Empirical Assessment of Sequence-Based Predictions of Intrinsically Disordered Regions Involved in Phase Separation
Abstract
1. Introduction
2. Materials and Methods
2.1. Survey and Selection of Predictors for Comparative Analysis
2.2. Collection and Annotation of Test Dataset
2.3. Assessment of Predictive Performance
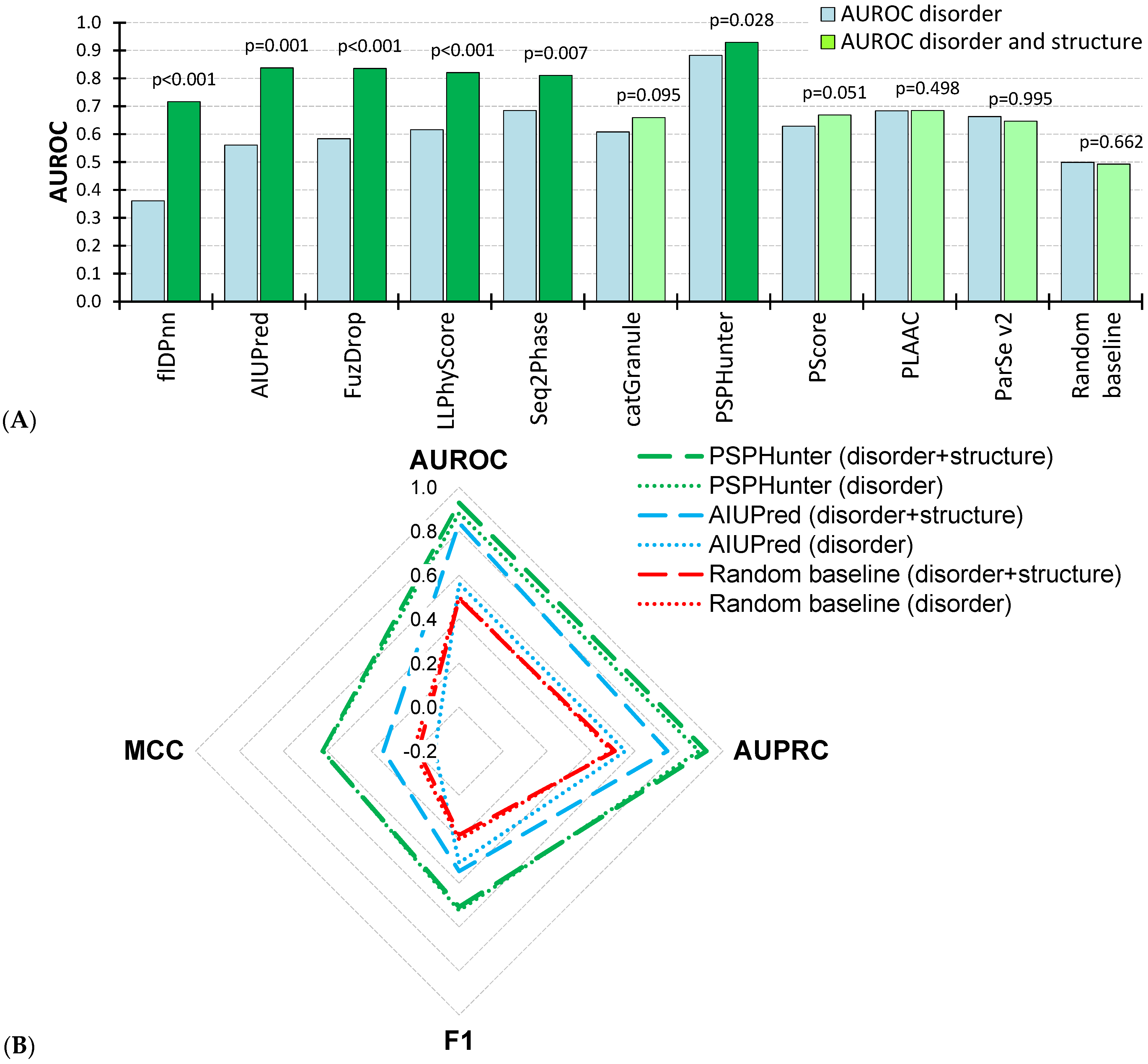
3. Results
3.1. Prediction of Phase Separation in Structured and Disordered Residues
3.2. Prediction of Phase Separation in Disordered Residues
3.3. Prediction of Phase Separation at the Protein Level
4. Conclusions and Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Gao, Y.; Li, X.; Li, P.; Lin, Y. A brief guideline for studies of phase-separated biomolecular condensates. Nat. Chem. Biol. 2022, 18, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Banjade, S.; Cheng, H.-C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.L.; Liu, Y.; Oldfield, C.J.; Uversky, V.N. Intrinsically Disordered Proteome of Human Membrane-Less Organelles. Proteomics 2017, 18, e1700193. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically Disordered Proteins in Overcrowded Milieu: Membrane-Less Organelles, Phase Separation, and Intrinsic Disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef]
- Chen, T.; Tang, G.; Li, T.; Yanghong, Z.; Hou, C.; Du, Z.; You, K.; Ma, L.; Li, T. Phasedis: A Manually Curated Database of Phase Separation-Disease Associations and Corresponding Small Molecules. Genom. Proteom. Bioinform. 2025, 23, qzaf014. [Google Scholar] [CrossRef]
- Mehta, S.; Zhang, J. Liquid–liquid phase separation drives cellular function and dysfunction in cancer. Nat. Rev. Cancer 2022, 22, 239–252. [Google Scholar] [CrossRef]
- Alberti, S.; Dormann, D. Liquid-Liquid Phase Separation in Disease. Annu. Rev. Genet. 2019, 53, 171–194. [Google Scholar] [CrossRef]
- Duan, D.; Koleske, A.J. Phase separation of microtubule-binding proteins—Implications for neuronal function and disease. J. Cell Sci. 2024, 137, jcs263470. [Google Scholar] [CrossRef]
- Vendruscolo, M.; Fuxreiter, M. Towards sequence-based principles for protein phase separation predictions. Curr. Opin. Chem. Biol. 2023, 75, 102317. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Chen, Z.; Yu, C.; Chen, T.; Shi, M.; Li, T. Computational Screening of Phase-Separating Proteins. Genom. Proteom. Bioinform. 2021, 19, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Vernon, R.M.; Forman-Kay, J.D. First-generation predictors of biological protein phase separation. Curr. Opin. Struct. Biol. 2019, 58, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Zhang, Y.; Qi, Y.; Zhang, Z. Evaluation of sequence-based predictors for phase-separating protein. Briefings Bioinform. 2023, 24, bbad213. [Google Scholar] [CrossRef]
- Kuechler, E.R.; Huang, A.; Bui, J.M.; Mayor, T.; Gsponer, J. Comparison of Biomolecular Condensate Localization and Protein Phase Separation Predictors. Biomolecules 2023, 13, 527. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Uversky, V.N.; Dunker, A.K.; Kurgan, L. Introduction to intrinsically disordered proteins and regions. In Intrinsically Disordered Proteins; Salvi, N., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 1–34. [Google Scholar]
- Ibrahim, A.Y.; Khaodeuanepheng, N.P.; Amarasekara, D.L.; Correia, J.J.; Lewis, K.A.; Fitzkee, N.C.; Hough, L.E.; Whitten, S.T. Intrinsically disordered regions that drive phase separation form a robustly distinct protein class. J. Biol. Chem. 2022, 299, 102801. [Google Scholar] [CrossRef]
- Paiz, E.A.; Allen, J.H.; Correia, J.J.; Fitzkee, N.C.; Hough, L.E.; Whitten, S.T. Beta turn propensity and a model polymer scaling exponent identify intrinsically disordered phase-separating proteins. J. Biol. Chem. 2021, 297, 101343. [Google Scholar] [CrossRef]
- Vendruscolo, M.; Fuxreiter, M. Sequence Determinants of the Aggregation of Proteins Within Condensates Generated by Liquid-liquid Phase Separation. J. Mol. Biol. 2022, 434, 167201. [Google Scholar] [CrossRef]
- Lancaster, A.K.; Nutter-Upham, A.; Lindquist, S.; King, O.D. PLAAC: A web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 2014, 30, 2501–2502. [Google Scholar] [CrossRef]
- Bolognesi, B.; Gotor, N.L.; Dhar, R.; Cirillo, D.; Baldrighi, M.; Tartaglia, G.G.; Lehner, B. A Concentration-Dependent Liquid Phase Separation Can Cause Toxicity upon Increased Protein Expression. Cell Rep. 2016, 16, 222–231. [Google Scholar] [CrossRef]
- Vernon, R.M.; Chong, P.A.; Tsang, B.; Kim, T.H.; Bah, A.; Farber, P.; Lin, H.; Forman-Kay, J.D. Pi-Pi Contacts Are an Overlooked Protein Feature Relevant to Phase Separation. Elife 2018, 7, e31486. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Vernon, R.M.; Forman-Kay, J.D. An Interpretable Machine-Learning Algorithm to Predict Disordered Protein Phase Separation Based on Biophysical Interactions. Biomolecules 2022, 12, 1131. [Google Scholar] [CrossRef] [PubMed]
- Hardenberg, M.; Horvath, A.; Ambrus, V.; Fuxreiter, M.; Vendruscolo, M. Widespread occurrence of the droplet state of proteins in the human proteome. Proc. Natl. Acad. Sci. USA 2020, 117, 33254–33262. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Hu, G.; Wu, Z.; Uversky, V.N.; Kurgan, L. Assessment of Disordered Linker Predictions in the CAID2 Experiment. Biomolecules 2024, 14, 287. [Google Scholar] [CrossRef]
- Zhang, F.; Kurgan, L. Evaluation of predictions of disordered binding regions in the CAID2 experiment. Comput. Struct. Biotechnol. J. 2024, 27, 78–88. [Google Scholar] [CrossRef]
- Del Conte, A.; Mehdiabadi, M.; Bouhraoua, A.; Monzon, A.M.; Tosatto, S.C.E.; Piovesan, D. Critical assessment of protein intrinsic disorder prediction (CAID)—Results of round 2. Proteins: Struct. Funct. Bioinform. 2023, 91, 1925–1934. [Google Scholar] [CrossRef]
- Necci, M.; Piovesan, D.; Predictors, C.; Curators, D.; Tosatto, S.C. Critical assessment of protein intrinsic disorder prediction. Nat. Methods 2021, 18, 5. [Google Scholar] [CrossRef]
- Katuwawala, A.; Kurgan, L. Comparative Assessment of Intrinsic Disorder Predictions with a Focus on Protein and Nucleic Acid-Binding Proteins. Biomolecules 2020, 10, 1636. [Google Scholar] [CrossRef]
- Fonin, A.V.; Antifeeva, I.A.; Kuznetsova, I.M.; Turoverov, K.K.; Zaslavsky, B.Y.; Kulkarni, P.; Uversky, V.N.; Mukhopadhyay, S. Biological soft matter: Intrinsically disordered proteins in liquid–liquid phase separation and biomolecular condensates. Essays Biochem. 2022, 66, 831–847. [Google Scholar] [CrossRef]
- Ma, H.; Liu, M.; Fu, R.; Feng, J.; Ren, H.; Cao, J.; Shi, M. Phase separation in innate immune response and inflammation-related diseases. Front. Immunol. 2023, 14, 1086192. [Google Scholar] [CrossRef]
- Agarwal, A.; Mukhopadhyay, S. Prion Protein Biology Through the Lens of Liquid-Liquid Phase Separation. J. Mol. Biol. 2022, 434, 167368. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.H.; Hsu, K.W.; Wu, K.J. Liquid-liquid phase separation (LLPS) in cellular physiology and tumor biology. Am. J. Cancer Res. 2021, 11, 3766–3776. [Google Scholar] [PubMed]
- Kolonko-Adamska, M.; Uversky, V.N.; Greb-Markiewicz, B. The Participation of the Intrinsically Disordered Regions of the bHLH-PAS Transcription Factors in Disease Development. Int. J. Mol. Sci. 2021, 22, 2868. [Google Scholar] [CrossRef] [PubMed]
- Biesaga, M.; Frigolé-Vivas, M.; Salvatella, X. Intrinsically disordered proteins and biomolecular condensates as drug targets. Curr. Opin. Chem. Biol. 2021, 62, 90–100. [Google Scholar] [CrossRef]
- Hou, S.; Hu, J.; Yu, Z.; Li, D.; Liu, C.; Zhang, Y. Machine learning predictor PSPire screens for phase-separating proteins lacking intrinsically disordered regions. Nat. Commun. 2024, 15, 1–13. [Google Scholar] [CrossRef]
- Aspromonte, M.C.; Nugnes, M.V.; Quaglia, F.; Bouharoua, A.; Tosatto, S.C.E.; Piovesan, D. DisProt in 2024: Improving function annotation of intrinsically disordered proteins. Nucleic Acids Res. 2023, 52, D434–D441. [Google Scholar] [CrossRef]
- Miyata, K.; Iwasaki, W.; Bateman, A. Seq2Phase: Language model-based accurate prediction of client proteins in liquid–liquid phase separation. Bioinform. Adv. 2023, 4, vbad189. [Google Scholar] [CrossRef]
- Sun, J.; Qu, J.; Zhao, C.; Zhang, X.; Liu, X.; Wang, J.; Wei, C.; Liu, X.; Wang, M.; Zeng, P.; et al. Precise prediction of phase-separation key residues by machine learning. Nat. Commun. 2024, 15, 1–18. [Google Scholar] [CrossRef]
- Nott, T.J.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.D.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase Transition of a Disordered Nuage Protein Generates Environmentally Responsive Membraneless Organelles. Mol. Cell 2015, 57, 936–947. [Google Scholar] [CrossRef]
- Wang, J.; Choi, J.-M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174, 688–699.e16. [Google Scholar] [CrossRef]
- Hughes, M.P.; Sawaya, M.R.; Boyer, D.R.; Goldschmidt, L.; Rodriguez, J.A.; Cascio, D.; Chong, L.; Gonen, T.; Eisenberg, D.S. Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science 2018, 359, 698–701. [Google Scholar] [CrossRef] [PubMed]
- Orlando, G.; Raimondi, D.; Tabaro, F.; Codicè, F.; Moreau, Y.; Vranken, W.F.; Hancock, J. Computational identification of prion-like RNA-binding proteins that form liquid phase-separated condensates. Bioinformatics 2019, 35, 4617–4623. [Google Scholar] [CrossRef] [PubMed]
- Kuechler, E.R.; Budzyńska, P.M.; Bernardini, J.P.; Gsponer, J.; Mayor, T. Distinct Features of Stress Granule Proteins Predict Localization in Membraneless Organelles. J. Mol. Biol. 2020, 432, 2349–2368. [Google Scholar] [CrossRef] [PubMed]
- Saar, K.L.; Morgunov, A.S.; Qi, R.; Arter, W.E.; Krainer, G.; Lee, A.A.; Knowles, T.P.J. Learning the molecular grammar of protein condensates from sequence determinants and embeddings. Proc. Natl. Acad. Sci. USA 2021, 118, e2019053118. [Google Scholar] [CrossRef]
- van Mierlo, G.; Jansen, J.R.; Wang, J.; Poser, I.; van Heeringen, S.J.; Vermeulen, M. Predicting protein condensate formation using machine learning. Cell Rep. 2021, 34, 108705. [Google Scholar] [CrossRef]
- Raimondi, D.; Orlando, G.; Michiels, E.; Pakravan, D.; Bratek-Skicki, A.; Bosch, L.V.D.; Moreau, Y.; Rousseau, F.; Schymkowitz, J.; Ponty, Y. In silico prediction of in vitro protein liquid–liquid phase separation experiments outcomes with multi-head neural attention. Bioinformatics 2021, 37, 3473–3479. [Google Scholar] [CrossRef]
- Kuechler, E.R.; Jacobson, M.; Mayor, T.; Gsponer, J. GraPES: The Granule Protein Enrichment Server for prediction of biological condensate constituents. Nucleic Acids Res. 2022, 50, W384–W391. [Google Scholar] [CrossRef]
- Mullick, P.; Trovato, A. Sequence-Based Prediction of Protein Phase Separation: The Role of Beta-Pairing Propensity. Biomolecules 2022, 12, 1771. [Google Scholar] [CrossRef]
- Chen, Z.; Hou, C.; Wang, L.; Yu, C.; Chen, T.; Shen, B.; Hou, Y.; Li, P.; Li, T. Screening membraneless organelle participants with machine-learning models that integrate multimodal features. Proc. Natl. Acad. Sci. USA 2022, 119, e2115369119. [Google Scholar] [CrossRef]
- Chu, X.; Sun, T.; Li, Q.; Xu, Y.; Zhang, Z.; Lai, L.; Pei, J. Prediction of liquid–liquid phase separating proteins using machine learning. BMC Bioinform. 2022, 23, 23. [Google Scholar] [CrossRef]
- Yu, K.; Liu, Z.; Cheng, H.; Li, S.; Zhang, Q.; Liu, J.; Ju, H.-Q.; Zuo, Z.; Zhao, Q.; Kang, S.; et al. dSCOPE: A software to detect sequences critical for liquid–liquid phase separation. Briefings Bioinform. 2022, 24, bbac550. [Google Scholar] [CrossRef]
- Zhou, S.; Zhou, Y.; Liu, T.; Zheng, J.; Jia, C. PredLLPS_PSSM: A novel predictor for liquid–liquid protein separation identification based on evolutionary information and a deep neural network. Briefings Bioinform. 2023, 24, bbad299. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Cai, R.; Lugo-Martinez, J.; Guo, Y. A hybrid positive unlabeled learning framework for uncovering scaffolds across human proteome by measuring the propensity to drive phase separation. Briefings Bioinform. 2023, 24, bbad009. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Lewis, K.A.; Fitzkee, N.C.; Hough, L.E.; Whitten, S.T. ParSe 2.0: A web tool to identify drivers of protein phase separation at the proteome level. Protein Sci. 2023, 32, e4756. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Zhang, Y.; Han, X.; Wang, T.; Wang, X.; Yan, Q.; Li, Q.; Qi, Y.; Zhang, Z. A sequence-based model for identifying proteins undergoing liquid–liquid phase separation/forming fibril aggregates via machine learning. Protein Sci. 2024, 33, e4927. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, S.; Bi, Y.; Zou, Q.; Jia, C. A two-task predictor for discovering phase separation proteins and their undergoing mechanism. Briefings Bioinform. 2024, 25, bbae528. [Google Scholar] [CrossRef]
- Saar, K.L.; Scrutton, R.M.; Bloznelyte, K.; Morgunov, A.S.; Good, L.L.; Lee, A.A.; Teichmann, S.A.; Knowles, T.P.J. Protein Condensate Atlas from predictive models of heteromolecular condensate composition. Nat. Commun. 2024, 15, 5418. [Google Scholar] [CrossRef]
- Liang, Q.; Peng, N.; Xie, Y.; Kumar, N.; Gao, W.; Miao, Y. MolPhase, an advanced prediction algorithm for protein phase separation. EMBO J. 2024, 43, 1898–1918. [Google Scholar] [CrossRef]
- Hadarovich, A.; Singh, H.R.; Ghosh, S.; Scheremetjew, M.; Rostam, N.; Hyman, A.A.; Toth-Petroczy, A. PICNIC accurately predicts condensate-forming proteins regardless of their structural disorder across organisms. Nat. Commun. 2024, 15, 10668. [Google Scholar] [CrossRef]
- Thompson, M.; Martín, M.; Olmo, T.S.; Rajesh, C.; Koo, P.K.; Bolognesi, B.; Lehner, B. Massive experimental quantification allows interpretable deep learning of protein aggregation. Sci. Adv. 2025, 11, eadt5111. [Google Scholar] [CrossRef]
- Song, J.; Kurgan, L.; Arighi, C. Availability of web servers significantly boosts citations rates of bioinformatics methods for protein function and disorder prediction. Bioinform. Adv. 2023, 3, vbad184. [Google Scholar] [CrossRef]
- Hou, C.; Wang, X.; Xie, H.; Chen, T.; Zhu, P.; Xu, X.; You, K.; Li, T. Phasepdb in 2022: Annotating Phase Separation-Related Proteins with Droplet States, Co-Phase Separation Partners and Other Experimental Information. Nucleic Acids Res. 2023, 51, D460–D465. [Google Scholar] [CrossRef]
- Hatos, A.; Hajdu-Soltész, B.; Monzon, A.M.; Palopoli, N.; Álvarez, L.; Aykac-Fas, B.; Bassot, C.; Benítez, G.I.; Bevilacqua, M.; Chasapi, A.; et al. DisProt: Intrinsic protein disorder annotation in 2020. Nucleic Acids Res. 2019, 48, D269–D276. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chao, H.; Chen, L.; Craig, P.A.; Crichlow, G.V.; Dalenberg, K.; Duarte, J.M.; et al. RCSB Protein Data Bank (RCSB.org): Delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2022, 51, D488–D508. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. Blast+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Hu, G.; Wu, Z.; Su, H.; Yang, J.; Kurgan, L. Comprehensive Survey and Comparative Assessment of RNA-Binding Residue Predictions with Analysis by RNA Type. Int. J. Mol. Sci. 2020, 21, 6879. [Google Scholar] [CrossRef]
- Miao, Z.; Westhof, E.; Chen, S.-J. A Large-Scale Assessment of Nucleic Acids Binding Site Prediction Programs. PLOS Comput. Biol. 2015, 11, e1004639. [Google Scholar] [CrossRef]
- Basu, S.; Kurgan, L. Taxonomy-Specific Assessment of Intrinsic Disorder Predictions at Residue and Region Levels in Higher Eukaryotes, Protists, Archaea, Bacteria and Viruses. Comput. Struct. Biotechnol. J. 2024, 23, 1968–1977. [Google Scholar] [CrossRef]
- Campen, A.; Williams, R.M.; Brown, C.J.; Meng, J.; Uversky, V.N.; Dunker, A.K. TOP-IDP-Scale: A New Amino Acid Scale Measuring Propensity for Intrinsic Disorder. Protein Pept. Lett. 2008, 15, 956–963. [Google Scholar] [CrossRef]
- Zhao, B.; Kurgan, L. Compositional Bias of Intrinsically Disordered Proteins and Regions and Their Predictions. Biomolecules 2022, 12, 888. [Google Scholar] [CrossRef]
- Katuwawala, A.; Oldfield, C.J.; Kurgan, L. Accuracy of protein-level disorder predictions. Briefings Bioinform. 2019, 21, 1509–1522. [Google Scholar] [CrossRef]
- Kurgan, L.; Hu, G.; Wang, K.; Ghadermarzi, S.; Zhao, B.; Malhis, N.; Erdős, G.; Gsponer, J.; Uversky, V.N.; Dosztányi, Z. Tutorial: A guide for the selection of fast and accurate computational tools for the prediction of intrinsic disorder in proteins. Nat. Protoc. 2023, 18, 3157–3172. [Google Scholar] [CrossRef]
- Erdős, G.; Dosztányi, Z. AIUPred: Combining energy estimation with deep learning for the enhanced prediction of protein disorder. Nucleic Acids Res. 2024, 52, W176–W181. [Google Scholar] [CrossRef]
- Wang, K.; Hu, G.; Wu, Z.; Kurgan, L. Accurate and Fast Prediction of Intrinsic Disorder Using flDPnn. In Prediction of Protein Secondary Structure; Kloczkowski, A., Kurgan, L., Faraggi, E., Eds.; Springer: New York, NY, USA, 2025; pp. 201–218. [Google Scholar]
- Hu, G.; Katuwawala, A.; Wang, K.; Wu, Z.; Ghadermarzi, S.; Gao, J.; Kurgan, L. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat. Commun. 2021, 12, 4438. [Google Scholar] [CrossRef] [PubMed]
- Murmu, S.; Chaurasia, H.; Rao, A.; Rai, A.; Jaiswal, S.; Bharadwaj, A.; Yadav, R.; Archak, S. PlantPathoPPI: An Ensemble-based Machine Learning Architecture for Prediction of Protein-Protein Interactions between Plants and Pathogens. J. Mol. Biol. 2025, 437, 169093. [Google Scholar] [CrossRef] [PubMed]
- Ghafoor, H.; Asim, M.N.; Ibrahim, M.A.; Dengel, A. ProSol-multi: Protein solubility prediction via amino acids multi-level correlation and discriminative distribution. Heliyon 2024, 10, e36041. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Wang, R.; Sun, Y.; Liu, J.; Chen, Z.; Zhang, Y.-J. Interpretable feature extraction and dimensionality reduction in ESM2 for protein localization prediction. Briefings Bioinform. 2024, 25, bbad534. [Google Scholar] [CrossRef]
- Elnaggar, A.; Heinzinger, M.; Dallago, C.; Rehawi, G.; Wang, Y.; Jones, L.; Gibbs, T.; Feher, T.; Angerer, C.; Steinegger, M.; et al. ProtTrans: Toward Understanding the Language of Life Through Self-Supervised Learning. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 7112–7127. [Google Scholar] [CrossRef]
- Alberti, S.; Halfmann, R.; King, O.; Kapila, A.; Lindquist, S. A Systematic Survey Identifies Prions and Illuminates Sequence Features of Prionogenic Proteins. Cell 2009, 137, 146–158. [Google Scholar] [CrossRef]
| Method Name [Reference] | Publication Year | Has Server | Has Code | Code or Server Is Working | Provides Per-Amino Acid Scores | Brief Description of Predictive Model and URL(s) of Code and/or Server for the Selected Predictors |
|---|---|---|---|---|---|---|
| PLAAC [20] | 2014 | Y | Y | Y | Y | Hidden Markov Model that identifies prion-like domains and non-prion-like domains https://github.com/whitehead/plaac (code); and http://plaac.wi.mit.edu/ (server) |
| Baldwin et al. [40] | 2015 | N | N | N | N | |
| catGranule [21] | 2016 | Y | N | Y | Y | Scoring function that combines propensity for RNA binding, intrinsic disorder, and content of selected amino acids, which are processed using a sliding window of size 50 http://service.tartaglialab.com/new_submission/catGRANULE (server) |
| R + Y [41] | 2018 | N | N | N | N | |
| LARKS [42] | 2018 | N | N | N | N | |
| Pscore [22] | 2018 | Y | Y | Y | Y | Scoring function that considers short-range, long-range, backbone, and sidechain pi-contact predictions, which are processed using three sliding windows of sizes 40, 80, and 120 https://github.com/haocai1992/PScore-online (code); https://pound.med.utoronto.ca/JFKlab/Software/psp.htm (server) |
| PSPer [43] | 2019 | Y | Y | Y | N | |
| MaGS [44] | 2020 | Y | N | Y | N | |
| DeePhase [45] | 2021 | Y | Y | Y | N | |
| PSAP [46] | 2021 | N | Y | Y | N | |
| Droppler [47] | 2021 | N | Y | Y | N | |
| ParSe [18] | 2021 | Y | Y | Y | Y | There is a newer version of this tool that was published in 2023 and which we used for the assessment. |
| MaGSeq [48] | 2022 | Y | N | Y | N | |
| PASTA + Pscore [49] | 2022 | N | N | N | N | |
| FuzDrop [19,24] | 2022 | Y | N | Y | Y | Logistic regression that combines predicted probability for intrinsic disorder and for disordered binding https://fuzdrop.bio.unipd.it (server) |
| PhaSePred [50] | 2022 | Y | N | Y | N | |
| PSPredictor [51] | 2022 | N | Y | Y | N | |
| LLPhyScore [23] | 2022 | N | Y | Y | Y | Scoring function that uses eight predictive inputs: residue-water, residue-carbon, pi-pi, and short-range electrostatic interactions, hydrogen bonds, predicted short and long intrinsic disorder, and kinked beta-strands, which are processed with a sliding window of size 3 https://github.com/julie-forman-kay-lab/LLPhyScore (code) |
| dSCOPE [52] | 2023 | N | N | N | N | |
| PredLLPS_PSSM [53] | 2023 | Y | Y | Y | N | |
| PULPS [54] | 2023 | Y | N | Y | N | |
| ParSe v2 [55] | 2023 | Y | N | Y | Y | Scoring regions in the 3-dimensional space defined by alpha-helix propensity, hydrophobicity, and vmodel values, which are computed using a sliding window of size 25 https://stevewhitten.github.io/Parse_v2_web/ (server) |
| FLFB [56] | 2024 | Y | Y | Y | N | |
| Opt_PredLLPS [57] | 2024 | N | Y | Y | N | |
| PSPire [36] | 2024 | N | Y | Y | N | |
| Knowles et al. [58] | 2024 | N | Y | Y | N | |
| MolPhase [59] | 2024 | Y | N | Y | N | |
| PICNIC [60] | 2024 | N | Y | Y | N | |
| PSPHunter [39] | 2024 | Y | Y | Y | Y | Random forest model that uses amino acid composition, evolutionary conservation, predicted secondary structure, solvent accessibility, intrinsic disorder, DNA and RNA binding, and selected posttranslational modification, protein–protein interaction, and sequence embedding generated with word2vec method as inputs http://psphunter.stemcellding.org/ (server); https://github.com/jsun9003/PSPHunter (code) |
| CANYA [61] | 2024 | N | Y | Y | N | |
| Seq2Phase [38] | 2024 | N | Y | Y | Y | An ensemble of random forest, support vector machine, gradient boosted decision tree and shallow feedforward neural network that uses hydrophobicity, content of charged residues, and low-complexity regions, Pscore prediction, embeddings with ProtTrans model, and prediction of intrinsic disorder as inputs, with the results processed with a sliding window of size 100. https://github.com/IwasakiLab/Seq2Phase (code) |
| AUROC | AUPRC | F1 | MCC | ||
|---|---|---|---|---|---|
| Predictors of phase separation | PSPHunter | 0.929 /+ | 0.925 /+ | 0.509 /+ | 0.423 /+ |
| FuzDrop | 0.836 −/+ | 0.769 −/+ | 0.311 −/+ | 0.176 −/+ | |
| LLPhyScore | 0.821 −/+ | 0.808 −/+ | 0.458 −/+ | 0.306 −/+ | |
| Seq2Phase | 0.811 −/+ | 0.820 −/+ | 0.278 −/+ | 0.253 −/+ | |
| PLAAC | 0.685 −/+ | 0.775 −/+ | 0.458 −/+ | 0.395 −/+ | |
| PScore | 0.669 −/+ | 0.724 −/+ | 0.456 −/+ | 0.314 −/+ | |
| catGranule | 0.660 −/+ | 0.723 −/+ | 0.445 −/+ | 0.246 −/+ | |
| ParSe v2 | 0.647 −/+ | 0.734 −/+ | 0.442 −/+ | 0.327 −/+ | |
| Predictors of intrinsic disorder | AIUPred | 0.838 −/+ | 0.747 −/+ | 0.346 −/+ | 0.145 −/+ |
| flDPnn | 0.717 −/+ | 0.646 −/+ | 0.537 −/+ | 0.273 −/+ | |
| Random baseline | 0.493 −/ | 0.508 −/ | 0.181 −/ | −0.014 −/ | |
| AUROC | AUPRC | F1 | MCC | ||
|---|---|---|---|---|---|
| Predictors of phase separation | PSPHunter | 0.882/+ | 0.891/+ | 0.525/+ | 0.420/+ |
| Seq2Phase | 0.685 −/+ | 0.710 −/+ | 0.267 −/+ | 0.185 −/+ | |
| PLAAC | 0.684 −/+ | 0.747 −/+ | 0.436 −/+ | 0.363 −/+ | |
| ParSe v2 | 0.663 −/+ | 0.722 −/+ | 0.452 −/+ | 0.324 −/+ | |
| PScore | 0.629 −/+ | 0.659 −/+ | 0.455 −/+ | 0.263 −/+ | |
| LLPhyScore | 0.616 −/+ | 0.660 −/+ | 0.405 −/+ | 0.168 −/+ | |
| catGranule | 0.608 −/+ | 0.694 −/+ | 0.459 =/+ | 0.137 −/+ | |
| FuzDrop | 0.584 −/= | 0.627 −/+ | 0.309 −/+ | 0.008 −/= | |
| Predictors of intrinsic disorder | AIUPred | 0.561 −/= | 0.556 −/= | 0.308 −/+ | −0.089 −/= |
| flDPnn | 0.361 −/= | 0.435 −/= | 0.421 =/+ | −0.180 −/= | |
| Random baseline | 0.499 −/ | 0.495 −/ | 0.201 −/ | 0.009 −/ | |
| AUROC | AUPRC | F1 | MCC | ||
|---|---|---|---|---|---|
| Predictors of phase separation | FuzDrop | 0.882/+ | 0.737 =/+ | 0.812/+ | 0.711/+ |
| PSPHunter | 0.863 =/+ | 0.804/+ | 0.793 =/+ | 0.694 =/+ | |
| Seq2Phase | 0.848 =/+ | 0.776 =/+ | 0.756 =/+ | 0.643 =/+ | |
| LLPhyScore | 0.825 −/+ | 0.676 −/+ | 0.715 −/+ | 0.567 −/+ | |
| PScore | 0.738 −/+ | 0.582 −/+ | 0.491 −/+ | 0.242 −/+ | |
| PLAAC | 0.700 −/+ | 0.722 −/+ | 0.619 −/+ | 0.424 −/+ | |
| catGranule | 0.680 −/+ | 0.516 −/+ | 0.508 −/+ | 0.255 −/+ | |
| ParSe v2 | 0.645 −/+ | 0.666 −/+ | 0.571 −/+ | 0.346 −/+ | |
| Predictors of intrinsic disorder | AIUPred | 0.826 −/+ | 0.602 −/+ | 0.723 −/+ | 0.574 −/+ |
| flDPnn | 0.680 −/+ | 0.468 −/+ | 0.582 −/+ | 0.353 −/+ | |
| Random baseline | 0.484 −/ | 0.351 −/ | 0.220 −/ | −0.203 −/ | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Wang, K.; Hu, G.; Kurgan, L. Empirical Assessment of Sequence-Based Predictions of Intrinsically Disordered Regions Involved in Phase Separation. Biomolecules 2025, 15, 1079. https://doi.org/10.3390/biom15081079
Wu X, Wang K, Hu G, Kurgan L. Empirical Assessment of Sequence-Based Predictions of Intrinsically Disordered Regions Involved in Phase Separation. Biomolecules. 2025; 15(8):1079. https://doi.org/10.3390/biom15081079
Chicago/Turabian StyleWu, Xuantai, Kui Wang, Gang Hu, and Lukasz Kurgan. 2025. "Empirical Assessment of Sequence-Based Predictions of Intrinsically Disordered Regions Involved in Phase Separation" Biomolecules 15, no. 8: 1079. https://doi.org/10.3390/biom15081079
APA StyleWu, X., Wang, K., Hu, G., & Kurgan, L. (2025). Empirical Assessment of Sequence-Based Predictions of Intrinsically Disordered Regions Involved in Phase Separation. Biomolecules, 15(8), 1079. https://doi.org/10.3390/biom15081079









