossRef]
- Kontur, C.; Jeong, M.; Cifuentes, D.; Giraldez, A.J. Ythdf m6A Readers Function Redundantly during Zebrafish Development. Cell Rep. 2020, 33, 108598. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, W.; Shen, F.; Yang, X.; Liu, H.; Dai, S.; Sun, X.; Huang, J.; Guo, Q. YTH Domain Proteins: A Family of m6A Readers in Cancer Progression. Front. Oncol. 2021, 11, 629560. [Google Scholar] [CrossRef]
- Zhou, Y.; Ćorović, M.; Hoch-Kraft, P.; Meiser, N.; Mesitov, M.; Körtel, N.; Back, H.; Vries, I.S.N.-D.; Katti, K.; Obrdlík, A.; et al. m6A sites in the coding region trigger translation-dependent mRNA decay. Mol. Cell 2024, 84, 4576–4593.e12. [Google Scholar] [CrossRef]
- Boo, S.H.; Ha, H.; Kim, Y.K. m1A and m6A modifications function cooperatively to facilitate rapid mRNA degradation. Cell Rep. 2022, 40, 111317. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Shi, Y.; Shen, H.; Xie, W. m6A-binding proteins: The emerging crucial performers in epigenetics. J. Hematol. Oncol. 2020, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Mathoux, J.; Henshall, D.C.; Brennan, G.P. Regulatory Mechanisms of the RNA Modification m6A and Significance in Brain Function in Health and Disease. Front. Cell. Neurosci. 2021, 15, 671932. [Google Scholar] [CrossRef] [PubMed]
- Ramesh-Kumar, D.; Guil, S. The IGF2BP family of RNA binding proteins links epitranscriptomics to cancer. Semin. Cancer Biol. 2022, 86, 18–31. [Google Scholar] [CrossRef]
- Edupuganti, R.R.; Geiger, S.; Lu, Z.; Wang, S.-Y.; Baltissen, M.P.A.; Jansen, P.W.T.C.; Rossa, M.; Müller, M.; Stunnenberg, H.G.; He, C.; et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 2017, 24, 870–878. [Google Scholar] [CrossRef]
- Qiao, Y.; Sun, Q.; Chen, X.; He, L.; Wang, D.; Su, R.; Xue, Y.; Sun, H.; Wang, H. Nuclear m6A reader YTHDC1 promotes muscle stem cell activation/proliferation by regulating mRNA splicing and nuclear export. eLife 2023, 12, e82703. [Google Scholar] [CrossRef]
- Zhou, K.I.; Shi, H.; Lyu, R.; Wylder, A.C.; Matuszek, Ż.; Pan, J.N.; He, C.; Parisien, M.; Pan, T. Regulation of Co-transcriptional Pre-mRNA Splicing by m6A through the Low-Complexity Protein hnRNPG. Mol. Cell 2019, 76, 70–81.e9. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Luo, G.-Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Hsu, P.J.; Shi, H.; Zhu, A.C.; Lu, Z.; Miller, N.; Edens, B.M.; Ma, Y.C.; He, C. The RNA-binding protein FMRP facilitates the nuclear export of N6-methyladenosine-containing mRNAs. J. Biol. Chem. 2019, 294, 19889–19895. [Google Scholar] [CrossRef]
- Hao, W.; Dian, M.; Zhou, Y.; Zhong, Q.; Pang, W.; Li, Z.; Zhao, Y.; Ma, J.; Lin, X.; Luo, R.; et al. Autophagy induction promoted by m6A reader YTHDF3 through translation upregulation of FOXO3 mRNA. Nat. Commun. 2022, 13, 5845. [Google Scholar] [CrossRef]
- Zhang, X.; Cong, T.; Wei, L.; Zhong, B.; Wang, X.; Sun, J.; Wang, S.; Xu, M.M.; Zhu, P.; Jiang, H.; et al. YTHDF3 modulates hematopoietic stem cells by recognizing RNA m6A modification on Ccnd1. Haematologica 2022, 107, 2381–2394. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wei, Q.; Jin, J.; Luo, Q.; Liu, Y.; Yang, Y.; Cheng, C.; Li, L.; Pi, J.; Si, Y.; et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020, 48, 3816–3831. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.A.; Lin, Y.; Duan, H.; Cheng, Y. eIF-Three to Tango: Emerging functions of translation initiation factor eIF3 in protein synthesis and disease. J. Mol. Cell Biol. 2020, 12, 403–409. [Google Scholar] [CrossRef]
- Smith, R.C.L.; Kanellos, G.; Vlahov, N.; Alexandrou, C.; Willis, A.E.; Knight, J.R.P.; Sansom, O.J. Translation initiation in cancer at a glance. J. Cell Sci. 2021, 134, jcs248476. [Google Scholar] [CrossRef]
- Lee, Y.; Choe, J.; Park, O.H.; Kim, Y.K. Molecular Mechanisms Driving mRNA Degradation by m6A Modification. Trends Genet. 2020, 36, 177–188. [Google Scholar] [CrossRef]
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Mol. Cell 2019, 74, 494–507.e8. [Google Scholar] [CrossRef]
- Zhang, F.; Kang, Y.; Wang, M.; Li, Y.; Xu, T.; Yang, W.; Song, H.; Wu, H.; Shu, Q.; Jin, P. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 2018, 27, 3936–3950. [Google Scholar] [CrossRef]
- Liu, Z.; Deng, K.; Su, Y.; Zhang, Z.; Shi, C.; Wang, J.; Fan, Y.; Zhang, G.; Wang, F. IGF2BP1-mediated the stability and protein translation of FGFR1 mRNA regulates myogenesis through the ERK signaling pathway. Int. J. Biol. Macromol. 2024, 280 Pt 3, 135989. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, L.; Kong, Y.; Li, K.; Li, J.; Xu, F.; Liang, S.; Chen, B. IGF2BP1 enhances the stability of SIK2 mRNA through m6A modification to promote non-small cell lung cancer progression. Biochem. Biophys. Res. Commun. 2023, 684, 149113. [Google Scholar] [CrossRef]
- Yoon, K.-J.; Ringeling, F.R.; Vissers, C.; Jacob, F.; Pokrass, M.; Jimenez-Cyrus, D.; Su, Y.; Kim, N.-S.; Zhu, Y.; Zheng, L.; et al. Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation. Cell 2017, 171, 877–889.e17. [Google Scholar] [CrossRef]
- Li, M.; Zhao, X.; Wang, W.; Shi, H.; Pan, Q.; Lu, Z.; Perez, S.P.; Suganthan, R.; He, C.; Bjørås, M.; et al. 2-mediated m6A mRNA clearance modulates neural development in mice. Genome Biol. 2018, 19, 69. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Li, A.; Sun, B.; Sun, J.-G.; Zhang, J.; Zhang, T.; Chen, Y.; Xiao, Y.; Gao, Y.; Zhang, Q.; et al. A novel m6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019, 29, 23–41. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Dzhashiashvili, Y.; Shah, A.; Kunjamma, R.B.; Weng, Y.-L.; Elbaz, B.; Fei, Q.; Jones, J.S.; Li, Y.I.; Zhuang, X.; et al. m6A mRNA Methylation Is Essential for Oligodendrocyte Maturation and CNS Myelination. Neuron 2020, 105, 293–309.e5. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Wang, S.; Li, J.; Wen, Y.; Cui, R.; Zhang, K.; Liu, Y.; Yang, X.; Zhang, L.; Xu, B.; et al. Protective role of mRNA demethylase FTO on axon guidance molecules of nigro-striatal projection system in manganese-induced parkinsonism. J. Hazard. Mater. 2022, 426, 128099. [Google Scholar] [CrossRef]
- Zhuang, M.; Li, X.; Zhu, J.; Zhang, J.; Niu, F.; Liang, F.; Chen, M.; Li, D.; Han, P.; Ji, S.-J. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 2019, 47, 4765–4777. [Google Scholar] [CrossRef]
- Widagdo, J.; Zhao, Q.-Y.; Kempen, M.-J.; Tan, M.C.; Ratnu, V.S.; Wei, W.; Leighton, L.; Spadaro, P.A.; Edson, J.; Anggono, V.; et al. Experience-Dependent Accumulation of N6 -Methyladenosine in the Prefrontal Cortex Is Associated with Memory Processes in Mice. J. Neurosci. 2016, 36, 6771–6777. [Google Scholar] [CrossRef]
- Walters, B.J.; Mercaldo, V.; Gillon, C.J.; Yip, M.; Neve, R.L.; Boyce, F.M.; Frankland, P.W.; Josselyn, S.A. The Role of The RNA Demethylase FTO (Fat Mass and Obesity-Associated) and mRNA Methylation in Hippocampal Memory Formation. Neuropsychopharmacology 2017, 42, 1502–1510. [Google Scholar] [CrossRef]
- Roy, B.; Ochi, S.; Dwivedi, Y. M6A RNA Methylation-Based Epitranscriptomic Modifications in Plasticity-Related Genes via miR-124-C/EBPα-FTO-Transcriptional Axis in the Hippocampus of Learned Helplessness Rats. Int. J. Neuropsychopharmacol. 2022, 25, 1037–1049. [Google Scholar] [CrossRef]
- Liu, S.-J.; Cai, T.-H.; Fang, C.-L.; Lin, S.-Z.; Yang, W.-Q.; Wei, Y.; Zhou, F.; Liu, L.; Luo, Y.; Guo, Z.-Y.; et al. Long-term exercise training down-regulates m6A RNA demethylase FTO expression in the hippocampus and hypothalamus: An effective intervention for epigenetic modification. BMC Neurosci. 2022, 23, 54. [Google Scholar] [CrossRef]
- Xue, A.; Huang, Y.; Li, M.; Wei, Q.; Bu, Q. Comprehensive Analysis of Differential m6A RNA Methylomes in the Hippocampus of Cocaine-Conditioned Mice. Mol. Neurobiol. 2021, 58, 3759–3768. [Google Scholar] [CrossRef]
- Sun, L.; Ma, L.; Zhang, H.; Cao, Y.; Wang, C.; Hou, N.; Huang, N.; von Deneen, K.M.; Zhao, C.; Shi, Y.; et al. Fto Deficiency Reduces Anxiety- and Depression-Like Behaviors in Mice via Alterations in Gut Microbiota. Theranostics 2019, 9, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, M.; Xie, D.; Huang, Z.; Zhang, L.; Yang, Y.; Ma, D.; Li, W.; Zhou, Q.; Yang, Y.-G.; et al. METTL3-mediated N6-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 2018, 28, 1050–1061. [Google Scholar] [CrossRef] [PubMed]
- Logan, R.W.; McClung, C.A. Rhythms of life: Circadian disruption and brain disorders across the lifespan. Nat. Rev. Neurosci. 2019, 20, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Fustin, J.-M.; Doi, M.; Yamaguchi, Y.; Hida, H.; Nishimura, S.; Yoshida, M.; Isagawa, T.; Morioka, M.S.; Kakeya, H.; Manabe, I.; et al. RNA-Methylation-Dependent RNA Processing Controls the Speed of the Circadian Clock. Cell 2013, 155, 793–806. [Google Scholar] [CrossRef]
- Wei, A.; Wu, H. Mammalian DNA methylome dynamics: Mechanisms, functions and new frontiers. Development 2022, 149, dev182683. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Y. Role of Mammalian DNA Methyltransferases in Development. Annu. Rev. Biochem. 2020, 89, 135–158. [Google Scholar] [CrossRef]
- Jones, P.A.; Takai, D. The Role of DNA Methylation in Mammalian Epigenetics. Science 2001, 293, 1068–1070. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef]
- Zhou, L.; Tian, S.; Qin, G. RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019, 20, 156. [Google Scholar] [CrossRef]
- Zhang, J.; Bai, R.; Li, M.; Ye, H.; Wu, C.; Wang, C.; Li, S.; Tan, L.; Mai, D.; Li, G.; et al. Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat. Commun. 2019, 10, 1858. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, L.; Deng, J.; Guo, B.; Li, F.; Wang, Y.; Wu, R.; Zhang, S.; Lu, J.; Zhou, Y. A Novel Micropeptide Encoded by Y-Linked LINC00278 Links Cigarette Smoking and AR Signaling in Male Esophageal Squamous Cell Carcinoma. Cancer Res. 2020, 80, 2790–2803. [Google Scholar] [CrossRef] [PubMed]
- Ramazi, S.; Allahverdi, A.; Zahiri, J. Evaluation of post-translational modifications in histone proteins: A review on histone modification defects in developmental and neurological disorders. J. Biosci. 2020, 45, 1–29. [Google Scholar] [CrossRef]
- Li, Y.; Xia, L.; Tan, K.; Ye, X.; Zuo, Z.; Li, M.; Xiao, R.; Wang, Z.; Liu, X.; Deng, M.; et al. N6-Methyladenosine co-transcriptionally directs the demethylation of histone H3K9me. Nat. Genet. 2020, 52, 870–877. [Google Scholar] [CrossRef]
- Wang, L.; Wen, M.; Cao, X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science 2019, 365, 6454. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Chen, C.; Chu, X. New insights into the regulatory role of microRNA in tumor angiogenesis and clinical implications. Mol. Cancer 2018, 17, 22. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef]
- Hu, X.; Peng, W.-X.; Zhou, H.; Jiang, J.; Zhou, X.; Huang, D.; Mo, Y.-Y.; Yang, L. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020, 27, 1782–1794. [Google Scholar] [CrossRef]
- Han, J.; Wang, J.Z.; Yang, X.; Yu, H.; Zhou, R.; Lu, H.C.; Yuan, W.B.; Lu, J.C.; Zhou, Z.J.; Lu, Q.; et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer 2019, 18, 110. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Y.; Qian, Z.; Zheng, W.; Wu, Q.; Chen, Y.; Zhu, G.; Liu, Y.; Bian, Z.; Xu, W.; et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 1091. [Google Scholar] [CrossRef]
- Cui, X.; Wang, Z.; Li, J.; Zhu, J.; Ren, Z.; Zhang, D.; Zhao, W.; Fan, Y.; Zhang, D.; Sun, R. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/β-catenin signalling pathway. Cell Prolif. 2020, 53, e12768. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Chen, Z.; Gao, W.; Zhang, Y.; Wang, J.; Wang, J.; Cao, M.; Cai, J.; Wu, J.; Wang, X. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. 2020, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Li, M.; Chang, H.; Wang, R.; Zhang, Z.; Zhang, J.; He, Y.; Ma, H. The m6A demethylase ALKBH5 promotes tumor progression by inhibiting RIG-I expression and interferon alpha production through the IKKε/TBK1/IRF3 pathway in head and neck squamous cell carcinoma. Mol. Cancer 2022, 21, 97. [Google Scholar] [CrossRef]
- Hu, Y.; Gong, C.; Li, Z.; Liu, J.; Chen, Y.; Huang, Y.; Luo, Q.; Wang, S.; Hou, Y.; Yang, S.; et al. Demethylase ALKBH5 suppresses invasion of gastric cancer via PKMYT1 m6A modification. Mol. Cancer 2022, 21, 34. [Google Scholar] [CrossRef]
- Yang, D.; Qiao, J.; Wang, G.; Lan, Y.; Li, G.; Guo, X.; Xi, J.; Ye, D.; Zhu, S.; Chen, W.; et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018, 46, 3906–3920. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef]
- Warda, A.S.; Kretschmer, J.; Hackert, P.; Lenz, C.; Urlaub, H.; Höbartner, C.; Sloan, K.E.; Bohnsack, M.T. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017, 18, 2004–2014. [Google Scholar] [CrossRef]
- Jin, D.; Guo, J.; Wu, Y.; Du, J.; Yang, L.; Wang, X.; Di, W.; Hu, B.; An, J.; Kong, L.; et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J. Hematol. Oncol. 2021, 14, 32. [Google Scholar] [CrossRef]
- Chen, L.-L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef]
- Zhou, C.; Molinie, B.; Daneshvar, K.; Pondick, J.V.; Wang, J.; Van Wittenberghe, N.; Xing, Y.; Giallourakis, C.C.; Mullen, A.C. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017, 20, 2262–2276. [Google Scholar] [CrossRef] [PubMed]
- Di Timoteo, G.; Dattilo, D.; Centrón-Broco, A.; Colantoni, A.; Guarnacci, M.; Rossi, F.; Incarnato, D.; Oliviero, S.; Fatica, A.; Morlando, M.; et al. Modulation of circRNA Metabolism by m6A Modification. Cell Rep. 2020, 31, 107641. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Xie, Y.; Yu, T.; Liu, N.; Wang, Z.; Woolsey, R.J.; Tang, Y.; Zhang, X.; Qin, W.; Zhang, Y.; et al. m6A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 2020, 30, 211–228. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.-X.; Chen, X.; Xia, L.-P.; Zhang, J.-X.; Pan, Z.-Z.; Ma, X.-D.; Han, K.; Chen, J.-W.; Judde, J.-G.; Deas, O.; et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019, 10, 4695. [Google Scholar] [CrossRef]
- Chen, Y.G.; Chen, R.; Ahmad, S.; Verma, R.; Kasturi, S.P.; Amaya, L.; Broughton, J.P.; Kim, J.; Cadena, C.; Pulendran, B.; et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol. Cell 2019, 76, 96–109.e9. [Google Scholar] [CrossRef]
- Jia, Z.; Wu, Q. Clustered Protocadherins Emerge as Novel Susceptibility Loci for Mental Disorders. Front. Neurosci. 2019, 14, 587819. [Google Scholar] [CrossRef]
- Mizutani, R.; Saiga, R.; Yamamoto, Y.; Uesugi, M.; Takeuchi, A.; Uesugi, K.; Terada, Y.; Suzuki, Y.; De Andrade, V.; De Carlo, F.; et al. Structural diverseness of neurons between brain areas and between cases. Transl. Psychiatry 2021, 11, 49. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Yue, M.; Wang, J.; Kumar, S.; Wechsler-Reya, R.J.; Zhang, Z.; Ogawa, Y.; Kellis, M.; Duester, G.; et al. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 2018, 21, 195–206. [Google Scholar] [CrossRef]
- Santomauro, D.F.; Herrera, A.M.M.; Shadid, J.; Zheng, P.; Ashbaugh, C.; Pigott, D.M.; Abbafati, C.; Adolph, C.; Amlag, J.O.; Aravkin, A.Y.; et al. Global prevalence and burden of depressive and anxiety disorders in 204 countries and territories in 2020 due to the COVID-19 pandemic. Lancet 2021, 398, 1700–1712. [Google Scholar] [CrossRef]
- Wu, T.; Jia, X.; Shi, H.; Niu, J.; Yin, X.; Xie, J.; Wang, X. Prevalence of mental health problems during the COVID-19 pandemic: A systematic review and meta-analysis. J. Affect. Disord. 2021, 281, 91–98. [Google Scholar] [CrossRef]
- Moller, C.I.; Davey, C.G.; Badcock, P.B.; Wrobel, A.L.; Cao, A.; Murrihy, S.; Sharmin, S.; Cotton, S.M. Correlates of suicidality in young people with depressive disorders: A systematic review. Aust. N. Z. J. Psychiatry 2022, 56, 910–948. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, S.; Kadkhoda, S.; Ghafouri-Fard, S. Synaptic plasticity and depression: The role of miRNAs dysregulation. Mol. Biol. Rep. 2022, 49, 9759–9765. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, Y.; Roy, B.; Kumar, N.; Mukhtar, M.S.; Dwivedi, Y. Molecular pathology associated with altered synaptic transcriptome in the dorsolateral prefrontal cortex of depressed subjects. Transl. Psychiatry 2021, 11, 73. [Google Scholar] [CrossRef]
- Tartt, A.N.; Mariani, M.B.; Hen, R.; Mann, J.J.; Boldrini, M. Dysregulation of adult hippocampal neuroplasticity in major depression: Pathogenesis and therapeutic implications. Mol. Psychiatry 2022, 27, 2689–2699. [Google Scholar] [CrossRef]
- Dean, J.; Keshavan, M. The neurobiology of depression: An integrated view. Asian J. Psychiatry 2017, 27, 101–111. [Google Scholar] [CrossRef]
- Grahek, I.; Shenhav, A.; Musslick, S.; Krebs, R.M.; Koster, E.H. Motivation and cognitive control in depression. Neurosci. Biobehav. Rev. 2019, 102, 371–381. [Google Scholar] [CrossRef]
- Niu, J.; Wang, B.; Wang, T.; Zhou, T. Mechanism of METTL3-mediated m6A modification in depression-induced cognitive deficits. Am. J. Med. Genetics Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genetics 2022, 189, 86–99. [Google Scholar] [CrossRef]
- Huang, R.; Zhang, Y.; Bai, Y.; Han, B.; Ju, M.; Chen, B.; Yang, L.; Wang, Y.; Zhang, H.; Zhang, H.; et al. N6-Methyladenosine Modification of Fatty Acid Amide Hydrolase Messenger RNA in Circular RNA STAG1-Regulated Astrocyte Dysfunction and Depressive-like Behaviors. Biol. Psychiatry 2020, 88, 392–404. [Google Scholar] [CrossRef]
- Vitovcova, B.; Skarkova, V.; Rudolf, K.; Rudolf, E. Biology of Glioblastoma Multiforme—Exploration of Mitotic Catastrophe as a Potential Treatment Modality. Int. J. Mol. Sci. 2020, 21, 5324. [Google Scholar] [CrossRef]
- Thomas, L.; Florio, T.; Perez-Castro, C. Extracellular Vesicles Loaded miRNAs as Potential Modulators Shared Between Glioblastoma, and Parkinson’s and Alzheimer’s Diseases. Front. Cell. Neurosci. 2020, 14, 590034. [Google Scholar] [CrossRef]
- Kondo, Y.; Katsushima, K.; Ohka, F.; Natsume, A.; Shinjo, K. Epigenetic dysregulation in glioma. Cancer Sci. 2014, 105, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chen, S.; Yu, J.; Gao, Z.; Sun, Z.; Yi, Y.; Long, T.; Zhang, C.; Li, Y.; Pan, Y.; et al. Interplay of m6A and histone modifications contributes to temozolomide resistance in glioblastoma. Clin. Transl. Med. 2021, 11, e553. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Tao, B.; Yang, F.; Xia, K.; Yang, X.; Chen, L.; Peng, T.; Xia, X.; Li, X.; Peng, L. Histone demethylase JMJD1C promotes the polarization of M1 macrophages to prevent glioma by upregulating miR-302a. Clin. Transl. Med. 2021, 11, e424. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Qin, S.; Liu, C.; Jiang, L.; Li, C.; Yang, J.; Zhang, S.; Yan, Z.; Liu, X.; Yang, J.; et al. m6A reader IGF2BP2-stabilized CASC9 accelerates glioblastoma aerobic glycolysis by enhancing HK2 mRNA stability. Cell Death Discov. 2021, 7, 292. [Google Scholar] [CrossRef]
- Wang, W.; Li, J.; Lin, F.; Guo, J.; Zhao, J. Identification of N6-methyladenosine-related lncRNAs for patients with primary glioblastoma. Neurosurg. Rev. 2021, 44, 463–470. [Google Scholar] [CrossRef]
- Zheng, P.; Zhang, X.; Ren, D.; Zhang, Y. RP11-552D4.1: A novel m6a-related LncRNA associated with immune status in glioblastoma. Aging 2022, 14, 7348–7363. [Google Scholar] [CrossRef]
- Xie, P.; Yan, H.; Gao, Y.; Li, X.; Zhou, D.-B.; Liu, Z.-Q. Construction of m6A-Related lncRNA Prognostic Signature Model and Immunomodulatory Effect in Glioblastoma Multiforme. Front. Oncol. 2022, 12, 920926. [Google Scholar] [CrossRef]
- Ming, Y.; Deng, Z.; Tian, X.; Jia, Y.; Ning, M.; Cheng, S. m6A Methyltransferase METTL3 Reduces Hippocampal Neuron Apoptosis in a Mouse Model of Autism Through the MALAT1/SFRP2/Wnt/β-catenin Axis. Psychiatry Investig. 2022, 19, 771–787. [Google Scholar] [CrossRef]
- Wang, L.; Yang, H.; Wang, Q.; Zhang, Q.; Wang, Z.; Zhang, Q.; Wu, S.; Li, H. Paraquat and MPTP induce alteration in the expression profile of long noncoding RNAs in the substantia nigra of mice: Role of the transcription factor Nrf. Toxicol. Lett. 2018, 291, 11–28. [Google Scholar] [CrossRef]
- Su, Q.; Chen, N.; Tang, J.; Wang, J.; Chou, W.-C.; Zheng, F.; Shao, W.; Yu, G.; Cai, P.; Guo, Z.; et al. Paraquat-induced oxidative stress regulates N6-methyladenosine (m6A) modification of long noncoding RNAs in Neuro-2a cells. Ecotoxicol. Environ. Saf. 2022, 237, 113503. [Google Scholar] [CrossRef]
- Allen, C.L.; Bayraktutan, U. Oxidative stress and its role in the pathogenesis of ischaemic stroke. Int. J. Stroke Off. J. Int. Stroke Soc. 2009, 4, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Mitchell, S.; Ciechanowicz, S.; Savage, S.; Wang, T.; Ji, X.; Ma, D. Argon protects against hypoxic-ischemic brain injury in neonatal rats through activation of nuclear factor (erythroid-derived 2)-like. Oncotarget 2016, 7, 25640–25651. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Li, Y.; Chen, J.-P.; Li, D.-Z.; Jiang, Q.; Wu, T.; Zhou, X.-Z. Oxygen glucose deprivation/re-oxygenation-induced neuronal cell death is associated with Lnc-D63785 m6A methylation and miR-422a accumulation. Cell Death Dis. 2020, 11, 816. [Google Scholar] [CrossRef]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.; et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef]
- Malacrida, A.; Rivara, M.; Di Domizio, A.; Cislaghi, G.; Miloso, M.; Zuliani, V.; Nicolini, G. 3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell line. Bioorganic Med. Chem. 2020, 28, 115300. [Google Scholar] [CrossRef]
- Takahashi, H.; Hase, H.; Yoshida, T.; Tashiro, J.; Hirade, Y.; Kitae, K.; Tsujikawa, K. Discovery of two novel ALKBH5 selective inhibitors that exhibit uncompetitive or competitive type and suppress the growth activity of glioblastoma multiforme. Chem. Biol. Drug Des. 2022, 100, 1–12. [Google Scholar] [CrossRef]
- Li, R.; Kuang, Y.; Niu, Y.; Zhang, S.; Chen, S.; Su, F.; Wang, J.; Lin, S.; Liu, D.; Shen, C.; et al. FTO-mediated RNA m6A methylation regulates synovial aggression and inflammation in rheumatoid arthritis. Biochim. et Biophys. Acta–-Mol. Basis Dis. 2024, 1870, 167341. [Google Scholar] [CrossRef]
- Shi, K.; Sa, R.; Dou, L.; Wu, Y.; Dong, Z.; Fu, X.; Yu, H. METTL3 exerts synergistic effects on m6A methylation and histone modification to regulate the function of VGF in lung adenocarcinoma. Clin. Epigenet. 2023, 15, 153. [Google Scholar] [CrossRef]
- Deng, J.; Chen, X.; Chen, A.; Zheng, X. m6A RNA methylation in brain injury and neurodegenerative disease. Front. Neurol. 2022, 13, 995747. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, C.; Jin, X.; Wang, H.; Xu, D. Crosstalk Between N6-Methyladenosine and Other Epigenetic Mechanisms in Central Nervous System Development and Disorders. Biomolecules 2025, 15, 1092. https://doi.org/10.3390/biom15081092
Qi C, Jin X, Wang H, Xu D. Crosstalk Between N6-Methyladenosine and Other Epigenetic Mechanisms in Central Nervous System Development and Disorders. Biomolecules. 2025; 15(8):1092. https://doi.org/10.3390/biom15081092
Chicago/Turabian StyleQi, Cuiping, Xiuping Jin, Hui Wang, and Dan Xu. 2025. "Crosstalk Between N6-Methyladenosine and Other Epigenetic Mechanisms in Central Nervous System Development and Disorders" Biomolecules 15, no. 8: 1092. https://doi.org/10.3390/biom15081092
APA StyleQi, C., Jin, X., Wang, H., & Xu, D. (2025). Crosstalk Between N6-Methyladenosine and Other Epigenetic Mechanisms in Central Nervous System Development and Disorders. Biomolecules, 15(8), 1092. https://doi.org/10.3390/biom15081092






