Comparative Analysis of Testicular Transcriptional and Translational Landscapes in Yak and Cattle–Yak: Implications for Hybrid Male Sterility
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Ribosome Footprint (RF) Recovery, Library Construction, and Sequencing
2.3. Data Filtering, Comparison, and Analysis of Ribosome Characteristics
2.4. Gene Abundance Quantification and Differentially Translated Gene (DTG) Analysis
2.5. Comparison of Transcriptional and Translational Differences
2.6. TE (Translation Efficiency) Calculation and Analysis
2.7. uORF Identification and Analysis
2.8. Protein Extraction and Determination
3. Results
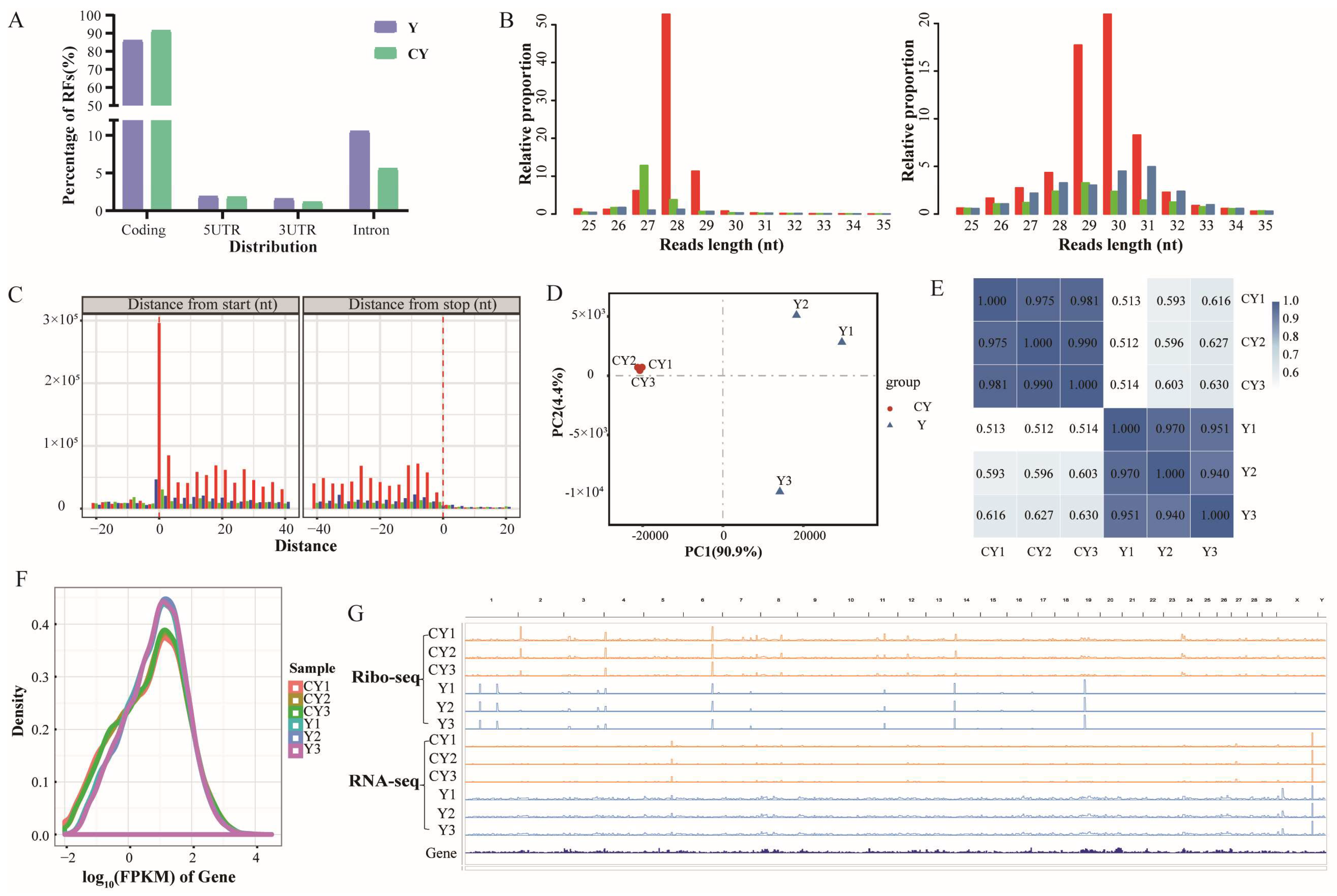
3.1. Read Quality, Alignment, and Ribosome Footprint (RF) Distribution According to Testicular Ribo-Seq
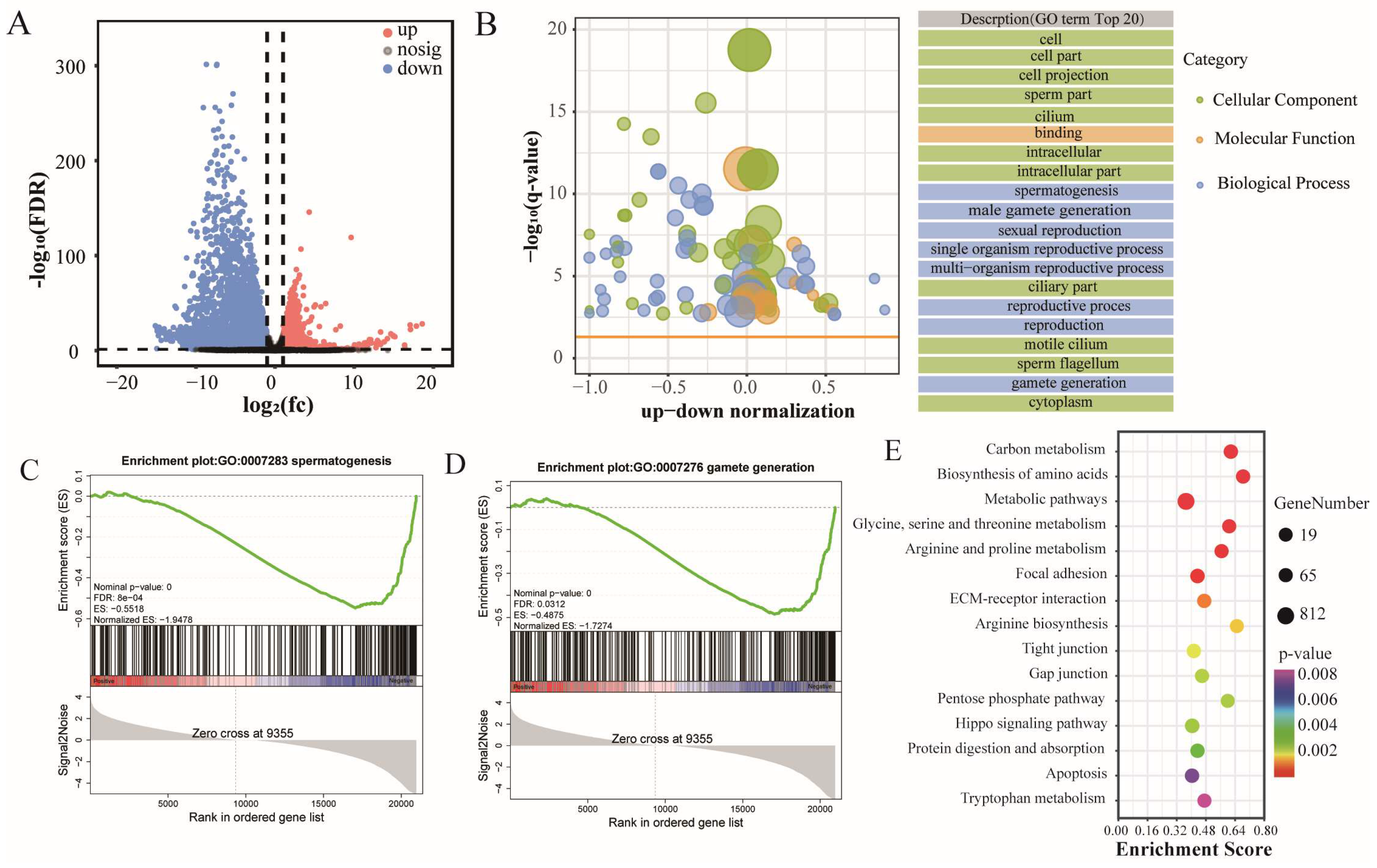
3.2. Translational Down-Regulation of Spermatogenesis Genes Is Prevalent in Cattle–YAK Testis
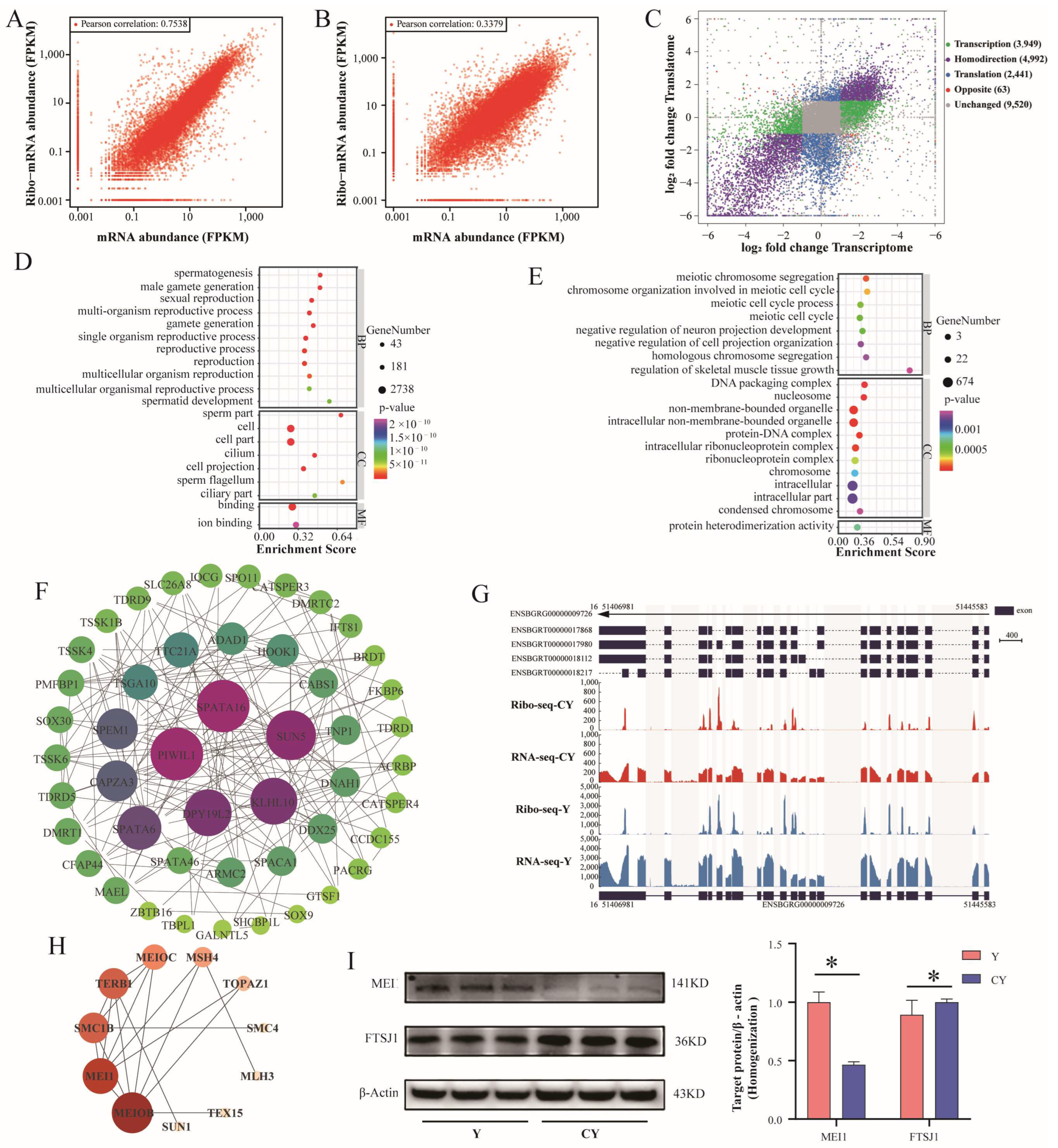
3.3. Transcription–Translation Coordination Controls Spermatogenesis
3.4. Identification of DTEGs and Analysis of CDS Characteristics
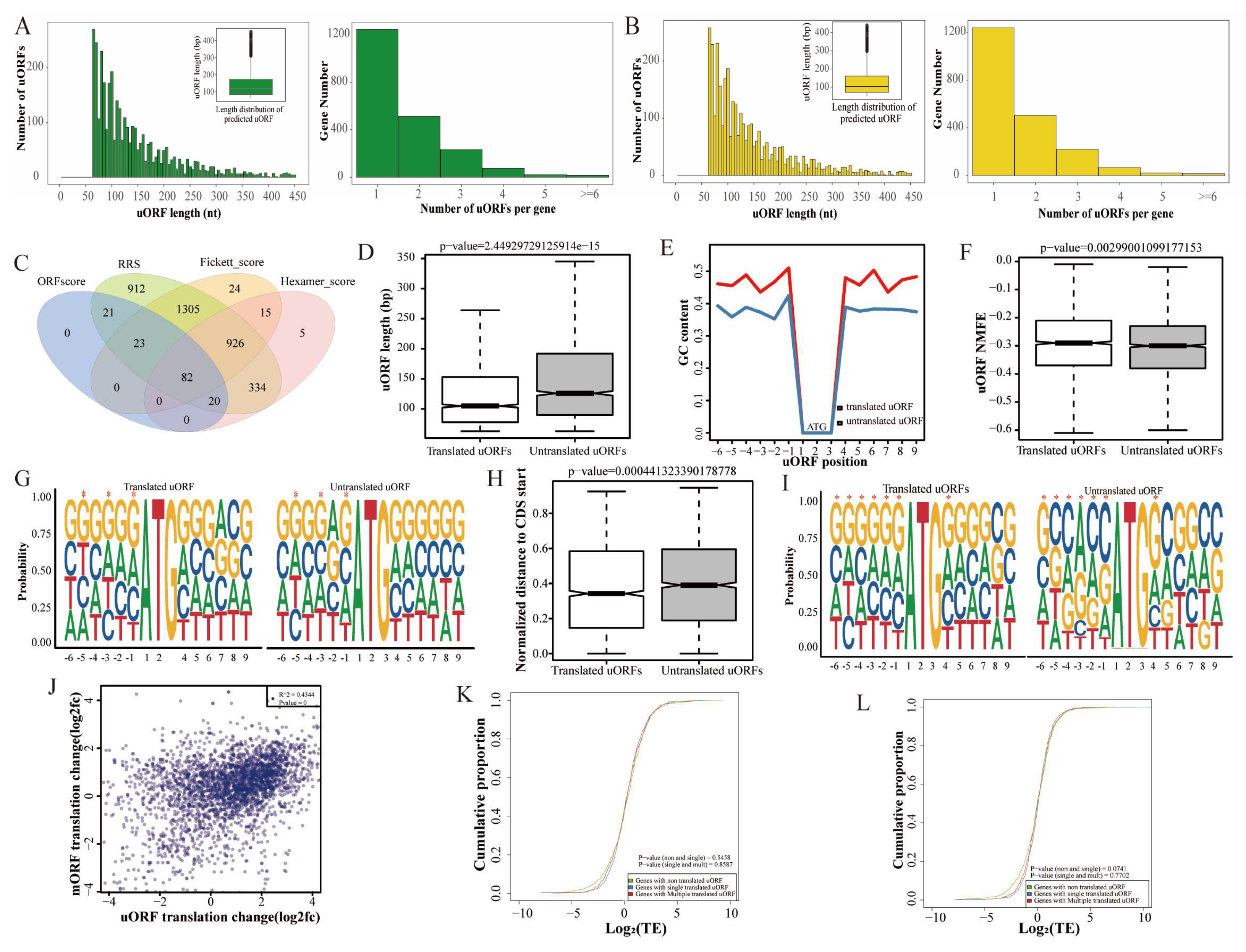
3.5. Characteristics of uORFs and Their Effect on mORF Translation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Niayale, R.; Cui, Y.; Adzitey, F. Male hybrid sterility in the cattle-yak and other bovines: A review. Biol. Reprod. 2021, 104, 495–507. [Google Scholar] [CrossRef]
- Barsila, S.R. Effect of parity in different grazing seasons on milk yield and composition of cattle × yak hybrids in the Himalayan alpines. J. Appl. Anim. Res. 2019, 47, 591–596. [Google Scholar] [CrossRef]
- Huang, C.; Ge, F.; Ma, X.; Dai, R.; Dingkao, R.; Zhaxi, Z.; Burenchao, G.; Bao, P.; Wu, X.; Guo, X.; et al. Comprehensive Analysis of mRNA, lncRNA, circRNA, and miRNA Expression Profiles and Their ceRNA Networks in the Longissimus Dorsi Muscle of Cattle-Yak and Yak. Front. Genet. 2021, 12, 772557. [Google Scholar] [CrossRef]
- Chang, X.; Xu, Y.; Cheng, L.; Yi, K.; Gu, X.; Luo, Z.; Zhang, J.; Wang, J.; Geng, F. Quantitative proteomic analysis of cattle-yak and yak longissimus thoracis provides insights into the differential mechanisms of meat quality. Food Res. Int. 2023, 173, 113253. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Wang, X.; Guo, S.; Kang, Y.; Pei, J.; Guo, X. F1 Male Sterility in Cattle-Yak Examined through Changes in Testis Tissue and Transcriptome Profiles. Animals 2022, 12, 2711. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.X.; Wang, G.W.; Fang, Y.G.; Chen, Y.W.; Jin, Y.Y.; Liu, X.T.; Jia, G.X.; Yang, Q.E. Transcriptome analysis reveals dysregulated gene expression networks in Sertoli cells of cattle-yak hybrids. Theriogenology 2023, 203, 33–42. [Google Scholar] [CrossRef]
- Wang, X.; Pei, J.; Xiong, L.; Kang, Y.; Guo, S.; Cao, M.; Ding, Z.; Bao, P.; Chu, M.; Liang, C.; et al. Single-cell RNA sequencing and UPHLC-MS/MS targeted metabolomics offer new insights into the etiological basis for male cattle-yak sterility. Int. J. Biol. Macromol. 2023, 253, 126831. [Google Scholar] [CrossRef]
- Mipam, T.; Chen, X.; Zhao, W.; Zhang, P.; Chai, Z.; Yue, B.; Luo, H.; Wang, J.; Wang, H.; Wu, Z.; et al. Single-cell transcriptome analysis and in vitro differentiation of testicular cells reveal novel insights into male sterility of the interspecific hybrid cattle-yak. BMC Genom. 2023, 24, 149. [Google Scholar] [CrossRef]
- Zhang, Y.W.; Wu, S.X.; Wang, G.W.; Wan, R.D.; Yang, Q.E. Single-cell analysis identifies critical regulators of spermatogonial development and differentiation in cattle-yak bulls. J. Dairy Sci. 2024, 107, 7317–7336. [Google Scholar] [CrossRef]
- Inada, T.; Winstall, E.; Tarun, S.Z., Jr.; Yates, J.R., 3rd; Schieltz, D.; Sachs, A.B. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA 2002, 8, 948–958. [Google Scholar] [CrossRef]
- Khan, Z.; Ford, M.J.; Cusanovich, D.A.; Mitrano, A.; Pritchard, J.K.; Gilad, Y. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science 2013, 342, 1100–1104. [Google Scholar] [CrossRef]
- Wu, L.; Candille, S.I.; Choi, Y.; Xie, D.; Jiang, L.; Li-Pook-Than, J.; Tang, H.; Snyder, M. Variation and genetic control of protein abundance in humans. Nature 2013, 499, 79–82. [Google Scholar] [CrossRef]
- Brar, G.A.; Weissman, J.S. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat. Rev. Mol. Cell Biol. 2015, 16, 651–664. [Google Scholar] [CrossRef]
- Ingolia, N.T. Ribosome Footprint Profiling of Translation throughout the Genome. Cell 2016, 165, 22–33. [Google Scholar] [CrossRef]
- Zou, D.; Li, K.; Su, L.; Liu, J.; Lu, Y.; Huang, R.; Li, M.; Mang, X.; Geng, Q.; Li, P.; et al. DDX20 is required for cell-cycle reentry of prospermatogonia and establishment of spermatogonial stem cell pool during testicular development in mice. Dev. Cell 2024, 59, 1707–1723. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Zheng, C.; Zhuang, Y.; Jin, P.; Wang, F. The m6A reader PRRC2A is essential for meiosis I completion during spermatogenesis. Nat. Commun. 2023, 14, 1636. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Gui, Y.; Ma, X.; Zhang, B.; Xiong, W.; Yang, S.; Cao, C.; Mo, S.; Shu, G.; Ye, J.; et al. N6-methyladenosine writer METTL16-mediated alternative splicing and translation control are essential for murine spermatogenesis. Genome Biol. 2024, 25, 193. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.K.; Yen, P. Differential translation of Dazap1 transcripts during spermatogenesis. PLoS ONE 2013, 8, e60873. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Leushkin, E.; Liechti, A.; Ovchinnikova, S.; Mößinger, K.; Brüning, T.; Rummel, C.; Grützner, F.; Cardoso-Moreira, M.; Janich, P.; et al. Transcriptome and translatome co-evolution in mammals. Nature 2020, 588, 642–647. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Brar, G.A.; Rouskin, S.; McGeachy, A.M.; Weissman, J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012, 7, 1534–1550. [Google Scholar] [CrossRef]
- Morlan, J.D.; Qu, K.; Sinicropi, D.V. Selective depletion of rRNA enables whole transcriptome profiling of archival fixed tissue. PLoS ONE 2012, 7, e42882. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Lauria, F.; Tebaldi, T.; Bernabò, P.; Groen, E.J.N.; Gillingwater, T.H.; Viero, G. riboWaltz: Optimization of ribosome P-site positioning in ribosome profiling data. PLoS Comput. Biol. 2018, 14, e1006169. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.; Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef]
- Zhong, Y.; Karaletsos, T.; Drewe, P.; Sreedharan, V.T.; Kuo, D.; Singh, K.; Wendel, H.G.; Rätsch, G. RiboDiff: Detecting changes of mRNA translation efficiency from ribosome footprints. Bioinformatics 2017, 33, 139–141. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Höner Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. AMB 2011, 6, 26. [Google Scholar] [CrossRef]
- Hao, Z.; Wang, J.; Luo, Y.; Hu, J.; Liu, X.; Li, S.; Li, M.; Shi, B.; Hu, L.; Liu, Y.; et al. MicroRNA-200c Affects Milk Fat Synthesis by Targeting PANK3 in Ovine Mammary Epithelial Cells. Int. J. Mol. Sci. 2022, 23, 15601. [Google Scholar] [CrossRef] [PubMed]
- van Heesch, S.; Witte, F.; Schneider-Lunitz, V.; Schulz, J.F.; Adami, E.; Faber, A.B.; Kirchner, M.; Maatz, H.; Blachut, S.; Sandmann, C.L.; et al. The Translational Landscape of the Human Heart. Cell 2019, 178, 242–260. [Google Scholar] [CrossRef]
- Jackson, R.; Standart, N. The awesome power of ribosome profiling. RNA 2015, 21, 652–654. [Google Scholar] [CrossRef]
- Zhou, Y.; Sun, Z.; Means, A.R.; Sassone-Corsi, P.; Bernstein, K.E. cAMP-response element modulator tau is a positive regulator of testis angiotensin converting enzyme transcription. Proc. Natl. Acad. Sci. USA 1996, 93, 12262–12266. [Google Scholar] [CrossRef]
- Guan, X.; Chen, P.; Zhao, X.; Hao, X.; Chen, F.; Ji, M.; Wen, X.; Lin, H.; Ye, L.; Chen, H. Characterization of stem cells associated with seminiferous tubule of adult rat testis for their potential to form Leydig cells. Stem Cell Res. 2019, 41, 101593. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Wu, K.; Yang, R. Physiological mechanism of normal sexual behavior in male cattle-yak based on diurnal variation of LH, T, P4 and 17β-E2 levels in blood. Grass-Feed. Livest. 1996, 2, 41–43+48. [Google Scholar] [CrossRef]
- Wu, S.; Mipam, T.; Xu, C.; Zhao, W.; Shah, M.A.; Yi, C.; Luo, H.; Cai, X.; Zhong, J. Testis transcriptome profiling identified genes involved in spermatogenic arrest of cattleyak. PLoS ONE 2020, 15, e0229503. [Google Scholar] [CrossRef]
- Ghasemi, N.; Azizi, H.; Skutella, T. Unraveling the Significance of Nanog in the Generation of Embryonic Stem-like Cells from Spermatogonia Stem Cells: A Combined In Silico Analysis and In Vitro Experimental Approach. Int. J. Mol. Sci. 2024, 25, 4833. [Google Scholar] [CrossRef]
- Petrusová, J.; Havalda, R.; Flachs, P.; Venit, T.; Darášová, A.; Hůlková, L.; Sztacho, M.; Hozák, P. Focal Adhesion Protein Vinculin Is Required for Proper Meiotic Progression during Mouse Spermatogenesis. Cells 2022, 11, 2013. [Google Scholar] [CrossRef]
- Su, L.; Mruk, D.D.; Lui, W.Y.; Lee, W.M.; Cheng, C.Y. P-glycoprotein regulates blood-testis barrier dynamics via its effects on the occludin/zonula occludens 1 (ZO-1) protein complex mediated by focal adhesion kinase (FAK). Proc. Natl. Acad. Sci. USA 2011, 108, 19623–19628. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Hsu, P.J.; Xing, X.; Fang, J.; Lu, Z.; Zou, Q.; Zhang, K.J.; Zhang, X.; Zhou, Y.; Zhang, T.; et al. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell Res. 2017, 27, 1216–1230. [Google Scholar] [CrossRef]
- Zhao, L.Y.; Song, J.; Liu, Y.; Song, C.X.; Yi, C. Mapping the epigenetic modifications of DNA and RNA. Protein Cell 2020, 11, 792–808. [Google Scholar] [CrossRef]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Sato, H.; Miyamoto, T.; Yogev, L.; Namiki, M.; Koh, E.; Hayashi, H.; Sasaki, Y.; Ishikawa, M.; Lamb, D.J.; Matsumoto, N.; et al. Polymorphic alleles of the human MEI1 gene are associated with human azoospermia by meiotic arrest. J. Hum. Genet. 2006, 51, 533–540. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Li, N.; Ji, Z.; Bai, H.; Ou, N.; Tian, R.; Li, P.; Zhi, E.; Huang, Y.; Zhao, J.; et al. Bi-allelic MEI1 variants cause meiosis arrest and non-obstructive azoospermia. J. Hum. Genet. 2023, 68, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wu, W.; Luo, H.; Liu, Z.; Liu, H.; Li, Q.; Pan, Z. Molecular characterization and epigenetic regulation of Mei1 in cattle and cattle-yak. Gene 2015, 573, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, Y.; Murtaza, G.; Zhou, J.; Jiao, Y.; Gong, C.; Hu, C.; Han, Q.; Zhang, H.; Zhang, Y.; et al. Whole-exome sequencing of consanguineous families with infertile men and women identifies homologous mutations in SPATA22 and MEIOB. Hum. Reprod. 2021, 36, 2793–2804. [Google Scholar] [CrossRef]
- Zhu, X.; Hu, K.; Cheng, H.; Wu, H.; Li, K.; Gao, Y.; Lv, M.; Xu, C.; Geng, H.; Shen, Q.; et al. Novel MEIOB pathogenic variants including a homozygous non-canonical splicing variant, cause meiotic arrest and human non-obstructive azoospermia. Clin. Genet. 2024, 105, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Ma, J.; Shen, G.; Jiang, X.; Wang, X.; Jiang, C.; Bai, H.; Zheng, Y.; Tian, K.; Yue, J.; et al. Identification of an SMC1B Mutation Associated with Necrozoospermia and Failure of Testi-ICSI: SMC1B Mutation Associated with Necrozoospermia. Reprod. Sci. 2025, 32, 1–14. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, F.; Chen, Q.; Li, M.; Lv, X.; Xiao, Y.; Zhang, Z.; Hou, L.; Lai, Y.; Zhang, Y.; et al. The piRNA pathway is essential for generating functional oocytes in golden hamsters. Nat. Cell Biol. 2021, 23, 1013–1022. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Li, Q.; Pan, Z.; Li, M.; Luo, H.; Xie, Z. Molecular cloning, gene expression and methylation status analysis of PIWIL1 in cattle-yaks and the parental generation. Anim. Reprod. Sci. 2013, 140, 131–137. [Google Scholar] [CrossRef]
- Yang, C.; Wang, X. Lysosome biogenesis: Regulation and functions. J. Cell Biol. 2021, 220, e202102001. [Google Scholar] [CrossRef]
- Yuan, J.; Ofengeim, D. A guide to cell death pathways. Nat. Rev. Mol. Cell Biol. 2024, 25, 379–395. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Shi, J.; Chen, J.; Zhang, M.; Sun, S.; Xie, S.; Li, X.; Zeng, B.; Peng, L.; Hauck, A.; et al. Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J. Cell Mol. Biol. 2015, 84, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Zhong, L.; Du, J.; Zhu, C.; Peng, X.; He, X.; Fu, J.; Ouyang, L.; Bian, J.; Hu, L.; et al. Ribosome profiling reveals the effects of nitrogen application translational regulation of yield recovery after abrupt drought-flood alternation in rice. Plant Physiol. Biochem. PPB 2020, 155, 42–58. [Google Scholar] [CrossRef]
- Awad, S.; Valleriani, A.; Chiarugi, D. A data-driven estimation of the ribosome drop-off rate in S. cerevisiae reveals a correlation with the genes length. NAR Genom. Bioinform. 2024, 6, lqae036. [Google Scholar] [CrossRef]
- Ma, R.; Cui, Y.; Yu, S.J.; Pan, Y.Y.; He, J.F.; Wang, Y.Y.; Zhao, L.; Bai, X.F.; Yang, S.S. Whole transcriptome sequencing revealed the gene regulatory network of hypoxic response in yak Sertoli cells. Sci. Rep. 2024, 14, 19903. [Google Scholar] [CrossRef]
- de Mateo, S.; Sassone-Corsi, P. Regulation of spermatogenesis by small non-coding RNAs: Role of the germ granule. Semin. Cell Dev. Biol. 2014, 29, 84–92. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Wu, X.; Tang, X.; Wu, C.; Lu, J. Determinants of genome-wide distribution and evolution of uORFs in eukaryotes. Nat. Commun. 2021, 12, 1076. [Google Scholar] [CrossRef]
- Wang, S.; Cai, Y.; Li, T.; Wang, Y.; Bao, Z.; Wang, R.; Qin, J.; Wang, Z.; Liu, Y.; Liu, Z.; et al. CWF19L2 is Essential for Male Fertility and Spermatogenesis by Regulating Alternative Splicing. Adv. Sci. 2024, 11, e2403866. [Google Scholar] [CrossRef] [PubMed]
- Calvo, S.E.; Pagliarini, D.J.; Mootha, V.K. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. USA 2009, 106, 7507–7512. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 1986, 44, 283–292. [Google Scholar] [CrossRef]
- Cao, X.; Slavoff, S.A. Non-AUG start codons: Expanding and regulating the small and alternative ORFeome. Exp. Cell Res. 2020, 391, 111973. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Lu, J. Function and Evolution of Upstream ORFs in Eukaryotes. Trends Biochem. Sci. 2019, 44, 782–794. [Google Scholar] [CrossRef]
- Johnstone, T.G.; Bazzini, A.A.; Giraldez, A.J. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J. 2016, 35, 706–723. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, Y.; Li, S.; Wen, J.; Zhu, L.; Yan, K.; Du, Y.; Li, S.; Yan, L.; Xie, Z.; et al. Ribosome footprint profiling enables elucidating the systemic regulation of fatty acid accumulation in Acer truncatum. BMC Biol. 2023, 21, 68. [Google Scholar] [CrossRef]
- Dever, T.E.; Ivanov, I.P.; Hinnebusch, A.G. Translational regulation by uORFs and start codon selection stringency. Genes Dev. 2023, 37, 474–489. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, M.; Guo, S.; Ding, Z.; Hu, L.; Xiong, L.; Ge, Q.; Pei, J.; Guo, X. Comparative Analysis of Testicular Transcriptional and Translational Landscapes in Yak and Cattle–Yak: Implications for Hybrid Male Sterility. Biomolecules 2025, 15, 1080. https://doi.org/10.3390/biom15081080
Cao M, Guo S, Ding Z, Hu L, Xiong L, Ge Q, Pei J, Guo X. Comparative Analysis of Testicular Transcriptional and Translational Landscapes in Yak and Cattle–Yak: Implications for Hybrid Male Sterility. Biomolecules. 2025; 15(8):1080. https://doi.org/10.3390/biom15081080
Chicago/Turabian StyleCao, Mengli, Shaoke Guo, Ziqiang Ding, Liyan Hu, Lin Xiong, Qianyun Ge, Jie Pei, and Xian Guo. 2025. "Comparative Analysis of Testicular Transcriptional and Translational Landscapes in Yak and Cattle–Yak: Implications for Hybrid Male Sterility" Biomolecules 15, no. 8: 1080. https://doi.org/10.3390/biom15081080
APA StyleCao, M., Guo, S., Ding, Z., Hu, L., Xiong, L., Ge, Q., Pei, J., & Guo, X. (2025). Comparative Analysis of Testicular Transcriptional and Translational Landscapes in Yak and Cattle–Yak: Implications for Hybrid Male Sterility. Biomolecules, 15(8), 1080. https://doi.org/10.3390/biom15081080





