Adventitious Root Formation in Cuttings: Insights from Arabidopsis and Prospects for Woody Plants
Abstract
1. Introduction
2. Cutting-Induced AR Formation
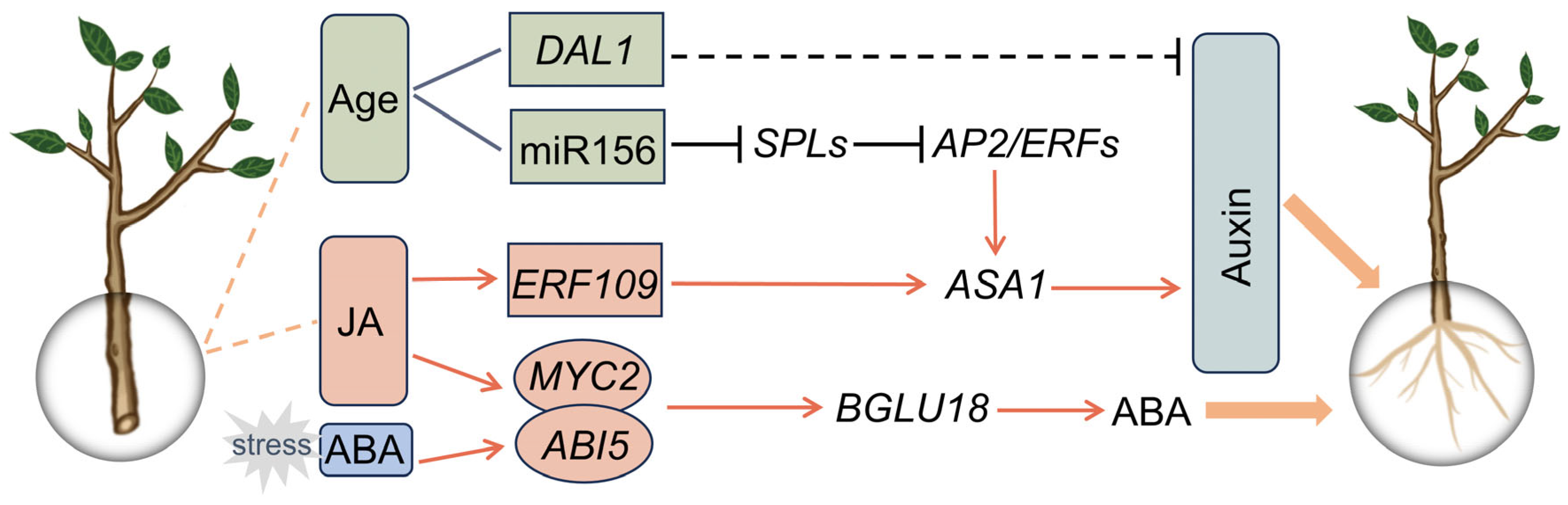
3. Key Factors in Cutting-Induced AR Formation
3.1. JA: A Master Trigger for AR Formation
3.2. Age: The Key Determinant Controlling AR Formation
3.3. Auxin: The Core Regulator Governing AR Formation
| Gene Name | Gene Family | Species | Roles in Adventitious Rooting | References |
|---|---|---|---|---|
| PagFBL1 | TIR1/AFB receptor family | Populus alba × P. glandulosa | Auxin receptor, interacts with IAA28 to promote AR formation | [63] |
| IAA4-1 | AUX/IAA family | Populus deltoides and P. euramericana | Represses ARF activity; negatively regulates AR formation | [64] |
| IAA4-2 | AUX/IAA family | Populus deltoides and P. euramericana | Represses ARF activity; negatively regulates AR formation | [64] |
| AtWOX11 | WUSCHEL-related homeobox gene family | Arabidopsis thaliana | Auxin-induced; activates root founder cells | [68] |
| AtWOX12 | WUSCHEL-related homeobox gene family | Arabidopsis thaliana | Auxin-induced; activates root founder cells | [68] |
| AtWOX5 | WUSCHEL-related homeobox gene family | Arabidopsis thaliana | Activates the transition of root founder cells into root primordia | [66] |
| AtWOX7 | WUSCHEL-related homeobox gene family | Arabidopsis thaliana | Activates the transition of root founder cells into root primordia | [66] |
| AtLBD16 | LBD family | Arabidopsis thaliana | Directly activated by AtWOX11/12, essential for AR formation | [67] |
| AtLBD29 | LBD family | Arabidopsis thaliana | Directly activated by AtWOX11/12, essential for AR formation | [67] |
| PtoWUSa | WUSCHEL-related homeobox gene family | Populus tomentosa | Alters polar auxin transport; reduces root length but increases AR number. | [70] |
| PagWOX11 | WUSCHEL-related homeobox gene family | Populus alba × P. glandulosa | Activates PagLBD16 to promote de novo root regeneration | [71] |
| PagLBD16 | LBD family | Populus alba × P. glandulosa | Acts downstream of PagWOX11 to mediate root regeneration | [71] |
| MdTCP17 | TCP family | Malus domestica (apple) | Interacts with MdWOX11 and blocks its binding to the MdLBD29 promoter | [72] |
| MdWOX11 | WUSCHEL-related homeobox gene family | Malus domestica (apple) | Activates MdLBD29; suppressed by MdTCP17 | [72] |
| AtARF6 | ARF family | Arabidopsis, Populus | Interacts with WOX11 to activate RGIs and LBD16 for adventitious root primordium | [68] |
| AtARF8 | ARF family | Arabidopsis, Populus | Interacts with WOX11 to activate RGIs and LBD16 for adventitious root primordium | [68] |
| miR167 | microRNA family | Populus deltoides × Populus euramericana | Suppresses AR formation by targeting ARF8 mRNA | [78] |
| miR160 | microRNA family | Populus deltoides × Populus euramericana | Promotes AR formation by regulating the activity of ARF17 | [79] |
| miR476a | microRNA family | Populus tomentosa | Represses RFL genes; activates PIN2/5b to promote auxin efflux and AR formation | [80] |
| MdARF8 | ARF family | Malus domestica (apple) | Enhances AR formation by modulating GH3 genes | [15] |
| MdBT2 | BTB-TAZ family | Malus domestica (apple) | Inhibits AR formation by interacting with MdARF8 and MdIAA3 | [15] |
| ArAuxIAA13 | AUX/IAA family | Acer rubrum | Repress AR formation through interaction with ARF proteins | [83] |
| ArAuxIAA16 | AUX/IAA family | Acer rubrum | Repress AR formation through interaction with ARF proteins | [83] |
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, S.; Li, X.; Xu, L.; Zhang, G. Hormone functions in adventitious root formation during cutting propagation of woody plants. J. Plant Res. 2024; ahead of print. [Google Scholar] [CrossRef]
- Zhou, C.; Gu, X.; Li, J.; Su, X.; Chen, S.; Tang, J.; Chen, L.; Cai, N.; Xu, Y. Physiological characteristics and transcriptomic responses of Pinus yunnanensis lateral branching to different shading environments. Plants 2024, 13, 1588. [Google Scholar] [CrossRef]
- Xu, L. De novo root regeneration from leaf explants: Wounding, auxin, and cell fate transition. Curr. Opin. Plant Biol. 2018, 41, 39–45. [Google Scholar] [CrossRef]
- Wang, S.; Sun, G.; Luo, Y.; Qian, W.; Fan, K.; Ding, Z.; Hu, J. Role of IAA and primary metabolites in two rounds of adventitious root formation in softwood cuttings of Camellia sinensis (L.). Agronomy 2022, 12, 2486. [Google Scholar] [CrossRef]
- Lou, X.; Wang, J.; Wang, G.; He, D.; Shang, W.; Song, Y.; Wang, Z.; He, S. Genome-wide analysis of the WOX family and its expression pattern in root development of Paeonia ostii. Int. J. Mol. Sci. 2024, 25, 7668. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Ni, R.; Wang, Y.; Sun, J.; Ma, M.; Bi, H. Effects of different growth regulators on the rooting of Catalpa bignonioides softwood cuttings. Life 2022, 12, 1231. [Google Scholar] [CrossRef] [PubMed]
- Ahkami, A.H.; Lischewski, S.; Haensch, K.T.; Porfirova, S.; Hofmann, J.; Rolletschek, H.; Melzer, M.; Franken, P.; Hause, B.; Druege, U.; et al. Molecular physiology of adventitious root formation in Petunia hybrida cuttings: Involvement of wound response and primary metabolism. New Phytol. 2009, 181, 613–625. [Google Scholar] [CrossRef] [PubMed]
- Ahkami, A.; Scholz, U.; Steuernagel, B.; Strickert, M.; Haensch, K.T.; Druege, U.; Reinhardt, D.; Nouri, E.; von Wirén, N.; Franken, P.; et al. Comprehensive transcriptome analysis unravels the existence of crucial genes regulating primary metabolism during adventitious root formation in Petunia hybrida. PLoS ONE 2014, 9, e100997. [Google Scholar] [CrossRef]
- Rasmussen, A.; Smith, T.E.; Hunt, M.A. Cellular stages of root formation, root system quality and survival of Pinus elliottii var. elliottii × P. caribaea var. hondurensis cuttings in different temperature environments. New For. 2009, 38, 285–294. [Google Scholar] [CrossRef]
- El-Banna, M.F.; Farag, N.B.B.; Massoud, H.Y.; Kasem, M.M. Exogenous IBA stimulated adventitious root formation of Zanthoxylum beecheyanum K. Koch stem cutting: Histo-physiological and phytohormonal investigation. Plant Physiol. Biochem. 2023, 197, 107639. [Google Scholar] [CrossRef]
- Legué, V.; Rigal, A.; Bhalerao, R.P. Adventitious root formation in tree species: Involvement of transcription factors. Physiol. Plant. 2014, 151, 192–198. [Google Scholar] [CrossRef]
- Wan, Q.; Zhai, N.; Xie, D.; Liu, W.; Xu, L. WOX11: The founder of plant organ regeneration. Cell Regen. 2023, 12, 1. [Google Scholar] [CrossRef]
- Chen, C.; Hu, Y.; Ikeuchi, M.; Jiao, Y.; Prasad, K.; Su, Y.H.; Xiao, J.; Xu, L.; Yang, W.; Zhao, Z.; et al. Plant regeneration in the new era: From molecular mechanisms to biotechnology applications. Sci. China Life Sci. 2024, 67, 1338–1367. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wang, C.; Zhang, M.; Wei, L.; Liao, W. Identification of key genes during ethylene-induced adventitious root development in Cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2022, 23, 12981. [Google Scholar] [CrossRef]
- Ji, X.L.; Li, H.L.; Qiao, Z.W.; Zhang, J.C.; Sun, W.J.; You, C.X.; Hao, Y.J.; Wang, X.F. The BTB protein MdBT2 recruits auxin signaling components to regulate adventitious root formation in apple. Plant Physiol. 2022, 189, 1005–1020. [Google Scholar] [CrossRef]
- Vilasboa, J.; da Costa, C.T.; Ransan, L.G.; Mariath, J.E.A.; Fett-Neto, A.G. Microcutting redox profile and anatomy in Eucalyptus spp. with distinct adventitious rooting competence. Front. Plant Sci. 2020, 11, 620832. [Google Scholar] [CrossRef]
- Chen, X.; Cheng, J.; Chen, L.; Zhang, G.; Huang, H.; Zhang, Y.; Xu, L. Auxin-independent NAC pathway acts in response to explant-specific wounding and promotes root tip emergence during de novo root organogenesis in Arabidopsis. Plant Physiol. 2016, 170, 2136–2145. [Google Scholar] [CrossRef]
- Klopotek, Y.; Franken, P.; Klaering, H.P.; Fischer, K.; Hause, B.; Hajirezaei, M.R.; Druege, U. A higher sink competitiveness of the rooting zone and invertases are involved in dark stimulation of adventitious root formation in Petunia hybrida cuttings. Plant Sci. 2016, 243, 10–22. [Google Scholar] [CrossRef]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef]
- Chen, L.; Tong, J.; Xiao, L.; Ruan, Y.; Liu, J.; Zeng, M.; Huang, H.; Wang, J.W.; Xu, L. YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 2016, 67, 4273–4284. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Murphy, A.S.; Peer, W.A.; Gan, L.; Li, Y.; Cheng, Z.-M. Physiological and molecular regulation of adventitious root formation. Crit. Rev. Plant Sci. 2015, 34, 506–521. [Google Scholar] [CrossRef]
- Zhang, Y.; Zang, Q.-L.; Qi, L.-W.; Han, S.-Y.; Li, W.-F. Effects of cutting, pruning, and grafting on the expression of age-related genes in Larix kaempferi. Forests 2020, 11, 218. [Google Scholar] [CrossRef]
- Vidoy-Mercado, I.; Narváez, I.; Palomo-Ríos, E.; Litz, R.E.; Barceló-Muñoz, A.; Pliego-Alfaro, F. Reinvigoration/Rejuvenation induced through micrografting of tree species: Signaling through Graft Union. Plants 2021, 10, 1197. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gao, Y.; Song, X.; Ma, Q.; Zhang, J.; Pei, D. A novel rejuvenation approach to induce endohormones and improve rhizogenesis in mature Juglans tree. Plant Methods 2018, 14, 13. [Google Scholar] [CrossRef]
- de Oliveira, L.S.; Leite, D.M.; Mendes, F.M.; Molinari, L.V.; Brondani, G.E.; Gonçalves, A.N.; de Almeida, M. Micropropagation and in vitro rejuvenation of Eucalyptus cloeziana F. Muell. 3 Biotech 2024, 14, 292. [Google Scholar] [CrossRef]
- Pizarro, A.; Díaz-Sala, C. Cellular dynamics during maturation-related decline of adventitious root formation in forest tree species. Physiol. Plant. 2019, 165, 73–80. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Z.; Wu, D.; Tian, Q.; Su, S.; Cheng, C.; Nie, J.; Yuan, Y.; Wang, Y.; Xu, X. DNA methylation variation is crucial to restore adventitious rooting ability during in vitro shoot culture-induced rejuvenation in apple rootstock. Plant J. 2023, 114, 554–569. [Google Scholar] [CrossRef]
- Rawat, S.S.; Laxmi, A. Rooted in communication: Exploring auxin-salicylic acid nexus in root growth and development. Plant Cell Environ. 2025, 48, 4140–4160. [Google Scholar] [CrossRef]
- Yan, S.P.; Yang, R.H.; Wang, F.; Sun, L.N.; Song, X.S. Effect of auxins and associated metabolic changes on cuttings of Hybrid Aspen. Forests 2017, 8, 117. [Google Scholar] [CrossRef]
- Bannoud, F.; Bellini, C. Adventitious rooting in populus species: Update and perspectives. Front. Plant Sci. 2021, 12, 668837. [Google Scholar] [CrossRef] [PubMed]
- Bettoni, J.C.; van der Walt, K.; Souza, J.A.; McLachlan, A.; Nadarajan, J. Sexual and asexual propagation of Syzygium maire, a critically endangered myrtaceae species of New Zealand. New Zeal J. Bot. 2024, 62, 35–52. [Google Scholar] [CrossRef]
- Rasmussen, A.; Hosseini, S.A.; Hajirezaei, M.R.; Druege, U.; Geelen, D. Adventitious rooting declines with the vegetative to reproductive switch and involves a changed auxin homeostasis. J. Exp. Bot. 2015, 66, 1437–1452. [Google Scholar] [CrossRef]
- Lischweski, S.; Muchow, A.; Guthörl, D.; Hause, B. Jasmonates act positively in adventitious root formation in petunia cuttings. BMC Plant Biol. 2015, 15, 229. [Google Scholar] [CrossRef]
- Fattorini, L.; Falasca, G.; Kevers, C.; Rocca, L.M.; Zadra, C.; Altamura, M.M. Adventitious rooting is enhanced by methyl jasmonate in tobacco thin cell layers. Planta 2009, 231, 155–168. [Google Scholar] [CrossRef]
- Wei, M.; Liu, Q.; Wang, Z.; Yang, J.; Li, W.; Chen, Y.; Lu, H.; Nie, J.; Liu, B.; Lv, K.; et al. PuHox52-mediated hierarchical multilayered gene regulatory network promotes adventitious root formation in Populus ussuriensis. New Phytol. 2020, 228, 1369–1385. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Liu, H.; Zhao, J.; Dong, Y.; Xu, Q.; Yu, Y. Hormone orchestrates a hierarchical transcriptional cascade that regulates Al-induced de novo root regeneration in tea nodal cutting. J. Agric. Food Chem. 2021, 69, 5858–5870. [Google Scholar] [CrossRef] [PubMed]
- Lakehal, A.; Dob, A.; Rahneshan, Z.; Novák, O.; Escamez, S.; Alallaq, S.; Strnad, M.; Tuominen, H.; Bellini, C. ETHYLENE RESPONSE FACTOR 115 integrates jasmonate and cytokinin signaling machineries to repress adventitious rooting in Arabidopsis. New Phytol. 2020, 228, 1611–1626. [Google Scholar] [CrossRef] [PubMed]
- Lakehal, A.; Dob, A.; Novák, O.; Bellini, C. A DAO1-mediated circuit controls auxin and jasmonate crosstalk robustness during adventitious root initiation in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 4428. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, W.; Gu, Z.; Wu, S.; E, Y.; Zhou, W.; Lin, J.; Xu, L. Roles of the wound hormone jasmonate in plant regeneration. J. Exp. Bot. 2023, 74, 1198–1206. [Google Scholar] [CrossRef]
- Zhang, G.; Zhao, F.; Chen, L.; Pan, Y.; Sun, L.; Bao, N.; Zhang, T.; Cui, C.X.; Qiu, Z.; Zhang, Y.; et al. Jasmonate-mediated wound signalling promotes plant regeneration. Nat. Plants 2019, 5, 491–497. [Google Scholar] [CrossRef]
- Lee, K.; Yoon, H.; Park, O.S.; Seo, P.J. ENHANCER OF SHOOT REGENERATION1 promotes de novo root organogenesis after wounding in Arabidopsis leaf explants. Plant Cell 2024, 36, 2359–2374. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Jing, S.; Jia, C.; Li, H.; Li, C.; He, Q.; Zhang, N.; Guo, Y.D. Integration of phytomelatonin signaling with jasmonic acid in wound-induced adventitious root regeneration. Adv. Sci. 2025, 12, e2413485. [Google Scholar] [CrossRef]
- Wan, Q.; Yao, R.; Zhao, Y.; Xu, L. JA and ABA signaling pathways converge to protect plant regeneration in stress conditions. Cell Rep. 2025, 44, 115423. [Google Scholar] [CrossRef]
- Kim, J.W.; Seo, P.J. The early hormone signaling network underlying wound-induced de novo root regeneration. J. Exp. Bot. 2025, 76, 1996–2004. [Google Scholar] [CrossRef]
- Dick, J.M.; Leakey, R.R.B. Differentiation of the dynamic variables affecting rooting ability in juvenile and mature cuttings of cherry (Prunus avium). J. Hortic. Sci. Biotechnol. 2006, 81, 296–302. [Google Scholar] [CrossRef]
- Aung, B.; Gao, R.; Gruber, M.Y.; Yuan, Z.C.; Sumarah, M.; Hannoufa, A. MsmiR156 affects global gene expression and promotes root regenerative capacity and nitrogen fixation activity in alfalfa. Transgenic Res. 2017, 26, 541–557. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Xu, Y.; Guo, C.; Zheng, J.; Zhou, B.; Zhang, Y.; Ding, Y.; Zhang, L.; Zhu, Z.; Wang, H.; et al. Modulation of miR156 to identify traits associated with vegetative phase change in tobacco (Nicotiana tabacum). J. Exp. Bot. 2016, 67, 1493–1504. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, X.; Hu, X.; Wu, T.; Wang, Y.; Xu, X.; Zhang, X.; Han, Z. High miR156 expression is required for auxin-induced adventitious root formation via MxSPL26 independent of PINs and ARFs in Malus xiaojinensis. Front. Plant Sci. 2017, 8, 1059. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zou, Z.; Zhang, J.; Zhang, Y.; Han, Q.; Hu, T.; Xu, X.; Liu, H.; Li, H.; Ye, Z. Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett. 2011, 585, 435–439. [Google Scholar] [CrossRef]
- Ye, B.B.; Shang, G.D.; Pan, Y.; Xu, Z.G.; Zhou, C.M.; Mao, Y.B.; Bao, N.; Sun, L.; Xu, T.; Wang, J.W. AP2/ERF Transcription Factors integrate age and wound signals for root regeneration. Plant Cell 2020, 32, 226–241. [Google Scholar] [CrossRef]
- Li, J.; Han, F.; Yuan, T.; Li, W.; Li, Y.; Wu, H.X.; Wei, H.; Niu, S. The methylation landscape of giga-genome and the epigenetic timer of age in Chinese pine. Nat. Commun. 2023, 14, 1947. [Google Scholar] [CrossRef]
- Ma, J.J.; Chen, X.; Song, Y.T.; Zhang, G.F.; Zhou, X.Q.; Que, S.P.; Mao, F.; Pervaiz, T.; Lin, J.X.; Li, Y.; et al. MADS-box transcription factors MADS11 and DAL1 interact to mediate the vegetative-to-reproductive transition in pine. Plant Physiol. 2021, 187, 247–262. [Google Scholar] [CrossRef]
- Li, X.Y.; Ye, Z.L.; Cheng, D.X.; Zang, Q.L.; Qi, L.W.; Li, W.F. LaDAL1 coordinates age and environmental signals in the life cycle of Larix kaempferi. Int. J. Mol. Sci. 2022, 24, 426. [Google Scholar] [CrossRef]
- Katahata, S.-I.; Futamura, N.; Igasaki, T.; Shinohara, K. Functional analysis of SOC1-like and AGL6-like MADS-box genes of the gymnosperm Cryptomeria japonica. Tree Genet. Genomes 2014, 10, 317–327. [Google Scholar] [CrossRef]
- Li, X.; Cheng, D.; Qi, L.; Zhan, J.; Li, W. Regulation of age-dependent expression patterns of five transcription factors in Larix kaempferi. For. Res. 2023, 3, 18. [Google Scholar] [CrossRef]
- Chen, Y.T.; Shen, C.H.; Lin, W.D.; Chu, H.A.; Huang, B.L.; Kuo, C.I.; Yeh, K.W.; Huang, L.C.; Chang, I.F. Small RNAs of Sequoia sempervirens during rejuvenation and phase change. Plant. Biol. 2013, 15, 27–36. [Google Scholar] [CrossRef]
- Thimann, K.V.; Koepfli, J.B. Identity of the growth promoting and root-forming substances of plants. Nature 1935, 135, 101–102. [Google Scholar] [CrossRef]
- Tütüncü, M. Application of machine learning in in vitro propagation of endemic Lilium akkusianum R. Gämperle. PLoS ONE 2024, 19, e0307823. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.M.; Huang, J.Z.; Hou, T.W.; Pan, I.C. Effects of light intensity and plant growth regulators on callus proliferation and shoot regeneration in the ornamental succulent Haworthia. Bot. Stud. 2019, 60, 10. [Google Scholar] [CrossRef]
- Guo, X.; Fu, X.; Zang, D.; Ma, Y. Effect of auxin treatments, cuttings’ collection date and initial characteristics on Paeonia ‘Yang Fei Chu Yu’ cutting propagation. Sci. Hortic. 2009, 119, 177–181. [Google Scholar] [CrossRef]
- Harfouche, A.; Baoune, N.; Merazga, H. Main and interaction effects of factors on softwood cutting of white poplar (Populus alba L.). Silvae Genet. 2007, 56, 287–294. [Google Scholar] [CrossRef]
- Roth, O.; Yechezkel, S.; Serero, O.; Eliyahu, A.; Vints, I.; Tzeela, P.; Carignano, A.; Janacek, D.P.; Peters, V.; Kessel, A.; et al. Slow release of a synthetic auxin induces formation of adventitious roots in recalcitrant woody plants. Nat. Biotechnol. 2024, 42, 1705–1716. [Google Scholar] [CrossRef] [PubMed]
- Shu, W.; Zhou, H.; Jiang, C.; Zhao, S.; Wang, L.; Li, Q.; Yang, Z.; Groover, A.; Lu, M.Z. The auxin receptor TIR1 homolog (PagFBL 1) regulates adventitious rooting through interactions with Aux/IAA28 in Populus. Plant Biotechnol. J. 2019, 17, 338–349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, X.; Cao, P.; Xiao, Z.; Zhan, C.; Liu, M.; Nvsvrot, T.; Wang, N. The bZIP53-IAA4 module inhibits adventitious root development in Populus. J. Exp. Bot. 2020, 71, 3485–3498. [Google Scholar] [CrossRef]
- Ribeiro, C.L.; Silva, C.M.; Drost, D.R.; Novaes, E.; Novaes, C.R.; Dervinis, C.; Kirst, M. Integration of genetic, genomic and transcriptomic information identifies putative regulators of adventitious root formation in Populus. BMC Plant Biol. 2016, 16, 66. [Google Scholar] [CrossRef]
- Hu, X.; Xu, L. Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 2016, 172, 2363–2373. [Google Scholar] [CrossRef]
- Sheng, L.; Hu, X.; Du, Y.; Zhang, G.; Huang, H.; Scheres, B.; Xu, L. Non-canonical WOX11-mediated root branching contributes to plasticity in Arabidopsis root system architecture. Development 2017, 144, 3126–3133. [Google Scholar] [CrossRef]
- Zhang, T.; Ge, Y.; Cai, G.; Pan, X.; Xu, L. WOX-ARF modules initiate different types of roots. Cell Rep. 2023, 42, 112966. [Google Scholar] [CrossRef]
- Liu, B.; Wang, L.; Zhang, J.; Li, J.; Zheng, H.; Chen, J.; Lu, M. WUSCHEL-related Homeobox genes in Populus tomentosa: Diversified expression patterns and a functional similarity in adventitious root formation. BMC Genom. 2014, 15, 296. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Sun, P.; Zhang, J.; Xia, Y.; Hu, J.; Wang, L.; Lu, M. The WUSCHELa (PtoWUSa) is involved in developmental plasticity of adventitious root in Poplar. Genes 2020, 11, 176. [Google Scholar] [CrossRef]
- Zhang, G.; Cai, G.; Zhang, Y.; Yilan, E.; Li, X.; Xu, L.; Lin, J. Transcriptome framework of root regeneration reveals the conserva-tion of the LBD16-mediated pathway in poplar cuttings. Innov. Life 2023, 1, 100007–100014. [Google Scholar] [CrossRef]
- Mao, J.; Niu, C.; Li, K.; Fan, L.; Liu, Z.; Li, S.; Ma, D.; Tahir, M.M.; Xing, L.; Zhao, C.; et al. Cytokinin-responsive MdTCP17 interacts with MdWOX11 to repress adventitious root primordium formation in apple rootstocks. Plant Cell 2023, 35, 1202–1221. [Google Scholar] [CrossRef]
- Zhao, Y.; Hu, Y.; Dai, M.; Huang, L.; Zhou, D.X. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 2009, 21, 736–748. [Google Scholar] [CrossRef]
- Zhang, T.; Xiang, Y.; Ye, M.; Yuan, M.; Xu, G.; Zhou, D.X.; Zhao, Y. The uORF-HsfA1a-WOX11 module controls crown root development in rice. New Phytol. 2025, 247, 760–773. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, C.; Chen, T.; Zha, L.; Zhang, J.; Huang, L. Identification and 3D gene expression patterns of WUSCEHEL-related homeobox (WOX) genes from Panax ginseng. Plant Physiol. Biochem. 2019, 143, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, G.; Zhang, S.; Chen, S.; Wang, Y.; Wen, P.; Ma, X.; Shi, Y.; Qi, R.; Yang, Y.; et al. Genomes of the Banyan Tree and Pollinator Wasp Provide Insights into Fig-Wasp Coevolution. Cell 2020, 183, 875–889.e17. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, J.; Wang, H.; Xu, H.; Zhou, Y.; Duan, L. MicroRNAs in plants development and stress resistance. Plant Cell Environ. 2025, 48, 5909–5929. [Google Scholar] [CrossRef]
- Cai, H.; Yang, C.; Liu, S.; Qi, H.; Wu, L.; Xu, L.A.; Xu, M. MiRNA-target pairs regulate adventitious rooting in Populus: A functional role for miR167a and its target auxin response factor 8. Tree Physiol. 2019, 39, 1922–1936. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yang, C.; Wu, L.; Cai, H.; Li, H.; Xu, M. The peu-miR160a-PeARF17.1/PeARF17.2 module participates in the adventitious root development of poplar. Plant Biotechnol. J. 2020, 18, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Tao, Y.; Fu, X.; Guo, L.; Xing, H.; Li, C.; Yang, Z.; Su, H.; Wang, X.; Hu, J.; et al. The microRNA476a-RFL module regulates adventitious root formation through a mitochondria-dependent pathway in Populus. New Phytol. 2021, 230, 2011–2028. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, J.; Wang, L.; Li, J.; Zheng, H.; Chen, J.; Lu, M. A survey of Populus PIN-FORMED family genes reveals their diversified expression patterns. J. Exp. Bot. 2014, 65, 2437–2448. [Google Scholar] [CrossRef]
- Wang, W.; Jiao, M.; Huang, X.; Liang, W.; Ma, Z.; Lu, Z.; Tian, S.; Gao, X.; Fan, L.; He, X.; et al. The auxin-responsive CsSPL9-CsGH3.4 module finely regulates auxin levels to suppress the development of adventitious roots in tea (Camellia sinensis). Plant J. 2024, 119, 2273–2287. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Li, H.; Yu, J.; Zhao, H.; Zhang, K.; Ge, W. Regulatory Mechanisms of ArAux/IAA13 and ArAux/IAA16 in the Rooting Process of Acer rubrum. Genes 2023, 14, 1206. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Fan, K.; Wang, Y.; Wang, H.; Ding, S.; Song, D.; Shen, J.; Li, H.; Song, Y.; Han, X.; et al. Red and blue light affect the formation of adventitious roots of tea cuttings (Camellia sinensis) by regulating hormone synthesis and signal transduction pathways of mature leaves. Front. Plant Sci. 2022, 13, 943662. [Google Scholar] [CrossRef]
- Pan, T.; Chen, X.L.; Hao, Y.P.; Jiang, C.W.; Wang, S.; Wang, J.S.; Wei, Q.; Chen, S.J.; Yu, X.S.; Cheng, F.; et al. Optimization of factors affecting the rooting of pine wilt disease resistant Masson pine (Pinus massoniana) stem cuttings. PLoS ONE 2021, 16, e0251937. [Google Scholar] [CrossRef]
- Tombesi, S.; Palliotti, A.; Poni, S.; Farinelli, D. Influence of light and shoot development stage on leaf photosynthesis and carbohydrate status during the adventitious root formation in cuttings of Corylus avellana L. Front. Plant Sci. 2015, 6, 973. [Google Scholar] [CrossRef]
- De Almeida, M.R.; Aumond, M., Jr.; Da Costa, C.T.; Schwambach, J.; Ruedell, C.M.; Correa, L.R.; Fett-Neto, A.G. Environmental control of adventitious rooting in Eucalyptus and Populus cuttings. Trees-Struct. Funct. 2017, 31, 1377–1390. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Zhang, S.; Wang, X.; Du, Y.; He, Q.; Zhang, Y.; Shen, L.; Hu, H.; Zhang, G.; Li, X. Adventitious Root Formation in Cuttings: Insights from Arabidopsis and Prospects for Woody Plants. Biomolecules 2025, 15, 1089. https://doi.org/10.3390/biom15081089
Liu P, Zhang S, Wang X, Du Y, He Q, Zhang Y, Shen L, Hu H, Zhang G, Li X. Adventitious Root Formation in Cuttings: Insights from Arabidopsis and Prospects for Woody Plants. Biomolecules. 2025; 15(8):1089. https://doi.org/10.3390/biom15081089
Chicago/Turabian StyleLiu, Peipei, Shili Zhang, Xinying Wang, Yuxuan Du, Qizhouhong He, Yingying Zhang, Lisha Shen, Hongfei Hu, Guifang Zhang, and Xiaojuan Li. 2025. "Adventitious Root Formation in Cuttings: Insights from Arabidopsis and Prospects for Woody Plants" Biomolecules 15, no. 8: 1089. https://doi.org/10.3390/biom15081089
APA StyleLiu, P., Zhang, S., Wang, X., Du, Y., He, Q., Zhang, Y., Shen, L., Hu, H., Zhang, G., & Li, X. (2025). Adventitious Root Formation in Cuttings: Insights from Arabidopsis and Prospects for Woody Plants. Biomolecules, 15(8), 1089. https://doi.org/10.3390/biom15081089





