The Memory Gene, Murashka, Is a Regulator of Notch Signalling and Controls the Size of the Drosophila Germline Stem Cell Niche
Abstract
1. Introduction
2. Materials and Methods
2.1. Drosophila Stocks and Culture
2.2. Molecular Biology
2.2.1. Primers
2.2.2. PCR of Genomic DNA
2.2.3. Construction of pUASTmura and pMTmura-V5 Vectors
2.3. Cell Culture Experiments
2.3.1. Luciferase Assays
2.3.2. Co-Immunopreciptiation
2.3.3. Immunofluorescence Staining of S2 Cells
2.4. Drosophila Tissue Dissection and Staining
2.4.1. Ovary and Gut Dissection and Immunostaining
2.4.2. Wing Imaginal Disc In Situ Hybridisation
2.4.3. Wing Imaginal Disc X-Gal Staining
2.4.4. Adult Wing Mounting
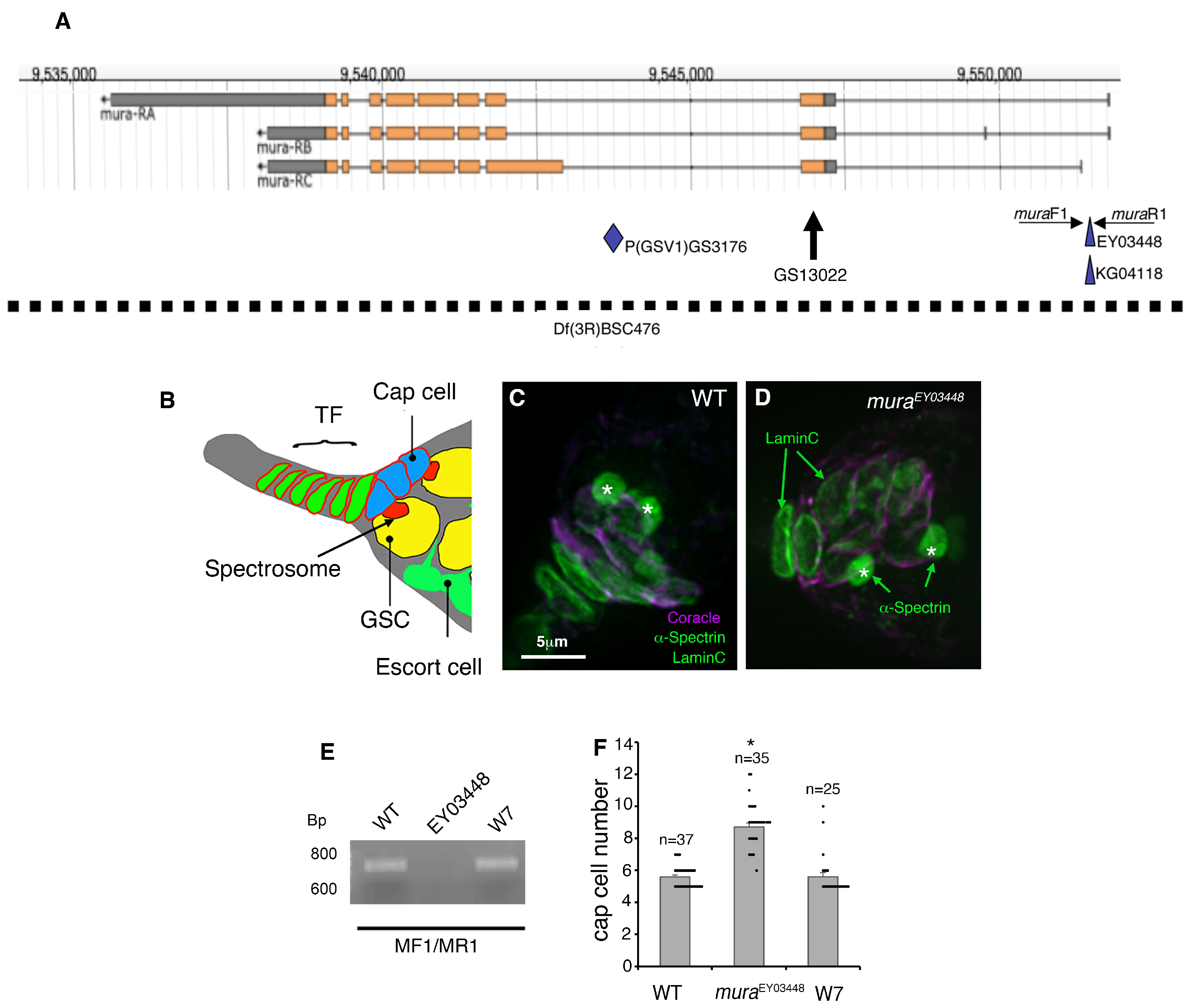
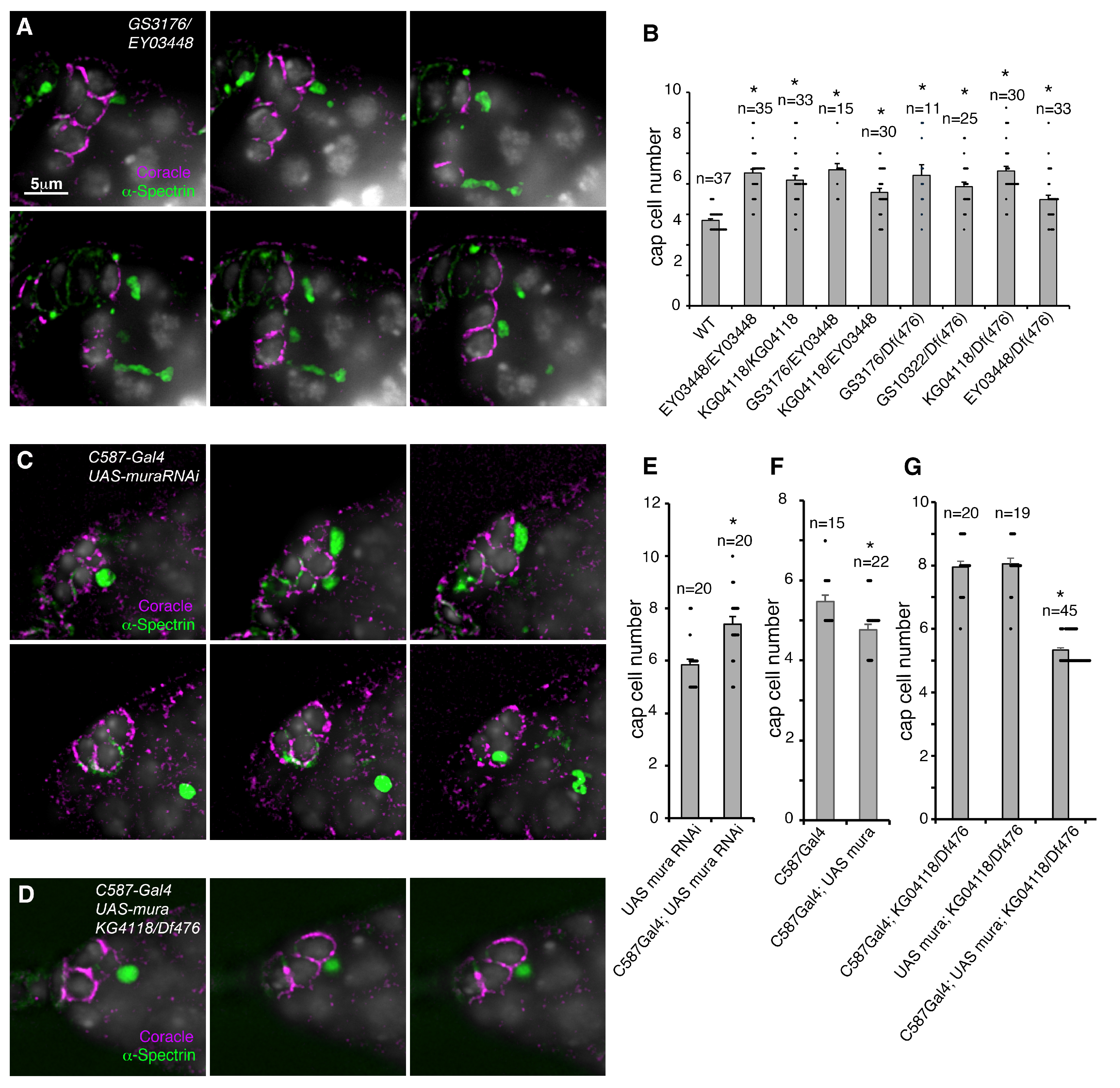
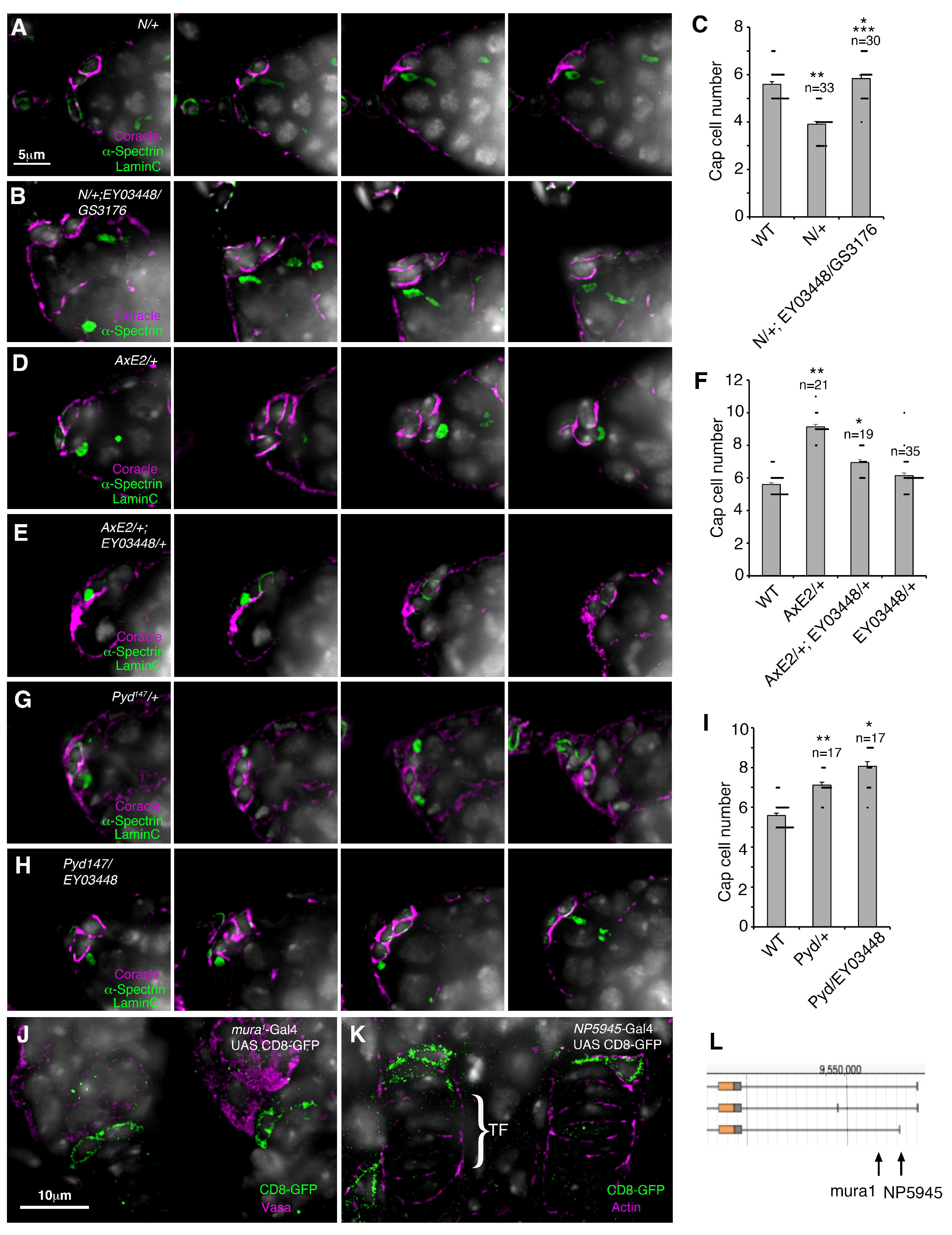
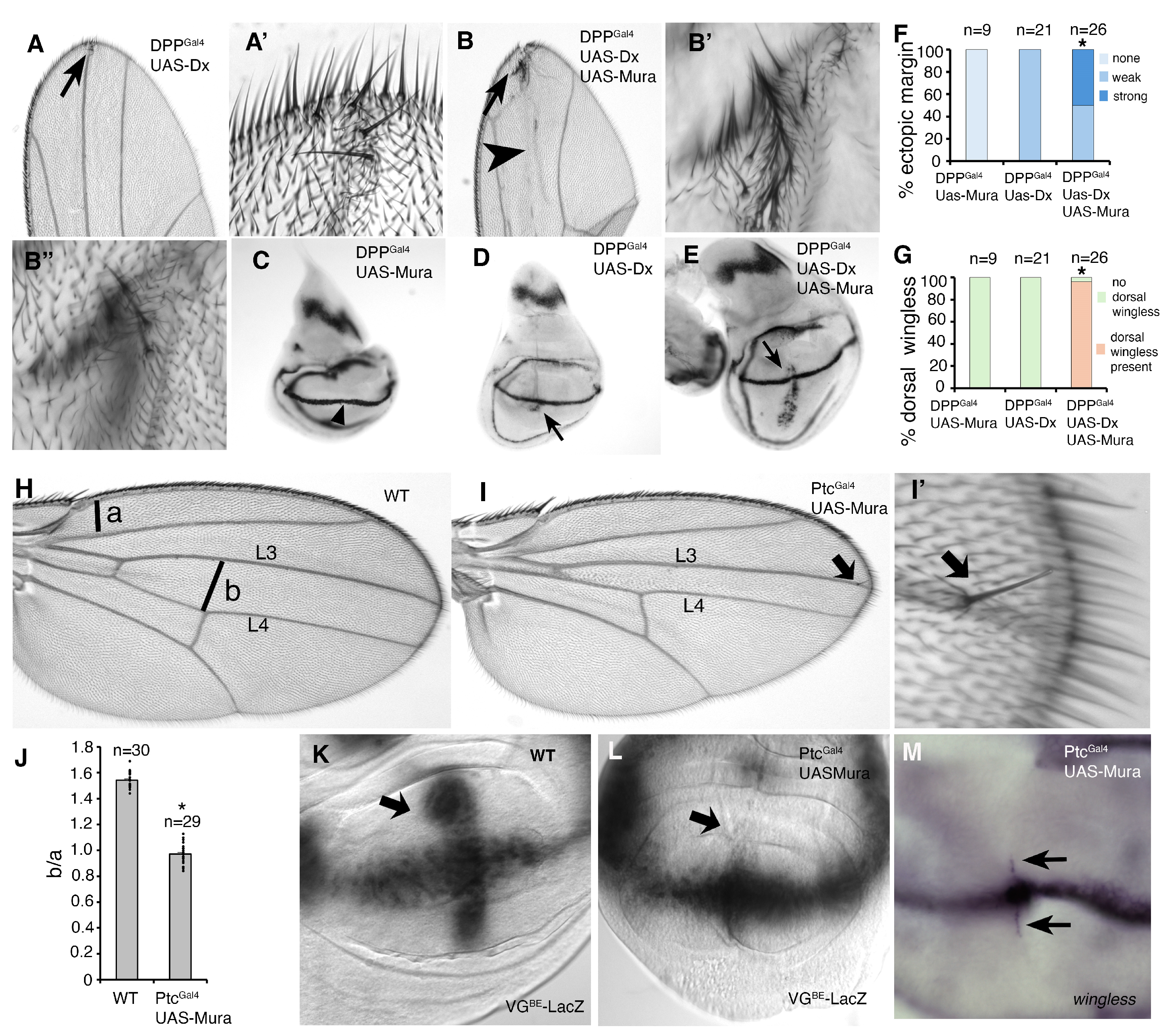
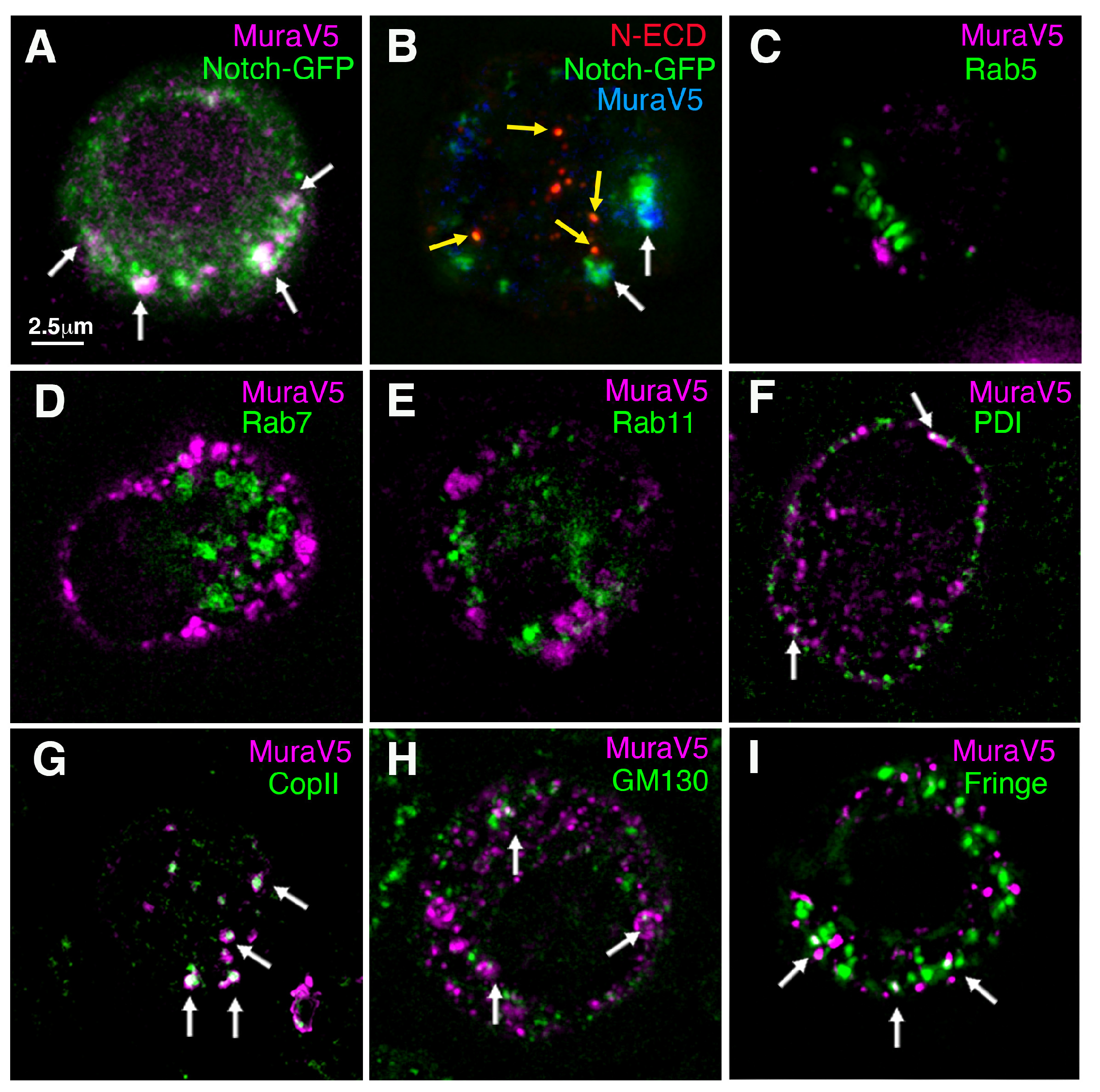
3. Results
4. Discussion
4.1. Mura Regulates the Size of the GSC Niche
4.2. Functional Interaction Between Mura and the Notch Signalling Pathway
4.3. Notch and Mura Interact in the Secretory Pathway
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andersson, E.R.; Sandberg, R.; Lendahl, U. Notch signaling: Simplicity in design, versatility in function. Development 2011, 138, 3593–3612. [Google Scholar] [CrossRef]
- Baron, M. An overview of the Notch signalling pathway. Semin. Cell Dev. Biol. 2003, 14, 113–119. [Google Scholar] [CrossRef]
- Seib, E.; Klein, T. The role of ligand endocytosis in notch signalling. Biol. Cell 2021, 113, 401–418. [Google Scholar] [CrossRef]
- Furriols, M.; Bray, S. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr. Biol. 2001, 11, 60–64. [Google Scholar] [CrossRef]
- Bray, S.J.; Gomez-Lamarca, M. Notch after cleavage. Curr. Opin. Cell Biol. 2018, 51, 103–109. [Google Scholar] [CrossRef]
- Wilkin, M.B.; Carbery, A.M.; Fostier, M.; Aslam, H.; Mazaleyrat, S.L.; Higgs, J.; Myat, A.; Evans, D.A.; Cornell, M.; Baron, M. Regulation of Notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr. Biol. 2004, 14, 2237–2244. [Google Scholar] [CrossRef]
- Wilkin, M.; Tongnok, P.; Gensch, N.; Clemence, S.; Motoki, M.; Yamada, K.; Hori, K.; Taniguchi-Kanai, M.; Franklin, E.; Matsuno, K.; et al. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of Notch in the endosomal trafficking pathway. Dev. Cell 2008, 15, 762–772. [Google Scholar] [CrossRef]
- Shimizu, H.; Woodcock, S.A.; Wilkin, M.B.; Trubenová, B.; Monk, N.A.; Baron, M. Compensatory flux changes within an endocytic trafficking network maintain thermal robustness of Notch signaling. Cell 2014, 157, 1160–1174. [Google Scholar] [CrossRef]
- Shimizu, H.; Hosseini-Alghaderi, S.; Woodcock, S.A.; Baron, M. Alternative mechanisms of Notch activation by partitioning into distinct endosomal domains. J. Cell Biol. 2024, 223, e202211041. [Google Scholar] [CrossRef]
- Oberg, C.; Li, J.; Pauley, A.; Wolf, E.; Gurney, M.; Lendahl, U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 2001, 276, 35847–36853. [Google Scholar] [CrossRef]
- Lan, K.; Verma, S.C.; Murakami, M.; Bajaj, B.; Kaul, R.; Robertson, E.S. Kaposi’s sarcoma herpesvirus-encoded latency-associated nuclear antigen stabilizes intracellular activated Notch by targeting the Sel10 protein. Proc. Natl. Acad. Sci. USA 2007, 104, 16287–16292. [Google Scholar] [CrossRef]
- Hori, K.; Fostier, M.; Ito, M.; Fuwa, T.j.; Go, M.J.; Okano, H.; Baron, M.; Matsuno, K. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development 2004, 131, 5527–5537. [Google Scholar] [CrossRef]
- Song, X.; Call, G.B.; Kirilly, D.; Xie, T. Notch signaling controls germline stem cell niche formation in the Drosophila ovary. Development 2007, 134, 1071–1080. [Google Scholar] [CrossRef]
- Hsu, H.J.; Drummond-Barbosa, D. Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proc. Natl. Acad. Sci. USA 2009, 106, 1117–1121. [Google Scholar] [CrossRef]
- Bonfini, A.; Wilkin, M.B.; Baron, M. Reversible regulation of stem cell niche size associated with dietary control of Notch signalling. BMC Dev. Biol. 2015, 15, 8. [Google Scholar] [CrossRef]
- Djiane, A.; Shimizu, H.; Wilkin, M.; Mazleyrat, S.; Jennings, M.D.; Avis, J.; Bray, S.; Baron, M. Su(dx) E3 ubiquitin ligase-dependent and -independent functions of polychaetoid, the Drosophila ZO-1 homologue. J. Cell Biol. 2011, 192, 189–200. [Google Scholar] [CrossRef]
- Shimizu, H.; Wilkin, M.B.; Woodcock, S.A.; Bonfini, A.; Hung, Y.; Mazaleyrat, S.; Baron, M. The Drosophila ZO-1 protein Polychaetoid suppresses Deltex-regulated Notch activity to modulate germline stem cell niche formation. Open Biol. 2017, 7, 160322. [Google Scholar] [CrossRef]
- Xie, T.; Spradling, A.C. Decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell 1998, 94, 251–260. [Google Scholar] [CrossRef]
- Xie, T.; Spradling, A.C. A niche maintaining germ line stem cells in the Drosophila ovary. Science 2000, 290, 328–330. [Google Scholar] [CrossRef]
- Casanueva, M.O.; Ferguson, E.L. Germline stem cell number in the Drosophila ovary is regulated by redundant mechanisms that control Dpp signaling. Development 2004, 131, 1881–1890. [Google Scholar] [CrossRef]
- Dubnau, J.; Chiang, A.S.; Tully, T. Neural substrates of memory: From synapse to system. J. Neurobiol. 2003, 54, 238–253. [Google Scholar] [CrossRef]
- Dubnau, J.; Chiang, A.S.; Grady, L.; Barditch, J.; Gossweiler, S.; McNeil, J.; Smith, P.; Buldoc, F.; Scott, R.; Certa, U.; et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 2003, 13, 286–296. [Google Scholar] [CrossRef]
- Berger, K.H.; Kong, E.C.; Dubnau, J.; Tully, T.; Moore, M.S.; Heberlein, U. Ethanol sensitivity and tolerance in long-term memory mutants of Drosophila melanogaster. Alcohol Clin. Exp. Res. 2008, 32, 895–908. [Google Scholar] [CrossRef]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar] [CrossRef]
- Staehling-Hampton, K.; Jackson, P.D.; Clark, M.J.; Brand, A.H.; Hoffmann, F.M. Specificity of bone morphogenetic protein-related factors: Cell fate and gene expression changes in Drosophila embryos induced by decapentaplegic but not 60A. Cell Growth Differ. 1994, 5, 585–593. [Google Scholar]
- Speicher, S.A.; Thomas, U.; Hinz, U.; Knust, E. The Serrate locus of Drosophila and its role in morphogenesis of the wing imaginal disc: Control of cell proliferation. Development 1994, 120, 535–544. [Google Scholar] [CrossRef]
- Zhu, C.H.; Xie, T. Clonal expansion of ovarian germline stem cells during niche formation in Drosophila. Development 2003, 130, 2579–2588. [Google Scholar] [CrossRef]
- Grimwade, B.G.; Muskavitch, M.A.; Welshons, W.J.; Yedvobnick, B.; Artavanis-Tsakonas, S. The molecular genetics of the Notch locus in Drosophila melanogaster. Dev. Biol. 1985, 107, 503–519. [Google Scholar] [CrossRef]
- Kelley, M.R.; Kidd, S.; Deutsch, W.A.; Young, M.W. Mutations altering the structure of epidermal growth factor-like coding sequences at the Drosophila Notch locus. Cell 1987, 51, 539–548. [Google Scholar] [CrossRef]
- Klein, T.; Martinez Arias, A. The vestigial gene product provides a molecular context for the interpretation of signals during the development of the wing in Drosophila. Development 1999, 126, 913–925. [Google Scholar] [CrossRef]
- Ivan, V.; de Voer, G.; Xanthakis, D.; Spoorendonk, K.M.; Kondylis, V.; Rabouille, C. Drosophila Sec16 mediates the biogenesis of tER sites upstream of Sar1 through an arginine-rich motif. Mol. Biol. Cell 2008, 19, 4352–4365. [Google Scholar] [CrossRef]
- Fehon, R.G.; Dawson, I.A.; Artavanis-Tsakonas, S. A Drosophila homologue of membrane-skeleton protein 4.1 is associated with septate junctions and is encoded by the coracle gene. Development 1994, 120, 545–557. [Google Scholar] [CrossRef]
- Broers, J.L.; Machiels, B.M.; Kuijpers, H.J.; Smedts, F.; van den Kieboon, R.; Raymond, Y.; Ramaekers, F.C. A- and B-type lamins are differentially expressed in normal human tissues. Histochem. Cell Biol. 1997, 107, 505–517. [Google Scholar] [CrossRef]
- Deng, W.; Lin, H. Spectrosomes and fusomes anchor mitotic spindles during asymmetric germ cell divisions and facilitate the formation of a polarized microtubule array for oocyte specification in Drosophila. Dev. Biol. 1997, 189, 79–94. [Google Scholar] [CrossRef]
- Engels, W.R.; Benz, W.K.; Preston, C.R.; Graham, P.L.; Phillis, R.W.; Robertson, H.M. Somatic effects of P element activity in Drosophila melanogaster: Pupal lethality. Genetics 1987, 117, 745–757. [Google Scholar] [CrossRef]
- Song, X.; Wong, M.D.; Kawase, E.; Xi, R.; Deng, B.C.; McCarthy, J.; Xie, T. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development 2004, 131, 1353–1364. [Google Scholar] [CrossRef]
- Weigel, A.V.; Chang, C.L.; Shtengel, G.; Xu, C.S.; Hoffman, D.P.; Freeman, M.; Iyer, N.; Aaron, J.; Khuon, S.; Bogovic, J.; et al. ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell 2021, 184, 2412–2429. [Google Scholar] [CrossRef]
- Chen, S.; Wang, S.; Xie, T. Restricting self-renewal signals within the stem cell niche: Multiple levels of control. Curr. Opin. Genet. Dev. 2011, 21, 684–689. [Google Scholar] [CrossRef]
- Wilkin, M.B.; Whiteford, R.; Akbar, T.; Hosseini-Alghaderi, S.; Revici, R.; Carbery, A.M.; Baron, M. The First Defined Null Allele of the Notch Regulator, a Suppressor of Deltex: Uncovering Its Novel Roles in Drosophila melanogaster Oogenesis. Biomolecules 2024, 14, 522. [Google Scholar] [CrossRef]
- Tseng, C.Y.; Kao, S.H.; Wan, C.L.; Cho, Y.; Tung, S.Y.; Hsu, H.J. Notch signaling mediates the age-associated decrease in adhesion of germline stem cells to the niche. PLoS Genet. 2014, 10, e1004888. [Google Scholar] [CrossRef]
- Reggiori, F.; Pelham, H.R. A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat. Cell Biol. 2002, 4, 117–123. [Google Scholar] [CrossRef]
- Progida, C.; Bakke, O. Bidirectional traffic between the Golgi and the endosomes—machineries and regulation. J. Cell Sci. 2016, 129, 3971–3982. [Google Scholar] [CrossRef]
- Janody, F.; Treisman, J.E. Requirements for mediator complex subunits distinguish three classes of notch target genes at the Drosophila wing margin. Dev. Dyn. 2011, 240, 2051–2059. [Google Scholar] [CrossRef]
- Hein, K.; Mittler, G.; Cizelsky, W.; Kühl, M.; Ferrante, F.; Liefke, R.; Berger, I.M.; Just, S.; Sträng, J.E.; Kestler, H.A.; et al. Site-specific methylation of Notch1 controls the amplitude and duration of the Notch1 response. Sci. Signal 2015, 8, ra30. [Google Scholar] [CrossRef]
- Guarani, V.; Deflorian, G.; Franco, C.A.; Krüger, M.; Phng, L.K.; Bentley, K.; Toussaint, L.; Dequiedt, F.; Mostoslavsky, R.; Schmidt, M.H.H.; et al. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature 2011, 473, 234–238. [Google Scholar] [CrossRef]
- Acar, M.; Jafar-Nejad, H.; Takeuchi, H.; Rajan, A.; Ibrani, D.; Rana, N.A.; Pan, H.; Haltiwanger, R.S.; Bellen, H.J. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell 2008, 132, 247–258. [Google Scholar] [CrossRef]
- Okajima, T.; Xu, A.; Lei, L.; Irvine, K. Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science 2005, 307, 1599–1603. [Google Scholar] [CrossRef]
- Sasamura, T.; Ishikawa, H.O.; Sasaki, N.; Higashi, S.; Kanai, M.; Nakao, S.; Ayukawa, T.; Toshiro, A.; Noda, K.; Miyoshi, E.; et al. The O-fucosyltransferase O-fut1 is an extracellular component that is essential for the constitutive endocytic trafficking of Notch in Drosophila. Development 2007, 134, 1347–1356. [Google Scholar] [CrossRef]
- Ishikawa, H.; Ayukawa, T.; Nakayama, M.; Higashi, S.; Kamiyama, S.; Nishihara, S.; Aoki, K.; Ishida, N.; Sania, Y.; Mastuno, K. Two pathways for importing GDP-fucose into the endoplasmic reticulum lumen function redundantly in the O-fucosylation of Notch in Drosophila. J. Biol. Chem. 2010, 285, 4122–4129. [Google Scholar] [CrossRef]
- Panin, V.M.; Payannopoulos, V.; Wilson, R.; Irvine, K.D. Fringe modulates Notch-ligand interactions. Nature 1997, 387, 908–912. [Google Scholar] [CrossRef]
- Cammarata-Mouchtouris, A.; Nguyen, X.H.; Acker, A.; Bonnay, F.; Goto, A.; Orian, A.; Fauvarque, M.O.; Boutros, M.; Reichhart, J.M.; Matt, N. Hyd ubiquitinates the NF-κB co-factor Akirin to operate an effective immune response in Drosophila. PLoS Pathog. 2020, 16, e1008458. [Google Scholar] [CrossRef]
- Ge, X.; Hannan, F.; Xie, Z.; Feng, C.; Tully, T.; Zhou, H.; Haimneg, X.; Zuoping, Z.Y. Notch signaling in Drosophila long-term memory formation. Proc. Natl. Acad. Sci. USA 2004, 101, 72–76. [Google Scholar] [CrossRef]
- Matsuno, M.; Horiuchi, J.; Tully, T.; Saitoe, M. The Drosophila cell adhesion molecule klingon is required for long-term memory formation and is regulated by Notch. Proc. Natl. Acad. Sci. USA 2009, 106, 310–315. [Google Scholar] [CrossRef]
- Lin, C.; Li, M.; Lin, N.; Zong, J.; Pan, J.; Ye, Y. RNF38 suppress growth and metastasis via ubiquitination of ACTN4 in nasopharyngeal carcinoma. BMC Cancer 2022, 22, 549. [Google Scholar] [CrossRef]
- Zhou, J.; Tang, Z.Y.; Sun, X.L. RNF38 inhibits osteosarcoma cell proliferation by binding to CRY1. Biochem. Cell Biol. 2021, 99, 629–635. [Google Scholar] [CrossRef]
- Pan, Y.; Zeng, W.; Liang, T.; Nie, X.; Liu, K.; Chen, H.; Luo, N.; Zhu, X.; Tian, K.; Chen, Y. RNF38 promotes gilteritinib resistance in acute myeloid leukemia via inducing autophagy by regulating ubiquitination of LMX1A. Cell Biol. Toxicol. 2024, 40, 105. [Google Scholar] [CrossRef]
- Li, Y.Y.; Wu, C.; Shah, S.S.; Chen, S.M.; Wangpaichitr, M.; Kuo, M.T.; Feun, L.G.; Han, X.; Suarez, M.; Prince, J.; et al. Degradation of AMPK-α1 sensitizes BRAF inhibitor-resistant melanoma cells to arginine deprivation. Mol. Oncol. 2017, 11, 1806–1825. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deen, T.; Shimizu, H.; Wilkin, M.B.; Baron, M. The Memory Gene, Murashka, Is a Regulator of Notch Signalling and Controls the Size of the Drosophila Germline Stem Cell Niche. Biomolecules 2025, 15, 1082. https://doi.org/10.3390/biom15081082
Deen T, Shimizu H, Wilkin MB, Baron M. The Memory Gene, Murashka, Is a Regulator of Notch Signalling and Controls the Size of the Drosophila Germline Stem Cell Niche. Biomolecules. 2025; 15(8):1082. https://doi.org/10.3390/biom15081082
Chicago/Turabian StyleDeen, Thifeen, Hideyuki Shimizu, Marian B. Wilkin, and Martin Baron. 2025. "The Memory Gene, Murashka, Is a Regulator of Notch Signalling and Controls the Size of the Drosophila Germline Stem Cell Niche" Biomolecules 15, no. 8: 1082. https://doi.org/10.3390/biom15081082
APA StyleDeen, T., Shimizu, H., Wilkin, M. B., & Baron, M. (2025). The Memory Gene, Murashka, Is a Regulator of Notch Signalling and Controls the Size of the Drosophila Germline Stem Cell Niche. Biomolecules, 15(8), 1082. https://doi.org/10.3390/biom15081082





