Abstract
Class IIa histone deacetylases (HDACs) are pleiotropic regulators of various differentiation pathways and adaptive responses. They form complexes with other co-repressors and can bind to DNA by interacting with selected transcription factors, with members of the Myocyte Enhancer Factor-2 (MEF2) family being the best characterized. A notable feature of class IIa HDACs is the substitution of tyrosine for histidine in the catalytic site, which has occurred over the course of evolution and has a profound effect on the efficiency of catalysis against acetyl-lysine. Another distinctive feature of this family of “pseudoenzymes” is the regulated nucleus–cytoplasm shuttling associated with several non-histone proteins that have been identified as potential substrates, including proteins localized in the cytosol. Within the complexity of class IIa HDACs, several aspects deserve further investigation. In the following, I will discuss some of the recent advances in our knowledge of class IIa HDACs.
1. Introduction
Acetylation of proteins is a widely used post-translation modification (PTM) that is exploited to influence a variety of cellular reactions. The amino group (NH2) attached to the ε-carbon of a lysine residue is the subject of such a PTM. Although the acetyl group is small, it can profoundly influence the protein surface by revoking the positive charge of the lysine. This can have a direct effect on the dynamics of local molecular interactions. In addition, acetylation of lysine can also maintain a local hydrophobic milieu that forms the binding site for the recruitment of proteins with specialized domains: the acetyl-lysine readers [1]. For these readers, the N-ε-acetylation of lysine represents a signal that is recognized by specific domains: the bromodomain, the YEATS domain, and the PHD fingers. These domains are characterized by the presence of a hydrophobic pocket suitable for the binding of peptide sequences containing an acetyl-lysine. These interactions modulate the assembly of multiprotein complexes on the DNA [2,3].
If the acetylated proteins are histones, this modification controls the accessibility to DNA and the assembly of multiprotein complexes on the chromatin, which activate gene transcription or modulate DNA repair [4]. The particular profile of histones acetylation can be maintained in the newly synthesized histones during DNA replication. Therefore, the memory of the differently accessible genome regions and the specific pattern of gene transcription can be inherited by the daughter cells after mitosis. This process is known as epigenetics [5,6].
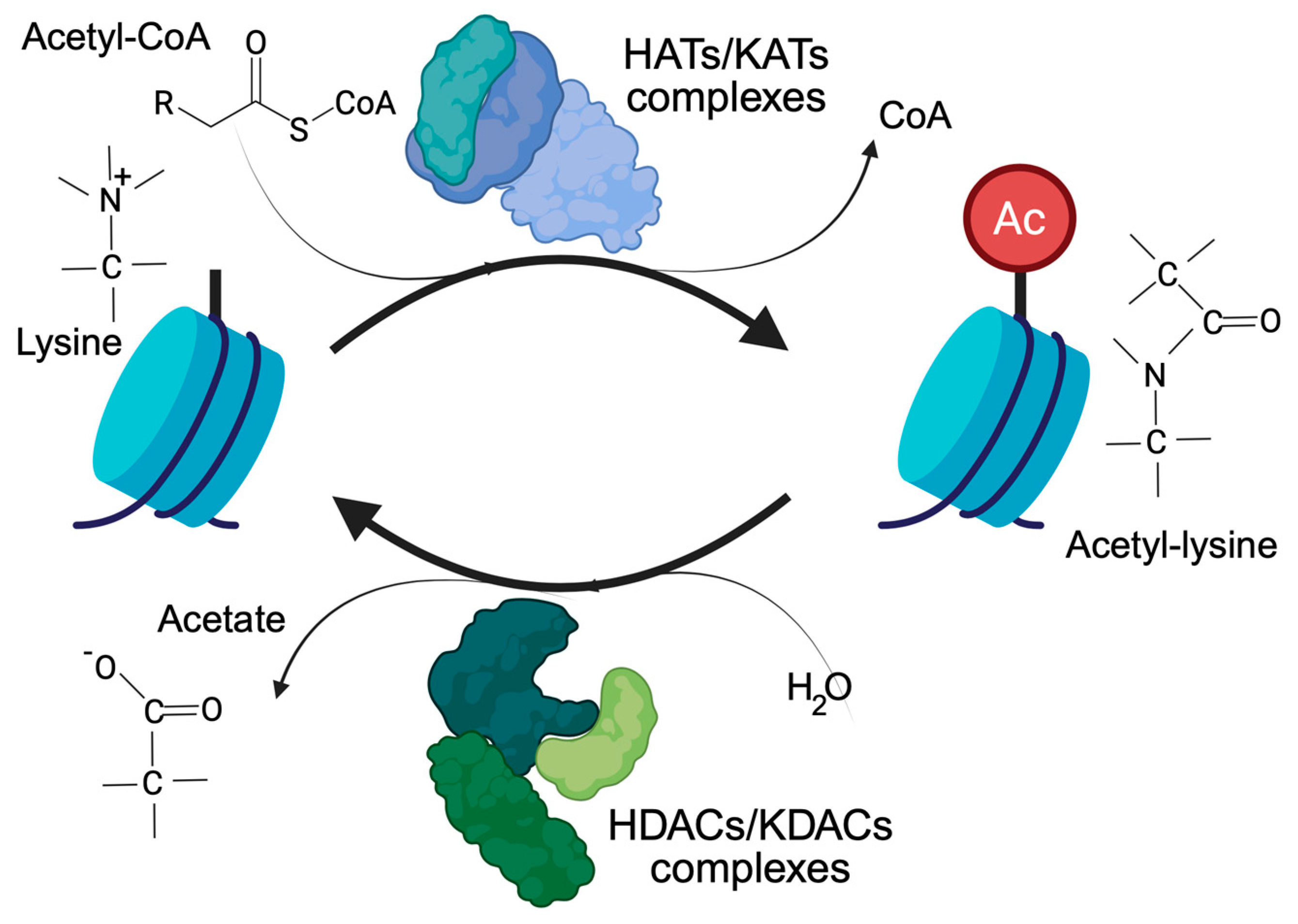
Histone acetylation, and more generally lysine acetylation, is a dynamic and reversible process controlled by two antagonistic enzyme families, the KATs/HATs (lysine/histone acetyl transferases) and the KDACs/HDACs (lysine/histone deacetylases). KATs use the acetyl-CoA as a donor of the acetyl group. KDACs, on the other hand, produce acetate [7], which can be converted back into acetyl-CoA by the acyl-CoA synthetase short-chain family member 2 (Figure 1). Cells have different ways of supplying the nucleus with the acetyl-CoA required to regulate the accessibility of chromatin [2,8]. The link between acetylation, epigenetics, and metabolism is strong and clearly recognizable. Histone acetylation and gene transcription are energy-consuming processes and require metabolically active cells with available energy (ATP) and metabolic substrates. In view of the large number of histone proteins, histone acetylation also represents an important storage of acetyl groups within the cell [8].
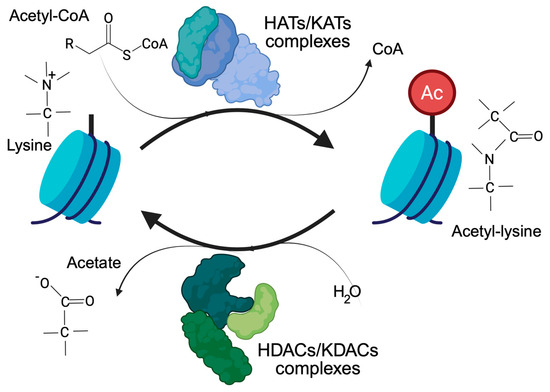
Figure 1.
The control of lysine acetylation in histones by HATs/KATs (histone acetyl-transferase/lysine acetyl-transferase) and HDACs/KDACs (histone deacetylase/lysine deacetylase). Acetate can be converted back to acetyl-CoA by ACSS2. Only the ε-carbon group of lysine is shown.
2. An Overview of the Various Types of HDACs/KDACs
Eighteen KDACs/HDACs are present in humans, which are divided into zinc-dependent and NAD+-dependent enzymes. Five distinct classes of vertebrate HDACs have been proposed based on structural homologies to yeast HDACs and other features. The zinc-dependent enzymes form classes I, IIa, IIb, and IV. The NAD+-dependent enzymes are summarized in class III. Class I includes HDAC1, 2, 3, and 8. HDAC1 and HDAC2 often form common repressive complexes [7,9,10]. HDAC3 forms a distinct repressive complex with the co-repressors NCOR1 and NCOR2 (nuclear receptor co-repressor), which are required for the maturation of the catalytic activity of HDAC3 [11]. The assembly into multiprotein complexes supports the full activation of the catalytic activity of these three HDACs [10]. HDAC1/2/3 are important regulators of histone acetylation. HDAC8 differs from the other members of the class I family in that it exhibits strong catalytic activity as an isolated protein. However, HDAC8 is also subject to modulation as it possesses an allosteric domain that is structured in the form of a helix-loop-helix. This “allosteric domain” is the target of regulatory PTMs that modulate the activity of HDAC8 [12,13].
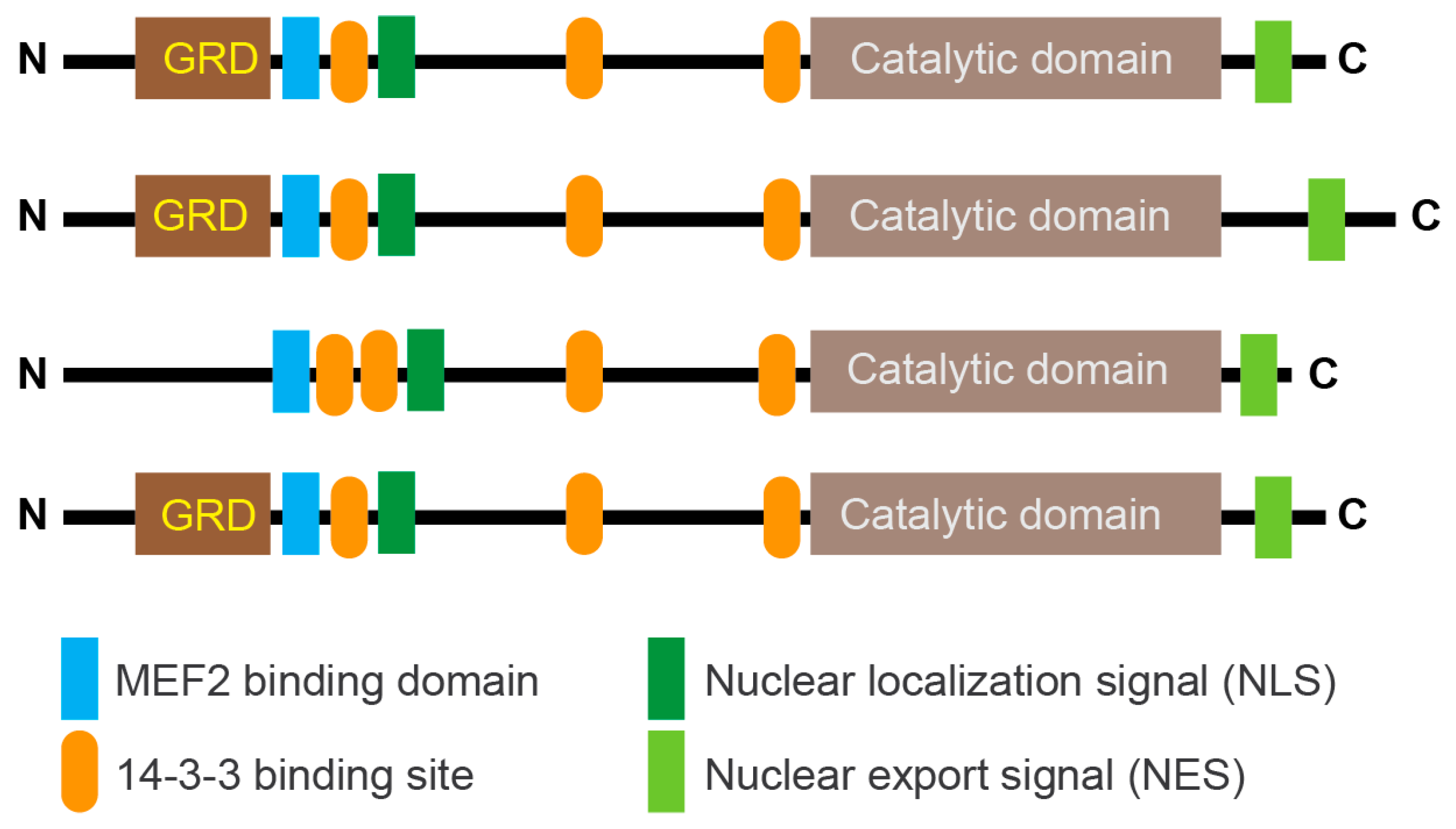
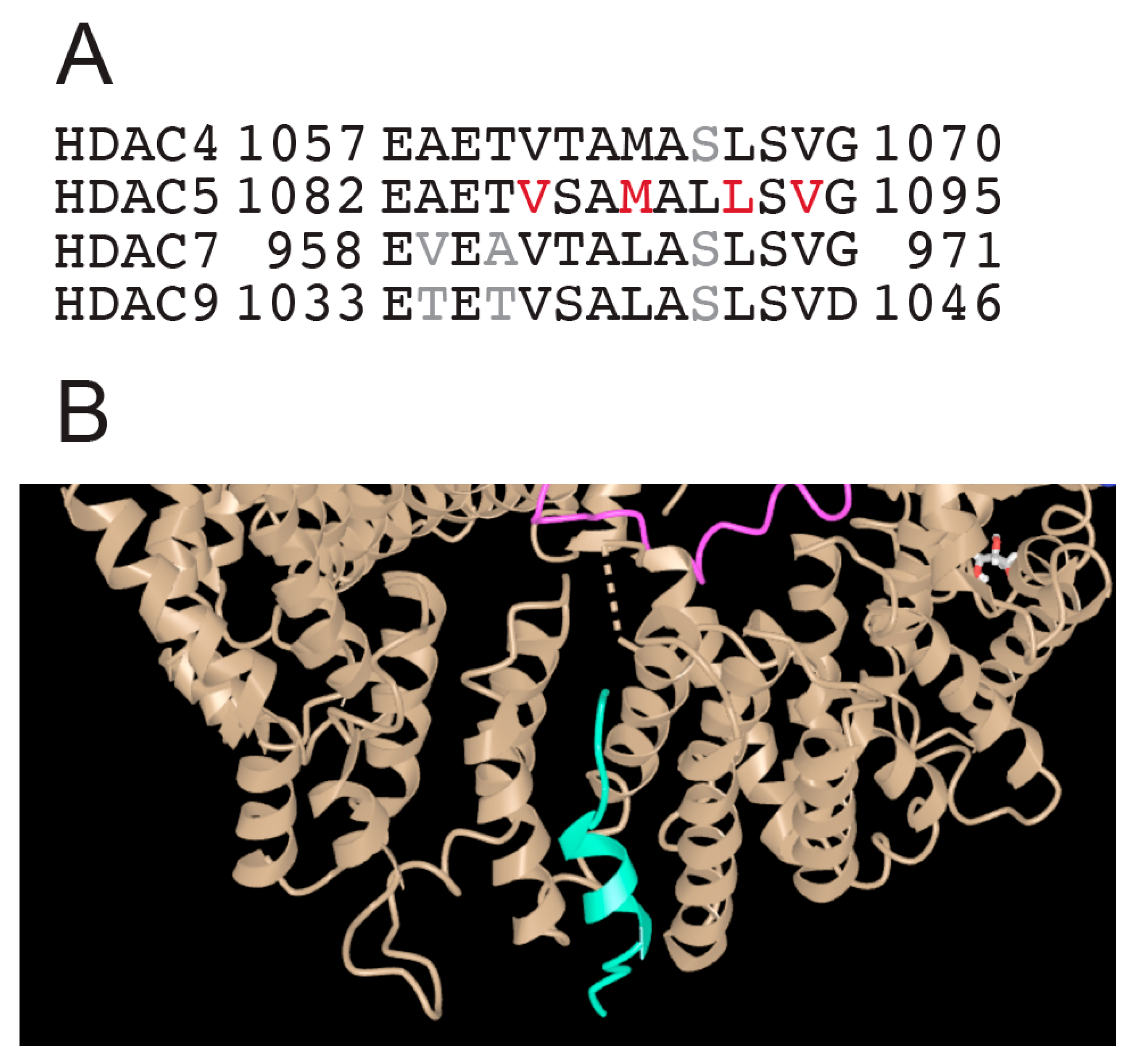
Class IIa groups HDAC4, HDAC5, HDAC7, and HDAC9. These KDACs have an extended amino-terminal region that is designed to interact with multiple partners, including transcription factors such as members of the MEF2 family and co-repressors. This region contains an NLS (nuclear localization sequence) and phosphorylation sites recognized by 14-3-3 chaperones (Figure 2). The carboxy-terminal region contains the deacetylase domain and the NES (nuclear export sequence). In vertebrates, the tyrosine in the catalytic pocket is replaced by a histidine residue that renders the reaction inefficient against acetyl-lysine. This modification may have converted the class IIa HDACs into acetyl-lysine readers or may have altered substrate specificity.
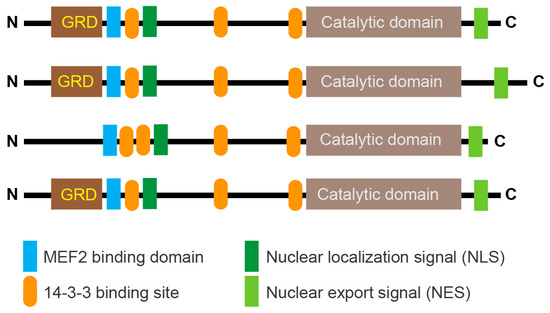
Figure 2.
Schematic representation of the main domain organization in class IIa HDACs. Binding to 14-3-3 binding sites highlights the major serine residues that are phosphorylated by various kinases and promote nuclear export of deacetylases. In vertebrates, the deacetylase domain exhibits very low enzymatic activity due to the His/Tyr substitution in the catalytic site. The glutamine-rich domain (GRD) is indicated. The control of lysine acetylation in histones by HATs/KATs and HDACs/KDACs. Acetate can be converted back to acetyl-CoA by ACSS2. Only the ε-carbon group of lysine is shown.
However, alternative substrates for class IIa HDACs have not yet been clearly defined.
The activities of class IIa HDACs are tightly controlled by nuclear/cytoplasmic shuttling, which is monitored by the specific phosphorylation of the various 14-3-3 binding sites. Although the activities of class IIa HDACs are often controlled by their subcellular localization, the cells can use several additional mechanisms [14]. For example, proteasomal-mediated degradation can control the level of class IIa HDACs depending on phosphorylation [15,16,17]. Consequently, class IIa ubiquitylation can be used to modulate various cellular responses. Viral proteins can promote the degradation of HDAC4/HDAC5 to limit interferon production and enable viral infection [18]. Proteasomal degradation is also involved in switching off the repressive activity of HDAC4 during senescence [19].
Class IIb includes HDAC6 and HDAC10. HDAC6 contains two catalytically active domains, CD1 and CD2. The CD2 domain acts as an α-tubulin deacetylase and controls several other substrates [20]. The CD1 domain has very few substrates and shows specificity for C-terminal acetyl-lysine residues [21]. HDAC10 functions as a polyamine deacetylase and can protect cancer cells from chemotherapy by activating autophagy [22,23]. Class IIb are cytosolic enzymes with low activity towards acetylated histones [24].
The NAD+-dependent deacetylates are known as sirtuins, which in humans consist of 7 members (Sirt1/2/3/4/5/6 and 7). The sirtuins form the class III family of HDACs [25]. They are homologous to Sir2 (silent information regulator) in yeast and are found in various cellular compartments, including the mitochondria. Targets of sirtuins are histones as well as various non-histone proteins [26,27].
Finally, HDAC11 alone forms class IV. Some peculiarities of the catalytic domain of HDAC11 have justified the creation of this additional class of KDACs. This zinc-dependent deacetylase exhibits a higher enzymatic activity towards long-chain acyl groups, which has led to the definition of HDAC11 as a lysine de-fatty acylase [28]. Unicellular eukaryotes do not appear to possess class IV HDACs [29].
3. Class IIa HDACs
Class IIa HDACs are involved in several differentiation pathways that lead to the formation of various tissues, including blood vessels, bone, muscle, adipose tissue, myelination, and the immune system. These “pseudoenzymes” are also involved in the control of various adaptive responses, including cell proliferation, cell death, metabolism, migration, addiction, tissue injury, and many others [14,30,31,32,33,34,35,36,37].
Several reviews have addressed the contribution of class IIa HDACs to specific cellular responses, and it is difficult to find a specific context in which their contribution can be excluded. The versatility of class IIa HDACs is evidenced by various biological processes under their influence. It must also be considered that the expression of class IIa HDACs can be regulated by interconnected circuits based on transcription factors whose activities can also be regulated by the same class IIa HDACs [38]. This strategy guarantees compensation mechanisms between the different members of the family and can limit our knowledge of the contribution of an individual member to a specific cell function [39,40]. In addition, different class IIa HDACs can also cooperate to maintain a particular differentiation fate [41]. Another level of complexity is exemplified by HDAC9. A structural variant (SV) is present in the coding sequence of HDAC9. This SV affects the functionality of the TWIST1 regulatory elements located within the HDAC9 sequence. In fact, the 3′-HDAC9 sequence functions as a critical TWIST1 regulatory region that includes craniofacial TWIST1 enhancers and CTCF sites. This SV is associated with craniosynostosis, a condition that alters postnatal skull and brain growth [42]. The complexity of class IIa HDACs can therefore also result from the inclusion of cis-regulatory elements that control the expression of other genes.
4. Structural Features of Class IIa HDACs
Although we have only partial information about the overall structure of class IIa, the definition of the structural characteristics of some specific domains/regions has provided insight into the possible mechanisms used by this gene family to modulate the various responses discussed here.
4.1. The N-Terminal Region
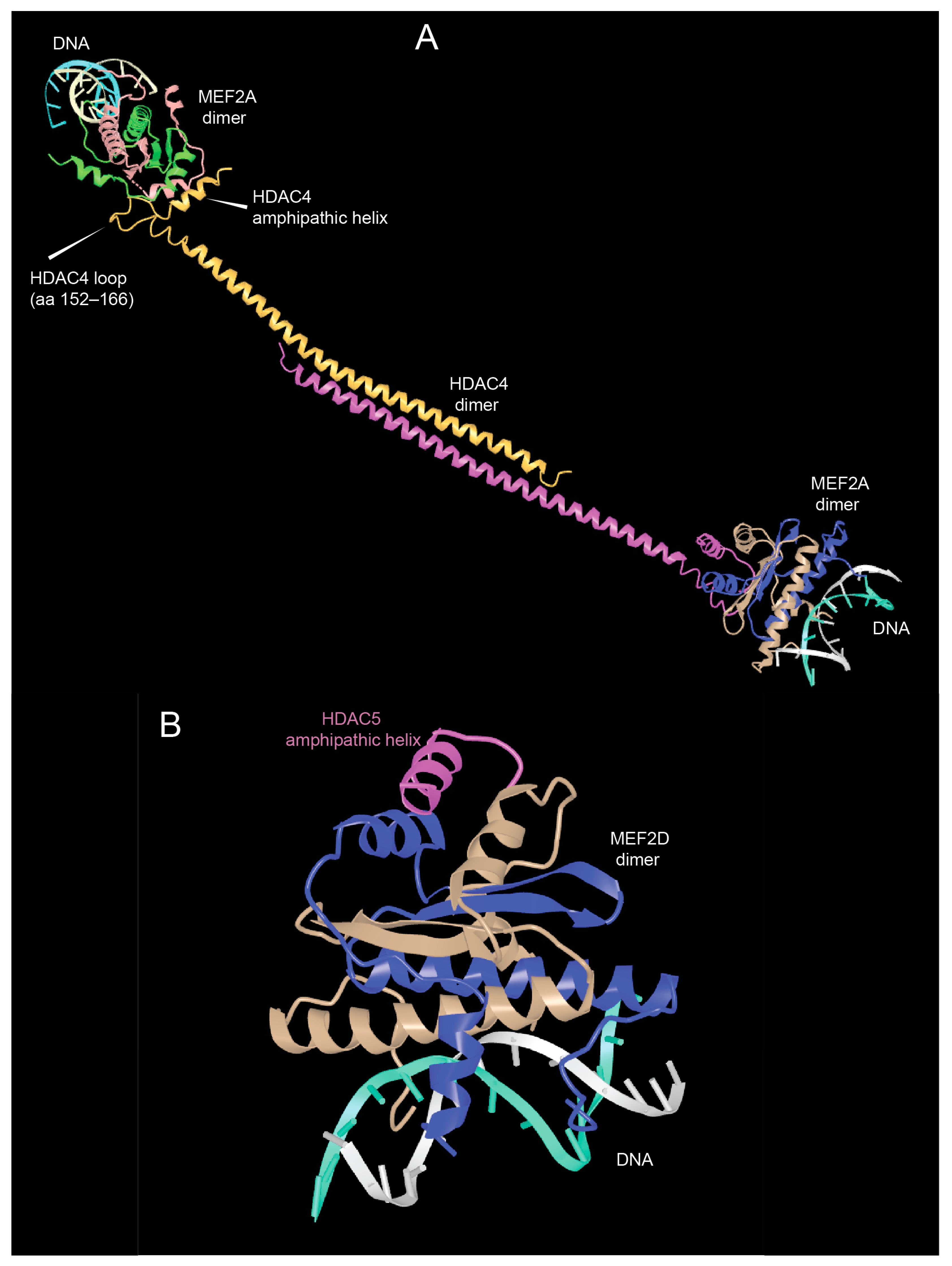
As shown in Figure 2, the N-terminal region of HDAC4/5 and 9 contains a glutamine-rich domain (GRD). Structural studies with the carboxy-terminal part of the HDAC4 GRD have shown that it can fold into a long helix that is dynamically balanced between dimer and tetramer [43]. This domain may also be involved in hetero-oligomerisation between the different members of the class IIa HDACs family. Recently, Dai et al. co-crystallized a complex between human HDAC4-GRD (aa 62–192) and the MADS box/MEF2s domain of MEF2A (aa 1–95) in the presence of a 15-mer DNA duplex with the MEF2 consensus sequence [44].
The analyzed region of HDAC4 consists of an amphipathic helix that binds MEF2 (see below), which exits the GRD after a loop. This loop also contributes to binding with MEF2A. In this new structure, the GRD is folded into an elongated helix, a conformational change possibly favored by MEF2A binding. The GRD enables the dimerization of HDAC4 and the coordination of two different MEF2A-DNA complexes (Figure 3A).
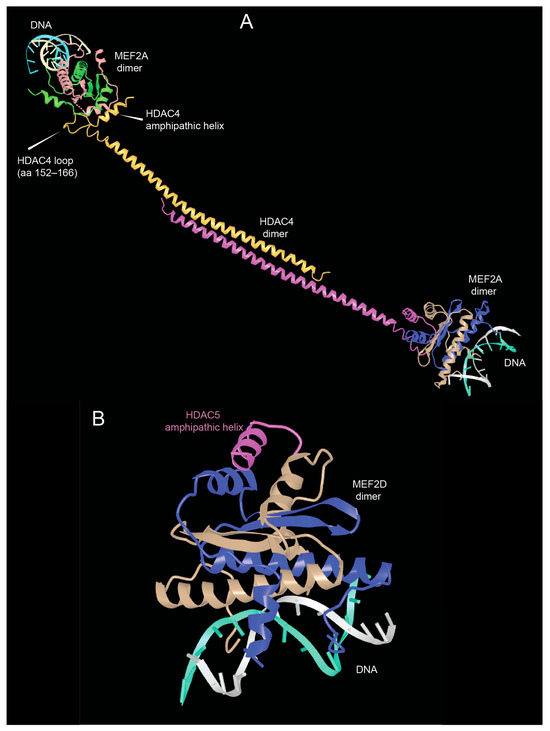
Figure 3.
The domains of the amino-terminal region of class IIa HDACs. (A) View of the glutamine-rich domain (GRD) of HDAC4 in complex with MEF2A. PDB ID: 7XUZ https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?&mmdbid=239077&bu=0&showanno=1&source=full-feature, 3 June 2025. (B) Crystal structure of the MADS box/MEF2 domain of MEF2D bound to dsDNA and the HDAC5 deacetylase peptide containing the MEF2 binding motif. PDB ID 8Q9P. https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?&mmdbid=245462&bu=0&showanno=1&source=full-feature, 3 June 2025. Each MEF2 dimer is shown with different colors (brown/blue and brown/green)—HDAC4-GDR monomer is shown with different colors (pink/yellow).
The formation of the complex between MEF2A and HDAC4 could also influence the ability of HDAC4-GRD to tetramerize and form higher order structures that favor the stabilization of dimers [43]. In addition, HDAC4-mediated dimerization of HDAC–MEF2A complexes may bridge to distant DNA sites to create a local repressive chromatin environment in a more productive manner [44].
Additional structural studies using class IIa HDACs peptides corresponding to the amphipathic helices that bind MEF2D have shown that these peptides are unstructured in solution and adopt a folded α-helical structure only after binding to MEF2D (Figure 3B). Furthermore, the adaptability of the hydrophobic MEF2 furrow that accommodates these class IIa peptides explains the multiple protein–protein interactions involving other transcriptional regulators that are recognized by MEF2 via this domain [45].
4.2. The Catalytic Domain
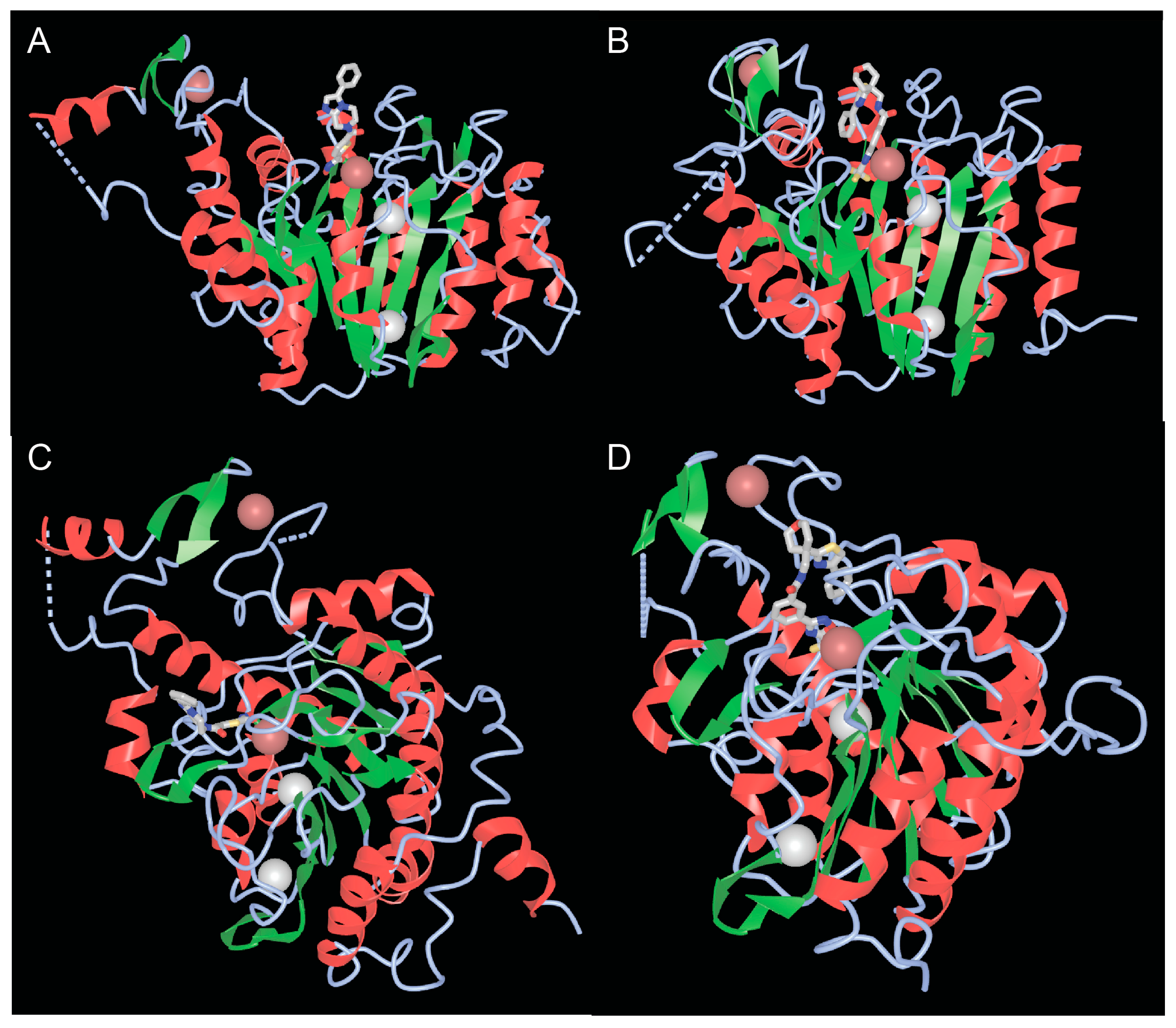
Well-defined functional activities embedded in the carboxy-terminal region of class IIa are the catalytic domain and the NES. The catalytic domain is structurally conserved between the four members [33,46]. The structure of the catalytic domains of HDAC4 and HDAC7, whether in complex with different inhibitors or not, has been solved [47,48]. These studies have revealed the homologies with the corresponding domain of class I HDACs, characterized by a central parallel β-sheet with eight β-strands (Figure 4A,B). In addition to the zinc ion, which is located within the catalytic domain, there are two potassium ions in the vicinity of the active sites, similar to class I HDACs (Figure 4A,B).
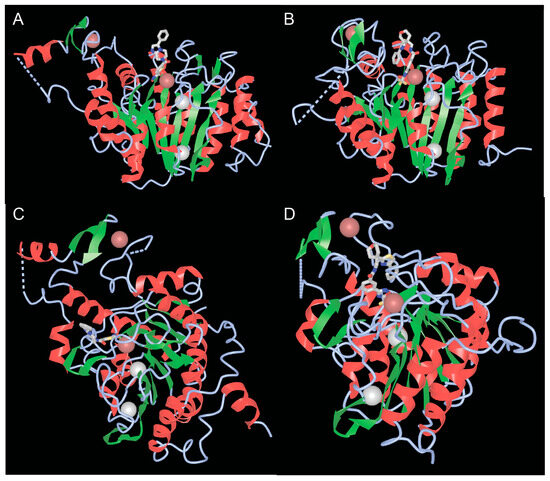
Figure 4.
The catalytic domains of class IIa HDACs. (A,B) View of the catalytic domains of HDAC4 (A) and HDAC7 (B) showing the catalytic pocket with the coordinated zinc and adjacent potassium ions. (C,D) Rotated view of the class IIa catalytic domain highlighting the second structural zinc ion and the structural zinc-binding domain (sZBD) of HDAC4 (C) and HDAC7 (D). PDB ID 2VQM. https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?&mmdbid=65359&bu=1&show-199%20anno=1&source=full-feature, 3 June 2025. The α-helices are shown in red and β-strands in green.
The low class IIa deacetylase activity in vertebrates depends on the substitution of tyrosine 976 (in HDAC4, Y843 in HDAC7) by a histidine in the catalytic pocket. This tyrosine is considered important for stabilizing the transition state by hydrogen bonding with the oxyanion intermediate, whereas the histidine, which is turned away from the active site (out conformation), makes the transition state less stable and deacetylation less efficient [47,48,49,50]. However, binding of some protein partners or substrates of class IIa HDACs (including sequences in addition to acetylated lysine) could promote inward rotation of the histidine side chain, orienting the histidine ring towards the catalytic site and the acetyl group of the substrate. An “in” conformation that could support catalysis involving a water molecule [47,48]. Recently, analyses of different hDAC4 crystal structures and computer simulations have shown that a rotational transition between the “in” and “out” conformation of H976 is possible due to the very high flexibility of the loop segments around this residue [51].
Another special feature of the catalytic domain of class IIa HDACs is the lack of a water-filled tunnel. In class I HDACs, this tunnel should allow the release of the acetate reaction product [47].
Although class IIa enzymatic activity can be measured on a non-physiological substrate (the trifluoroacetyl-lysine), a detailed study using a peptide library was performed to determine the substrate specificities of class IIa HDACs. With respect to trifluoroacetyl-lysine, the aa were mapped at positions −3 and +3. Interestingly, class IIa HDACs show a strong preference for bulky aromatic acids flanking the central trifluoroacetyl-lysine, while positively charged residues and proline are not favored [52]. These results could help to further improve the development of specific class IIa inhibitors.
The question of possible alternative class IIa substrates has not yet been resolved. Recently, de-crotonylation activity has been associated with HDAC7. During leucine deprivation-induced autophagy, 14-3-3 proteins are crotonylated. This PTM reduces interactions with partners, including protein phosphatase 1B (PPM1B), which stimulates autophagy via ULK1. Specific class IIa inhibitors increase the crotonylation of PPM1B [53]. However, further studies are needed to determine whether HDAC7 can function as decrotonylases or whether this activity is mediated by an associated class I HDAC [54].
4.3. The Second Structural Zinc Ion
A third important feature of the catalytic domain of class IIa HDACs is the presence of a second zinc ion that fulfills structural roles and is also of interest as a potential target for selective class IIa inhibitors (Figure 4C,D). The structural zinc ion is crucial for the connection of two segments of HDAC4. The first segment is a 17 amino acid loop (Lα1-α2) containing three residues (His665, Cys667, and His678), which chelate zinc. The other segment is a helix-turn-helix motif of 35 amino acids (α6-α7-β3-β4), followed by a β-hairpin with the fourth residue that can chelate zinc (Cys751). This domain is known as the structural zinc-binding domain (sZBD).
By comparing the inhibitor-free and inhibitor-bound structures of the catalytic domain of HDAC4, Bottomley and co-authors proposed that the inhibitor can influence the conformation of the sZBD. An open conformation is stabilized by the inhibitor, while the closed conformation is the native conformation. The authors concluded that different conformations of the sZBD may be required to allow conformational flexibility and may play a role in regulating HDAC4 function. Indeed, this structural domain in the closed conformation is also involved in regulating the interaction with the repressive complex HDAC3-NCOR1/NCOR2 [47].
Recent studies have shown that the sZBD plays a key role in the global structural integrity and stability of HDAC4, but these studies have also challenged the open conformation hypothesis and questioned its existence. Therefore, the authors suggest that only the closed conformation should be considered for the development of HDACs-specific class IIa-inhibitors [55].
4.4. NCOR1 and NCOR2
NCOR (nuclear receptor co-repressor) 1 and 2, the latter originally called SMRT (silencing mediator of retinoic acid and thyroid hormone receptor), are pleiotropic transcriptional co-regulators. Their activities, which are not redundant, are crucial for mouse development [56,57,58]. Although the multiprotein complexes that assemble with NCOR1 and 2 change in different contexts, some partners are stably observed. These include the following: HDAC3, which is the major partner and requires NCORs to exert full enzymatic activity; the G protein pathway suppressor (GPS2), the transducing β-like 1 (TBL1) and its homolog, TBL-related 1 (TBLR1). Under certain conditions, the NCOR complex can also associate with HDAC1 [59,60]. NCOR proteins can be considered as large platforms with an average weight of about 270 kDa that support interaction with multiple partners. Different NCOR domains mediate interactions with specific partners. At the amino terminus, the RDs (repressive domains), which are highly conserved between NCOR1 and 2, mediate interaction with GPS2 and TBL1 or other co-repressors, including class IIa HDACs recognized by RD3. The SANT-like domains are important for deacetylation and one of them includes the deacetylase activation domain (DAD), which binds HDAC3. One of the SANT domains is part of the deacetylase activation domain (DAD). Finally, there are 3 receptor interaction domains (RIDs) that mediate the interaction with nuclear receptors [58].
A conserved repetition of eight amino acid motifs with the consensus sequence G-S-I-t/s-q-G-t-P characterizes RD3. In particular, the repetition of the GSI motif was hypothesized to be critical for the interaction with class IIa HDACs. The GSITQGTP corresponding to sequence 1450-1469 of NCOR2 is sufficient to bind the catalytic domain of class IIa HDACs. Within this sequence, the GSI and the G and T at position 6 and 7, respectively, are absolutely required for the interaction. On the other hand, mutations of critical zinc chelating residues within the sZBD of HDAC4 have shown that the peptide and thus the NCORs recognize these surface loops near the catalytic site and in closed conformation [61].
4.5. The Nuclear Export Signal (NES)
The regulation of class IIa HDACs nuclear export plays a central role in various cellular states from cell proliferation/survival to differentiation and in many adaptive responses [14,25,31,32,33,34,62,63].
The export of proteins from the nucleus is often mediated by a leucine-rich NES sequence. In general, this sequence is a short stretch of 8–15 amino acids with regularly spaced hydrophobic residues. NESs are the consensus sequence recognized and bound by the export karyopherin [64]. Many of the identified leucine-rich NESs deviate significantly from the generally accepted loose consensus L-x(2,3)-[LIVFM]-x(2,3)-L-x-[LI] [65]. This has led to the classification of NES into different subclasses [66]. An NES binds to the hydrophobic pocket (labeled P0–P4) in the hydrophobic groove defined by the HEAT repeats 11 and 12 of CRM1/Exportin-1 [66,67].
At the carboxy terminus, after the deacetylase domain, all class IIa HDACs contain an NES. This sequence is highly conserved among the different members, with the only difference that in HDAC4/5 a methionine has been replaced by a leucine, in HDAC7/9 as the third key hydrophobic residue of the motif (Figure 5A). In these sequences, the positions of the hydrophobic residues do not indicate the formation of an amphipathic helix. However, the corresponding peptide of HDAC5, which was used to define the complex with CRM1/exportin-1, partially folds into an α-helix (Figure 5B) [68].
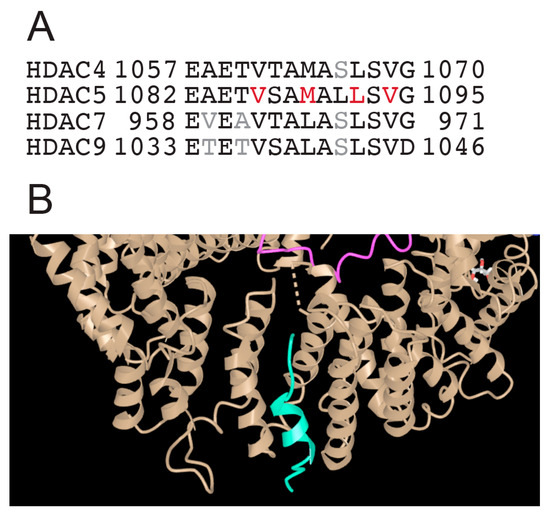
Figure 5.
The NES of class IIa HDACs. (A) Alignment of the class IIa carboxy-terminal sequence with the different nuclear export signals (NESs). (B) Enlarged view of the structure of the HDAC5 NES peptide (green) in complex with CRM1/Exportin-1 (brown). The original molecular complex also contains RAN (pink) and RAN-GAP (blue). PDB-ID 5UWI. https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?&mmdbid=148948&bu=1&showanno=1&source=full-feature, 3 June 2025.
Phosphorylation of 14-3-3 binding sites affects NES activity, and an integral NES is required for the export of class IIa HDACs even in the presence of 14-3-3 binding [69,70,71]. In the case of HDAC7, the forced expression of CRM1 can trigger the nuclear export of deacetylase even in the presence of mutations at the 14-3-3 binding sites [72].
5. Class IIa HDACs as Pleiotropic Regulators of Both Differentiating and Adaptive Responses
Class IIa HDACs are considered pleiotropic genes that are involved in the regulation of a broad spectrum of biological responses. One example discussed here is HDAC4.
In the posterior hypothalamus, Hdac4 and Hdac5 regulate non-rapid eye movement sleep (NREMS). Phosphorylation of Hdac4 and Hdac5, which occurs via the Lkb1-Sik3 signaling pathway, is associated with an increased need for sleep, and Hdac4 haploinsufficiency in mice increases sleep [73,74]. In humans, both haploinsufficiency of HDAC4 and point mutations that impair binding to 14-3-3 proteins are associated with sleep disorders. These point mutations in HDAC4 are responsible for several additional alterations, including dysmorphic facial features, dysphagia and/or drooling, congenital hip dislocation, and progressive kyphoscoliosis [75]. Haploinsufficiency of HDAC4, on the other hand, is responsible for 2q37 deletion syndrome, a disorder characterized by brachydactyly type E (BDE) and typical facial features. The haploinsufficiency is not completely penetrant and additional defects vary in severity between patients [33,76,77].
HDAC4 is abundantly expressed in bone and cartilage, and osteoblast lineage-specific knock-outs have demonstrated its key role in skeletal growth. The most obvious defects in these mice are premature ossification of developing bone [78,79]. In particular, conditional knock-out of Hdac4 in osteoprogenitors is associated with increased susceptibility to dwarfism, growth plate closure, and osteoporosis progression [79].
Further studies in mice in which Hdac4 was silenced in a tissue-specific manner have revealed a contribution of deacetylase in various contexts, including the mediation of inflammatory pain [80], proliferation and differentiation of satellite cells [81] or protection of the diabetic heart [82]. It has also been reported that the cytoplasmic functions of HDAC4 are critical for plasma membrane repair in a mouse model of Duchenne muscular dystrophy [83].
In summary, genetic studies and animal models have clearly demonstrated the involvement of HDAC4 in a variety of cellular responses that require modulation of gene expression and readjustment of epigenetic profiles. However, epigenetically independent effects of HDAC4 have also been observed. The example of HDAC4 briefly discussed here shows the pleiotropic nature of class IIa HDACs, which is probably underestimated given the redundancy and compensatory circuits that characterize the regulation of this enzyme family.
6. Recent Advances in Class IIa HDACs Biology
The contribution of class IIa HDACs to various differentiation and adaptation responses has been discussed in many reviews [14,31,32,33,34,62,63]. Therefore, in the following sections I will focus on the new results that have been published in recent years.
6.1. DNA Repair
The contribution of class IIa to DNA repair was initially investigated in a few studies, but only recently have details of the mechanisms involved become known [84,85,86]. Repair of DNA double-strand breaks (DSBs) can occur mainly via two pathways: non-homologous end joining and homologous recombination. Alternative native end-joining repair mechanisms can also be used [87].
HDAC4 may act as an epigenetic regulator of H2BK120 acetylation at the sites of DSBs through a complex with HDAC1/HDAC2. This activity is important for the proper recruitment of DNA repair enzymes, including BRCA1, which is required for homology-directed repair [88]. Interestingly, class IIa HDACs can also repress the expression of genes involved in non-homologous end joining in an MEF2-dependent manner [89].
Additionally, class IIa HDACs can influence DNA repair by controlling the acetylation of non-histone proteins. HDAC5 can deacetylate PARP1 at Lys498 and Lys521. Acetylation of these residues limits PARP1 PARylation and DNA repair [90]. As a result, depletion of HDAC4 and HDAC5 increases the amount of unrepaired DNA, as indicated by yH2AX positivity. HDAC4 and HDAC5 may also indirectly contribute to DNA repair by downregulating gene transcription through acetylation of ATF9, a component of the elongation machinery. The authors propose that the export of class IIa HDACs to the cytoplasm upon DNA damage, as driven by CamKII, favors acetylation of ATF9 [91]. For both examples of PARP1 and ATF9 deacetylation, it remains to be clarified whether class I HDACs in complex with HDAC5 can contribute to enzymatic activity.
6.2. Senescence
It has recently been shown that class IIa HDAC can inhibit senescence in certain cellular models [19,92,93,94]. Since senescence can also be triggered by the accumulation of DNA damage [95,96,97,98], the influence of class IIa HDACs on the efficiency of DNA repair and the accumulation of DNA damage could be the cause of the senescent phenotype. Certainly, the transcriptional control exerted by class IIa can also help to antagonize the epigenetic and transcriptomic profiles that characterize senescence, particularly through the control of super-enhancers [19,92,93].
6.3. Metabolism
Class IIa HDACs are known substrates of SIKs (Salt-inducible kinases) that control their nuclear export [14]. In a brown mouse cell line, Hdac4, but not other class IIa HDACs, is required for the expression of the key factor for thermogenesis, uncoupling protein 1 (Ucp1). The activity of Hdac4 is explained by the deacetylation and activation of PGC1α (peroxisome proliferator-activated receptor gamma (PPARγ) co-activator-1α), an important co-activator of gene expression [99]. Mitochondrial metabolism is also influenced by HDAC7, but in a different context and in a different way. In renal cell carcinoma, HDAC7 suppresses the expression of enzymes involved in the tricarboxylic acid cycle (TCA). In this malignant disease, HDAC7 is activated by TGF-β signaling to alter TCA metabolism [100].
6.4. Class IIa HDACs and Differentiation
The effect of class IIa on epigenome rearrangement can be dramatic. During terminal erythroid differentiation, chromatin condensation is followed by nuclear polarization and extrusion of the condensed nucleus [101]. In an in vitro model of erythroid differentiation, HDAC5 expression is upregulated, and its knockdown impairs differentiation, nuclear condensation, and enucleation. This effect correlates with a specific defect in H4K12 acetylation, which is not reduced during erythroid terminal differentiation in the absence of HDAC5 [102].
In other contexts, single class IIa affects the epigenome mainly on regions subject to dynamic regulation and buffers the acetylation level of accessible regions. It is less clear whether class IIa may play a role in maintaining the heterochromatic regions of the genome [102,103,104,105]. Whether the simultaneous removal of the different family members could affect the opening of heterochromatic regions is an open question that deserves further investigation.
The role of class IIa in specific differentiation pathways could imply a radical change in the epigenetic status of cells to allow transcription of differentiation genes and repression of unnecessary genes. There are several examples of differentiation pathways regulated by class IIa [14,31,32,33,34,62,63].
Recently, a correlative analysis has pointed to the possible role of Mef2a and Hdac9 in determining a subpopulation of olfactory sensory neurons in the mouse [106]. Phosphorylation of 14-3-3 binding motifs is associated with the reprogramming of fibroblasts into cardiomyocyte-like cells. Various TFs are involved in this reprogramming, including Mef2c. Phosphorylation of 14-3-3 binding motifs in Hdac4 resolves nuclear condensates, and the phosphatase PP2A is important for controlling phosphorylation turnover and these condensates define transcriptionally repressed regions. Disruption of Hdac4 condensates stimulates cardiac reprogramming [107]. The observation of HDAC4 as nuclear speckles/foci/aggregates/condensates has also been reported in previous studies, particularly when analyzing the localization of ectopically expressed proteins mutated at 14-3-3 binding sites [15,85,108]. In principle, the formation of these HDAC4 condensates could depend on the GRD, but the class IIa-specific inhibitor TMP269 was sufficient to dissociate Hdac4 condensates in induced cardiomyocyte-like cells, suggesting the existence of some alternative mechanisms [107].
In epithelial cells, HDAC7 represses genes involved in cell junction assembly and membrane organization—an activity that alters tissue architecture by inhibiting cell polarity, cell differentiation, and the formation of primary cilia and lumen [109]. It is assumed that the activity of HDAC7 is regulated by the transmembrane protein complex Crumbs/PATJ/Lin-7 [110]. Whether the inhibition occurs through sequestration of HDAC7 into the PATJ protein complex requires further investigation.
From these few examples it is clear that the role of class IIa in differentiation is pleiotropic, sometimes antagonistic, but in other cases also promoting differentiation, and that the cells use several mechanisms to regulate their activities.
6.5. Differentiation and the Immune System
HDAC7 has several functions in the differentiation of immune cells. It is required for the development of both T cells and B cells and controls the differentiation and activation of lymphocytes as well as inflammatory signaling in macrophages [32,111,112,113,114,115]. This complexity can also be regulated by alternative splicing [116]. Recent advances have drawn attention to the contribution of HDAC7 in controlling the final stages of effector T cell differentiation. HDAC7 is stabilized by phosphorylation of salt-inducible kinase 1 (SIK1), accumulates in cell nuclei, where it reduces histone 3-lysine 27 acetylation (H3K27ac) at specific cytokine loci, thereby reducing their expression [117]. The effects of SIKs on the class IIa nuclear/cytoplasmic shuttling, with some lineage dependent peculiarities, have been documented in several studies [63,118,119]. Normally, class IIa phosphorylation by SIKs is associated with nuclear export. Indeed, SIK1 controls HDAC7 in cardiomyocytes by promoting its stabilization and cytoplasmic localization. In contrast to the other class IIa HDACs, here, HDAC7 promotes cardiac hypertrophy by controlling c-Myc expression [120].
In the context of T cell subpopulations, Hdac4 and Hdac7 can modulate the differentiation of Th17 cells. These cells are a subset of pro-inflammatory CD4+ helper T cells [121]. Hdac4 and Hdac7 are upregulated during Th17 differentiation and are highly expressed compared to other T cell subsets (Th1, Th2 and Treg). The promoters of Hdac4 and Hdac7 are occupied by Th17 lineage-specific TFs and H3K27ac is increased. What is special about this differentiation is the role of Hdac4, which promotes the transcription of lineage-specific genes via an interaction with JunB. In contrast, Hdac7 in complex with Aiolos and the co-repressors Ncor1/Ncor2/Hdac3 represses the non-lineage-specific genetic programs [41]. Previous studies have provided evidence for the complexity of the class II contribution in T cells. Hdac5 is required for the efficient generation of T regulatory (Treg) cells and also for the full activity of CD8(+) T cells [122]. In Treg cells, Hdac9 controls the acquisition of an effector phenotype by repressing Mef2d transcriptional activity [123].
In the differentiation of B cells, Hdac7 plays an important role in the transition from the pro-B to the pre-B cell stage. Deletion of Hdac7 leads to chromatin decondensation and changes in gene expression. The genes suppressed by Hdac7 include the enzyme ten-eleven translocation 2 (TET), which triggers DNA demethylation by converting 5-methylcytosine (5-mC) into 5-hydroxymethylcytosine (5-hmC). Hdac7 binds to both the promoter and an enhancer that controls Tet transcription. Changes in 5-hmC lead to aberrant transcription of miRNAs and transposable elements LINE-1 [115]. An effect on repetitive elements was also observed for HDAC4 during senescence, leading to an interferon response due to the accumulation of dsRNAs [94].
6.6. Class IIa HDACs and Inflammation
There are several indications of a contribution of HDAC9 to the development of cardiovascular disease (CVD) and the formation of atherosclerotic plaques [34]. Genetic variants in the HDAC9 locus are associated with stroke, myocardial infarction or CVD [34,124,125]. These variants are located within cis-regulatory elements (CREs) that control the expression of HDAC9 itself and possibly other genes. The contribution of HDAC9 to the development of vascular diseases has been investigated and several pathological mechanisms have been observed and proposed. HDAC9 may induce a dysregulated inflammatory response in both macrophages and endothelial cells by controlling the acetylation and activation of IKK (inhibitory kappa B kinase)-α and β [126]. HDAC9 may also be involved in endothelial–mesenchymal transition, a feature associated with CVD [127].
A mouse model was developed in which the CRE variant rs2107595, which regulates HDAC9 expression and is associated with chronic inflammation and atherosclerosis, was deleted [128]. Deletion of this CRE leads to upregulation of Hdac9 expression in a cell lineage-specific manner. Hdac9 was upregulated in bone marrow-derived macrophages and myeloid cells, but not in T cells, smooth muscle cells, and endothelial cells. By transplanting cells, the authors were able to show that the upregulation of Hdac9 in myeloid cells exacerbates atherosclerosis and worsens plaque destabilization. This effect is associated with an increased production of inflammatory cytokines. A direct influence of HDAC9 on the inflammasome and the activation of Caspase-1 is suspected as a mechanism [128]. Inflammasomes are cytosolic multiprotein complexes that recognize pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) to initiate inflammatory responses [129]. NOD-like receptor pyrin domain-containing 3 (NLRP3) is a member of the major family of receptors involved in inflammasome activation. HDAC9 interacts with NLRP3 and mediates its deacetylation, which promotes oligomerization and activation of Caspase-1. Caspase-1 can then process Gasdermin D (GSDMD) to initiate pyroptosis [128]. These data point to a new function of HDAC9 that may also have profound implications for CVD from a therapeutic perspective. However, there are still some open questions, especially related to the very low catalytic activity of HDAC9. Are class I HDACs in complex with HDAC9 involved in this process? The molecular details of the interaction between HDAC9 and NLRP3 also require further investigation. In addition, the link between class IIa HDACs and pyroptosis may be even more complicated. Another study suggested that HDAC4 directly deacetylates GSDMD, specifically at lysine 248, which acts as an inhibitor of pyroptosis [130]. This study also needs to better define whether associated class I HDACs contribute to the enzymatic activity proposed for HDAC4.
6.7. Functions in the Central Nervous System (CNS)
In addition to the sleep modulation described above, class IIa HDACs are involved in various adaptive responses in the CNS. Interest in Hdac5 was sparked by its role in cocaine-induced behaviors [131,132]. In the nucleus accumbens, Hdac5 also limits relapse-associated behavior after operant heroin self-administration and forced abstinence [133]. Hdac5 activity is described in two distinct populations of medium spiny neurons that suppress cue-induced heroin craving and heroin-induced drug craving. Hdac5 acts epigenetically to influence neuronal excitability, thereby reducing the formation of strong, relapse-inducing drug memories [133]. The importance of HDAC5 in the regulation of brain functions and plasticity finds important confirmation from an evolutionary perspective. A search for human-specific deletions in conserved regions (hCONDELs) compared to the chimpanzee genome identified a single base deletion in an active enhancer of HDAC5. It is hypothesized that this deletion restricts the expression of HDAC5. A new condition that could affect neurogenesis in the human brain [134] and that was confirmed by preliminary experimental data [135].
7. The Genomic Landscape Under the Influence of Class IIa HDACs
Although class IIa can also affect the acetylation of non-histone proteins, their effect as epigenetic regulators has been observed in several studies and further strengthened by defining their binding to specific genomic regions [19,41,103,104,105,131,136,137,138]. There are three main strategies by which class IIa can bind to DNA. First, they can be recruited as partners of selected TFs that confer the DNA-binding motif. An example is represented by the MEF2 family of TFs, well-known partners of class IIa HDACs [139]. Alternatively, they could be recruited by other epigenetic regulators that contain subunits that act as readers of histone PTMs. The third possibility is that the class IIa HDACs themselves act as readers of acetyl-lysine via the “pseudocatalytic” domain and serve as epigenetic regulators via the positioning of the class I HDACs. In fact, the dissociation constants of class IIa HDACs for acetyl-lysine are within the range of binding affinities observed for bromodomains (Kd = 10–100 μm) [140,141,142]. In addition, in vitro studies have shown that binding to acetylated peptides can affect the interaction with NCOR1 and thus the associated deacetylase activity of HDAC3 [142]. Although this study is limited by the use of ectopically overexpressed proteins, it is conceivable in principle that once class IIa HDACs recognize an acetylated histone, disassembly of the NCORs/HDAC3 complexes could allow deacetylation of the surrounding nucleosomes, leaving the nucleosome bound by class IIa protected from deacetylation. A small region of acetylated histone could represent some sort of memory of a previous acetylated region. Of course, further studies are needed to prove this hypothesis.
However, FRAP experiments indicate that class IIa shows a highly dynamic interaction with chromatin with high on/off rates [71]. There is an indication of highly transient interactions with DNA as a possible consequence of the various PTMs, mainly phosphorylations, controlling the repressive activities of class IIa HDACs. An alternative hypothesis could consider the involvement of an associated deacetylase activity that removes the acetyl group shortly after the class IIa HDACs recognize an acetyl-lysine. On the other hand, ChIP-seq experiments have shown that class IIa HDACs can accumulate in specific genomic regions [19,41,103,104,105,131,136,137,138]. It is noteworthy that these regions are acetylated regions which are often analyzed for H3K27ac. This observation suggests that class IIa may be part of an epigenetic sensor that constantly acts on regions that are highly dynamic in terms of acetylation status and gene transcription. If the expression of class IIa HDACs is disrupted, this affects the acetylation level of these bound regions and the expression of the genes they regulate.
The influence of class IIa on histones is not limited to the control of acetylation status. Other PTMs can also be regulated to achieve a more stable definition of chromatin state. In a study aimed at exploring the contribution of alternative splicing of HDAC7 to epigenetic control, mass spectrometry studies have demonstrated the effect of HDAC7 not only on H3K4ac and H3K76ac, but also on the methylation status of H3K27 and H3K36 residues [116], a regulation that is important for controlling the expression of T cell surface markers [116]. In cardiomyocytes, HDAC4 can also act as a scaffold for regulated histone methyl-transferase activities to complete a repressive signature, as in the case of H3K9m3, which can be subverted by cardiac preload and HDAC4-induced nuclear export [143].
The activity of class IIa HDACs as triggers/supervisors of various histone PTMs to repress gene transcription has also been reported in other studies, although the molecular complexes involved are not clearly defined. In cardiomyocytes, deletion of HDAC4 promotes the acquisition of an open chromatin state at cardiomyocyte-related genes defined by histone marks of active transcription (H3K4me3, H3K9ac, and H3K27ac) around the relative TSS. These regions are characterized by the presence of MEF2 binding sites [144].
Surprisingly, however, the corresponding genes were not upregulated in cells in which HDAC4 was knocked out. These promoters are poised. Physical exercise activates the binding of MEF2, which removes H3K9me2-mediated repression and allows gene transcription to occur.
8. Conclusions
The ability of class IIa HDACs to modulate so many different biological processes is remarkable. This complexity is compounded by the alteration of the catalytic site, which no longer effectively triggers the deacetylation of lysine residues. These changes, which have occurred during vertebrate evolution, implicate the ability of class IIa to recruit other HDACs. This has led to the evolution of class IIa as a platform for the coordination of multiprotein complexes, which may explain some of these pleiotropic effects. To understand the mechanisms employed by class IIa in different contexts, it is important to define the molecular composition of these complexes. A challenge that requires intensive studies as well as better reagents and most likely more accurate cellular models that come closer to physiological in vivo conditions. Sophisticated mass spectrometric approaches should be used to dissect the class IIa HDACs-specific interactomes and clearly distinguish between true partners and contaminants [88,130,145].
In recent years, many biological functions have been characterized under the supervision of class IIa. Now it is time to better understand how class IIa HDACs work.
Funding
The research in the lab of Epigenomics is funded by AIRC under IG 2021—ID. 26200 project; Interreg Italia-Osterreich ITAT11-018 SENECA; CIB “Nuove tendenze per le applicazioni biotecnologiche”; MUR-PRIN Bando 2022 Prot. 20228A7JM7 “Targeting class IIa HDACs to reset super-enhancers activity in cancer cells: towards selective epigenetic therapies in the 3D reality”.
Acknowledgments
I would like to thank the members of the Epigenomics Lab—Alessio, Martina, Martina, Elenora Gabriele, and Emanuele—for their constant commitment to experimental work on Class IIa HDACs. Thanks also to all the former members of the lab—the list would be very long—and special thanks to Eros Di Giorgio for his dedication and enthusiasm. Some of the figures in this manuscript were created using BioRender (https://www.biorender.com/).
Conflicts of Interest
The author declares no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CRE | Cis-regulatory elements |
| CVD | Cardiovascular disease |
| GRD | Glutamine-rich domain |
| HDAC | Histone deacetylase |
| HAT | Histone-acetyl transferase |
| KAT | Lysine-acetyl transferase |
| MEF2 | Myocyte Enhancer Factor |
| NCOR | Nuclear receptor co-repressor |
| NES | Nuclear export signal |
| NLS | Nuclear localization signal |
| PTM | Post translational modification |
| sZBD | structural zinc-binding domain |
References
- Chen, Y.C.; Koutelou, E.; Dent, S.Y.R. Now open: Evolving insights to the roles of lysine acetylation in chromatin organization and function. Mol. Cell 2022, 82, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Charidemou, E.; Kirmizis, A. A two-way relationship between histone acetylation and metabolism. Trends Biochem. Sci. 2024, 49, 1046–1062. [Google Scholar] [CrossRef] [PubMed]
- Schulze, J.M.; Wang, A.Y.; Kobor, M.S. Reading chromatin: Insights from yeast into YEATS domain structure and function. Epigenetics 2010, 5, 573–577. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mohan, C.; Das, C.; Tyler, J. Histone and Chromatin Dynamics Facilitating DNA repair. DNA Repair. 2021, 107, 103183. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Nodelman, I.M.; Zhou, H.; Tsukiyama, T.; Bowman, G.D.; Zhang, Z. H3K56 acetylation regulates chromatin maturation following DNA replication. Nat. Commun. 2025, 16, 134. [Google Scholar] [CrossRef] [PubMed]
- Popova, L.V.; Nagarajan, P.; Lovejoy, C.M.; Sunkel, B.D.; Gardner, M.L.; Wang, M.; Freitas, M.A.; Stanton, B.Z.; Parthun, M.R. Epigenetic regulation of nuclear lamina-associated heterochromatin by HAT1 and the acetylation of newly synthesized histones. Nucleic Acids Res. 2021, 49, 12136–12151. [Google Scholar] [CrossRef] [PubMed]
- Porter, N.J.; Christianson, D.W. Structure, mechanism, and inhibition of the zinc-dependent histone deacetylases. Curr. Opin. Struct. Biol. 2019, 59, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Nirello, V.D.; Rodrigues de Paula, D.; Araújo, N.V.P.; Varga-Weisz, P.D. Does chromatin function as a metabolite reservoir? Trends Biochem. Sci. 2022, 47, 732–735. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef] [PubMed]
- Brancolini, C.; Gagliano, T.; Minisini, M. HDACs and the epigenetic plasticity of cancer cells: Target the complexity. Pharmacol. Ther. 2022, 238, 108190. [Google Scholar] [CrossRef] [PubMed]
- Emmett, M.J.; Lazar, M.A. Integrative regulation of physiology by histone deacetylase 3. Nat. Rev. Mol. Cell Biol. 2019, 20, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Deardorff, M.A.; Porter, N.J.; Christianson, D.W. Structural aspects of HDAC8 mechanism and dysfunction in Cornelia de Lange syndrome spectrum disorders. Protein Sci. 2016, 25, 1965–1976. [Google Scholar] [CrossRef] [PubMed]
- Werbeck, N.D.; Shukla, V.K.; Kunze, M.B.A.; Yalinca, H.; Pritchard, R.B.; Siemons, L.; Mondal, S.; Greenwood, S.O.R.; Kirkpatrick, J.; Marson, C.M.; et al. A distal regulatory region of a class I human histone deacetylase. Nat. Commun. 2020, 11, 3841. [Google Scholar] [CrossRef] [PubMed]
- Di Giorgio, E.; Brancolini, C. Regulation of class IIa HDAC activities: It is not only matter of subcellular localization. Epigenomics 2016, 8, 251–269. [Google Scholar] [CrossRef] [PubMed]
- Cernotta, N.; Clocchiatti, A.; Florean, C.; Brancolini, C. Ubiquitin-dependent degradation of HDAC4, a new regulator of random cell motility. Mol. Biol. Cell 2011, 22, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Griffin, E.A., Jr.; Melas, P.A.; Zhou, R.; Li, Y.; Mercado, P.; Kempadoo, K.A.; Stephenson, S.; Colnaghi, L.; Taylor, K.; Hu, M.C.; et al. Prior alcohol use enhances vulnerability to compulsive cocaine self-administration by promoting degradation of HDAC4 and HDAC5. Sci. Adv. 2017, 3, e1701682. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Song, S.; Liu, Y.; Ko, S.H.; Kao, H.Y. Phosphorylation of the histone deacetylase 7 modulates its stability and association with 14-3-3 proteins. J. Biol. Chem. 2004, 279, 34201–34208. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhao, Y.; Gao, C.; Suresh, S.; Men, J.; Sawyers, A.; Smith, G.L. HDAC5 enhances IRF3 activation and is targeted for degradation by protein C6 from orthopoxviruses including Monkeypox virus and Variola virus. Cell Rep. 2024, 43, 113788. [Google Scholar] [CrossRef] [PubMed]
- Di Giorgio, E.; Paluvai, H.; Dalla, E.; Ranzino, L.; Renzini, A.; Moresi, V.; Minisini, M.; Picco, R.; Brancolini, C. HDAC4 degradation during senescence unleashes an epigenetic program driven by AP-1/p300 at selected enhancers and super-enhancers. Genome Biol. 2021, 22, 129. [Google Scholar] [CrossRef] [PubMed]
- Haggarty, S.J.; Koeller, K.M.; Wong, J.C.; Grozinger, C.M.; Schreiber, S.L. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl. Acad. Sci. USA 2003, 100, 4389–4394. [Google Scholar] [CrossRef] [PubMed]
- Christianson, D.W. Chemical Versatility in Catalysis and Inhibition of the Class IIb Histone Deacetylases. Acc. Chem. Res. 2024, 57, 1135–1148. [Google Scholar] [CrossRef] [PubMed]
- Oehme, I.; Linke, J.P.; Böck, B.C.; Milde, T.; Lodrini, M.; Hartenstein, B.; Wiegand, I.; Eckert, C.; Roth, W.; Kool, M.; et al. Histone deacetylase 10 promotes autophagy-mediated cell survival. Proc. Natl. Acad. Sci. USA 2013, 110, E2592–E2601. [Google Scholar] [CrossRef] [PubMed]
- Hai, Y.; Shinsky, S.A.; Porter, N.J.; Christianson, D.W. Histone deacetylase 10 structure and molecular function as a polyamine deacetylase. Nat. Commun. 2017, 8, 15368. [Google Scholar] [CrossRef] [PubMed]
- Lambona, C.; Zwergel, C.; Fioravanti, R.; Valente, S.; Mai, A. Histone deacetylase 10: A polyamine deacetylase from the crystal structure to the first inhibitors. Curr. Opin. Struct. Biol. 2023, 82, 102668. [Google Scholar] [CrossRef] [PubMed]
- Clocchiatti, A.; Florean, C.; Brancolini, C. Class IIa HDACs: From important roles in differentiation to possible implications in tumourigenesis. J. Cell. Mol. Med. 2011, 15, 1833–1846. [Google Scholar] [CrossRef] [PubMed]
- Tharayil, J.S.; Kandettu, A.; Chakrabarty, S. The curious case of mitochondrial sirtuin in rewiring breast cancer metabolism: Mr Hyde or Dr Jekyll? Biochim. Biophys. Acta Mol. Basis Dis. 2025, 1871, 167691. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, F.; Mai, A.; Rotili, D. The role of structural biology in the design of sirtuin activators. Curr. Opin. Struct. Biol. 2023, 82, 102666. [Google Scholar] [CrossRef] [PubMed]
- Núñez-Álvarez, Y.; Suelves, M. HDAC11: A multifaceted histone deacetylase with proficient fatty deacylase activity and its roles in physiological processes. FEBS J. 2022, 289, 2771–2792. [Google Scholar] [CrossRef] [PubMed]
- Milazzo, G.; Mercatelli, D.; Di Muzio, G.; Triboli, L.; De Rosa, P.; Perini, G.; Giorgi, F.M. Histone Deacetylases (HDACs): Evolution, Specificity, Role in Transcriptional Complexes, and Pharmacological Actionability. Genes 2020, 11, 556. [Google Scholar] [CrossRef] [PubMed]
- Mathias, R.A.; Guise, A.J.; Cristea, I.M. Post-translational modifications regulate class IIa histone deacetylase (HDAC) function in health and disease. Mol. Cell. Proteom. 2015, 14, 456–470. [Google Scholar] [CrossRef] [PubMed]
- Guttzeit, S.; Backs, J. Post-translational modifications talk and crosstalk to class IIa histone deacetylases. J. Mol. Cell. Cardiol. 2022, 162, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Abrol, R.; Mak, J.Y.W.; Das Gupta, K.; Ramnath, D.; Karunakaran, D.; Fairlie, D.P.; Sweet, M.J. Histone deacetylase 7: A signalling hub controlling development, inflammation, metabolism and disease. FEBS J. 2023, 290, 2805–2832. [Google Scholar] [CrossRef] [PubMed]
- Cuttini, E.; Goi, C.; Pellarin, E.; Vida, R.; Brancolini, C. HDAC4 in cancer: A multitasking platform to drive not only epigenetic modifications. Front. Mol. Biosci. 2023, 10, 1116660. [Google Scholar] [CrossRef] [PubMed]
- Brancolini, C.; Di Giorgio, E.; Formisano, L.; Gagliano, T. Quis Custodiet Ipsos Custodes (Who Controls the Controllers)? Two Decades of Studies on HDAC9. Life 2021, 11, 90. [Google Scholar] [CrossRef] [PubMed]
- Sando, R., 3rd; Gounko, N.; Pieraut, S.; Liao, L.; Yates, J., 3rd; Maximov, A. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell 2012, 151, 821–834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Y.; Yang, F.; Liu, X.; Zhan, P.; Wu, J.; Wang, X.; Wang, Z.; Tang, W.; Sun, Y.; et al. HDAC9-mediated epithelial cell cycle arrest in G2/M contributes to kidney fibrosis in male mice. Nat. Commun. 2023, 14, 3007. [Google Scholar] [CrossRef] [PubMed]
- Veerapaneni, P.; Goo, B.; Ahmadieh, S.; Shi, H.; Kim, D.S.; Ogbi, M.; Cave, S.; Chouhaita, R.; Cyriac, N.; Fulton, D.J.; et al. Transgenic Overexpression of HDAC9 Promotes Adipocyte Hypertrophy, Insulin Resistance and Hepatic Steatosis in Aging Mice. Biomolecules 2024, 14, 494. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Aviles, S.; Patel, N.; Casillas-Bajo, A.; Frutos-Rincón, L.; Velasco, E.; Gallar, J.; Arthur Farraj, P.; Gomez-Sanchez, J.A.; Cabedo, H. A genetic compensatory mechanism regulated by Jun and Mef2d modulates the expression of distinct class IIa Hdacs to ensure peripheral nerve myelination and repair. Elife 2022, 11, e72917. [Google Scholar] [CrossRef] [PubMed]
- Mielcarek, M.; Seredenina, T.; Stokes, M.P.; Osborne, G.F.; Landles, C.; Inuabasi, L.; Franklin, S.A.; Silva, J.C.; Luthi-Carter, R.; Beaumont, V.; et al. HDAC4 does not act as a protein deacetylase in the postnatal murine brain in vivo. PLoS ONE 2013, 8, e80849. [Google Scholar] [CrossRef] [PubMed]
- Price, V.; Wang, L.; D’Mello, S.R. Conditional deletion of histone deacetylase-4 in the central nervous system has no major effect on brain architecture or neuronal viability. J. Neurosci. Res. 2013, 91, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.L.; Zhao, L.; Sharma, R.; Ghosh, A.A.; Appiah, M.; Sun, Y.; Jaganathan, A.; Hu, Y.; LeJeune, A.; Xu, F.; et al. Class IIa HDAC4 and HDAC7 cooperatively regulate gene transcription in Th17 cell differentiation. Proc. Natl. Acad. Sci. USA 2024, 121, e2312111121. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, N.; Dahan, I.; D’haene, E.; Avni, M.; Vergult, S.; Vidal-García, M.; Magini, P.; Graziano, C.; Severi, G.; Bonora, E.; et al. HDAC9 structural variants disrupting TWIST1 transcriptional regulation lead to craniofacial and limb malformations. Genome Res. 2022, 32, 1242–1253. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Han, A.; Bates, D.L.; Cao, J.; Chen, L. Crystal structure of a conserved N-terminal domain of histone deacetylase 4 reveals functional insights into glutamine-rich domains. Proc. Natl. Acad. Sci. USA 2007, 104, 4297–4302. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Guo, L.; Dey, R.; Guo, M.; Zhang, X.; Bates, D.; Cayford, J.; Jiang, L.; Wei, H.; Chen, Z.; et al. Structural insights into the HDAC4-MEF2A-DNA complex and its implication in long-range transcriptional regulation. Nucleic Acids Res. 2024, 52, 2711–2723. [Google Scholar] [CrossRef] [PubMed]
- Chinellato, M.; Perin, S.; Carli, A.; Lastella, L.; Biondi, B.; Borsato, G.; Di Giorgio, E.; Brancolini, C.; Cendron, L.; Angelini, A. Folding of Class IIa HDAC Derived Peptides into α-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA. J. Mol. Biol. 2024, 436, 168541. [Google Scholar] [CrossRef] [PubMed]
- Minisini, M.; Mascaro, M.; Brancolini, C. HDAC-driven mechanisms in anticancer resistance: Epigenetics and beyond. Cancer Drug Resist. 2024, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- Bottomley, M.J.; Lo Surdo, P.; Di Giovine, P.; Cirillo, A.; Scarpelli, R.; Ferrigno, F.; Jones, P.; Neddermann, P.; De Francesco, R.; Steinkühler, C.; et al. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J. Biol. Chem. 2008, 283, 26694–26704. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, A.; Min, J.; Allali-Hassani, A.; Schapira, M.; Shuen, M.; Loppnau, P.; Mazitschek, R.; Kwiatkowski, N.P.; Lewis, T.A.; Maglathin, R.L.; et al. Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity. J. Biol. Chem. 2008, 283, 11355–11363. [Google Scholar] [CrossRef] [PubMed]
- Finnin, M.S.; Donigian, J.R.; Cohen, A.; Richon, V.M.; Rifkind, R.A.; Marks, P.A.; Breslow, R.; Pavletich, N.P. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 1999, 401, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Di Giorgio, E.; Gagliostro, E.; Brancolini, C. Selective class IIa HDAC inhibitors: Myth or reality. Cell. Mol. Life Sci. 2015, 72, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Schweipert, M.; Nehls, T.; Wurster, E.; Böltner, J.; Anton, K.; Lammer, P.; Lermyte, F.; Meyer-Almes, F.J. The pivotal role of histidine 976 in human histone deacetylase 4 for enzyme function and ligand recognition. Bioorganic Chem. 2024, 153, 107883. [Google Scholar] [CrossRef] [PubMed]
- Kutil, Z.; Meleshin, M.; Baranova, P.; Havlinova, B.; Schutkowski, M.; Barinka, C. Characterization of the class IIa histone deacetylases substrate specificity. FASEB J. 2022, 36, e22287. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Yan, G.; Li, X.; Fei, Y.; Sun, L.; Yu, H.; Niu, Y.; Gao, W.; Zhong, Q.; Yan, X. Lysine crotonylation regulates leucine-deprivation-induced autophagy by a 14-3-3ε-PPM1B axis. Cell Rep. 2022, 41, 111850. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Liu, X.; Chen, J.; Gao, S.; Lu, L.; Zhang, H.; Ding, G.; Wang, Z.; Chen, Z.; Shi, T.; et al. Class I histone deacetylases are major histone decrotonylases: Evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017, 27, 898–915. [Google Scholar] [CrossRef] [PubMed]
- Schweipert, M.; Nehls, T.; Frühauf, A.; Debarnot, C.; Kumar, A.; Knapp, S.; Lermyte, F.; Meyer-Almes, F.J. The catalytic domain of free or ligand bound histone deacetylase 4 occurs in solution predominantly in closed conformation. Protein Sci. 2024, 33, e4917. [Google Scholar] [CrossRef] [PubMed]
- Jepsen, K.; Solum, D.; Zhou, T.; McEvilly, R.J.; Kim, H.J.; Glass, C.K.; Hermanson, O.; Rosenfeld, M.G. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 2007, 450, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Jepsen, K.; Hermanson, O.; Onami, T.M.; Gleiberman, A.S.; Lunyak, V.; McEvilly, R.J.; Kurokawa, R.; Kumar, V.; Liu, F.; Seto, E.; et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 2000, 102, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Mottis, A.; Mouchiroud, L.; Auwerx, J. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes Dev. 2013, 27, 819–835. [Google Scholar] [CrossRef] [PubMed]
- Cartron, P.F.; Blanquart, C.; Hervouet, E.; Gregoire, M.; Vallette, F.M. HDAC1-mSin3a-NCOR1, Dnmt3b-HDAC1-Egr1 and Dnmt1-PCNA-UHRF1-G9a regulate the NY-ESO1 gene expression. Mol. Oncol. 2013, 7, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, R.; Picon-Ruiz, M.; Sho, M.; Van Booven, D.; Nunes de Paiva, V.; Diaz-Ruano, A.B.; Ince, T.A.; Slingerland, J. Estrone, the major postmenopausal estrogen, binds ERa to induce SNAI2, epithelial-to-mesenchymal transition, and ER+ breast cancer metastasis. Cell Rep. 2022, 41, 111672. [Google Scholar] [CrossRef] [PubMed]
- Hudson, G.M.; Watson, P.J.; Fairall, L.; Jamieson, A.G.; Schwabe, J.W.R. Insights into the Recruitment of Class IIa Histone Deacetylases (HDACs) to the SMRT/NCoR Transcriptional Repression Complex. J. Biol. Chem. 2015, 290, 18237–18244. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Croteau, S.; Hardy, P. Histone deacetylase (HDAC) 9: Versatile biological functions and emerging roles in human cancer. Cell. Oncol. 2021, 44, 997–1017. [Google Scholar] [CrossRef] [PubMed]
- Jagannath, A.; Taylor, L.; Ru, Y.; Wakaf, Z.; Akpobaro, K.; Vasudevan, S.; Foster, R.G. The multiple roles of salt-inducible kinases in regulating physiology. Physiol Rev. 2023, 103, 2231–2269. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Grishin, N.V.; Chook, Y.M. NESdb: A database of NES-containing CRM1 cargoes. Mol. Biol. Cell 2012, 23, 3673–3676. [Google Scholar] [CrossRef] [PubMed]
- la Cour, T.; Gupta, R.; Rapacki, K.; Skriver, K.; Poulsen, F.M.; Brunak, S. NESbase version 1.0: A database of nuclear export signals. Nucleic Acids Res. 2003, 31, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Fung, H.Y.; Fu, S.C.; Brautigam, C.A.; Chook, Y.M. Structural determinants of nuclear export signal orientation in binding to exportin CRM1. Elife 2015, 4, e10034. [Google Scholar] [CrossRef] [PubMed]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Fung, H.Y.; Fu, S.C.; Chook, Y.M. Nuclear export receptor CRM1 recognizes diverse conformations in nuclear export signals. Elife 2017, 6, e23961. [Google Scholar] [CrossRef] [PubMed]
- McKinsey, T.A.; Zhang, C.L.; Olson, E.N. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol. 2001, 21, 6312–6321. [Google Scholar] [CrossRef] [PubMed]
- Kao, H.Y.; Verdel, A.; Tsai, C.C.; Simon, C.; Juguilon, H.; Khochbin, S. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 2001, 276, 47496–47507. [Google Scholar] [CrossRef] [PubMed]
- Paroni, G.; Fontanini, A.; Cernotta, N.; Foti, C.; Gupta, M.P.; Yang, X.J.; Fasino, D.; Brancolini, C. Dephosphorylation and caspase processing generate distinct nuclear pools of histone deacetylase 4. Mol. Cell Biol. 2007, 27, 6718–6732. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Li, X.; Lam, M.; Liu, Y.; Chakraborty, S.; Kao, H.Y. CRM1 mediates nuclear export of HDAC7 independently of HDAC7 phosphorylation and association with 14-3-3s. FEBS Lett. 2006, 580, 5096–5104. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Wang, G.; Li, Q.; Meng, F.; Liu, C.; Gan, R.; Ju, D.; Liao, M.; Xu, J.; Sang, D.; et al. A signalling pathway for transcriptional regulation of sleep amount in mice. Nature 2022, 612, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Hotta-Hirashima, N.; Asano, F.; Kitazono, T.; Iwasaki, K.; Nakata, S.; Komiya, H.; Asama, N.; Matsuoka, T.; Fujiyama, T.; et al. Kinase signalling in excitatory neurons regulates sleep quantity and depth. Nature 2022, 612, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Wakeling, E.; McEntagart, M.; Bruccoleri, M.; Shaw-Smith, C.; Stals, K.L.; Wakeling, M.; Barnicoat, A.; Beesley, C.; DDD Study; Hanson-Kahn, A.K.; et al. Missense substitutions at a conserved 14-3-3 binding site in HDAC4 cause a novel intellectual disability syndrome. Hum. Genet. Genom. Adv. 2021, 2, 100015. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.R.; Aldred, M.A.; Der Kaloustian, V.M.; Halal, F.; Gowans, G.; McLeod, D.R.; Zondag, S.; Toriello, H.V.; Magenis, R.E.; Elsea, S.H. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am. J. Hum. Genet. 2010, 87, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, P.G.; Huang, D.; Dai, Z. Haploinsufficiency of HDAC4 does not cause intellectual disability in all affected individuals. Am. J. Med. Genet. Part A 2014, 164A, 1826–1829. [Google Scholar] [CrossRef] [PubMed]
- Vega, R.B.; Matsuda, K.; Oh, J.; Barbosa, A.C.; Yang, X.; Meadows, E.; McAnally, J.; Pomajzl, C.; Shelton, J.M.; Richardson, J.A.; et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 2004, 119, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, R.; Dong, Z.; Wang, W.; Guo, L.; Sun, J.; Rong, X.; Li, P. Loss of Hdac4 in osteoprogenitors impairs postnatal trabecular and cortical bone formation, resulting in a dwarfism and osteopenia phenotype in mice. J. Biol. Chem. 2024, 300, 107941. [Google Scholar] [CrossRef] [PubMed]
- Crow, M.; Khovanov, N.; Kelleher, J.H.; Sharma, S.; Grant, A.D.; Bogdanov, Y.; Wood, J.N.; McMahon, S.B.; Denk, F. HDAC4 is required for inflammation-associated thermal hypersensitivity. FASEB J. 2015, 29, 3370–3378. [Google Scholar] [CrossRef] [PubMed]
- Marroncelli, N.; Bianchi, M.; Bertin, M.; Consalvi, S.; Saccone, V.; De Bardi, M.; Puri, P.L.; Palacios, D.; Adamo, S.; Moresi, V. HDAC4 regulates satellite cell proliferation and differentiation by targeting P21 and Sharp1 genes. Sci. Rep. 2018, 8, 3448. [Google Scholar] [CrossRef] [PubMed]
- Kronlage, M.; Dewenter, M.; Grosso, J.; Fleming, T.; Oehl, U.; Lehmann, L.H.; Falcão-Pires, I.; Leite-Moreira, A.F.; Volk, N.; Gröne, H.J.; et al. O-GlcNAcylation of Histone Deacetylase 4 Protects the Diabetic Heart from Failure. Circulation 2019, 140, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Renzini, A.; Marroncelli, N.; Cavioli, G.; Di Francescantonio, S.; Forcina, L.; Lambridis, A.; Di Giorgio, E.; Valente, S.; Mai, A.; Brancolini, C.; et al. Cytoplasmic HDAC4 regulates the membrane repair mechanism in Duchenne muscular dystrophy. J. Cachexia Sarcopenia Muscle 2022, 13, 1339–1359. [Google Scholar] [CrossRef] [PubMed]
- Geng, L.; Cuneo, K.C.; Fu, A.; Tu, T.; Atadja, P.W.; Hallahan, D.E. Histone deacetylase (HDAC) inhibitor LBH589 increases duration of gamma-H2AX foci and confines HDAC4 to the cytoplasm in irradiated non-small cell lung cancer. Cancer Res. 2006, 66, 11298–11304. [Google Scholar] [CrossRef] [PubMed]
- Kao, G.D.; McKenna, W.G.; Guenther, M.G.; Muschel, R.J.; Lazar, M.A.; Yen, T.J. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J. Cell Biol. 2003, 160, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhou, M.; Ze, K.; Sun, X.; Zhao, C.; Li, Z.; Lu, H.; Jiao, Y.; Wang, T.; Li, S.; et al. Protective role of histone deacetylase 4 from ultraviolet radiation-induced DNA lesions. Mol. Carcinog. 2020, 59, 1292–1301. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Di Giorgio, E.; Dalla, E.; Tolotto, V.; D’Este, F.; Paluvai, H.; Ranzino, L.; Brancolini, C. HDAC4 influences the DNA damage response and counteracts senescence by assembling with HDAC1/HDAC2 to control H2BK120 acetylation and homology-directed repair. Nucleic Acids Res. 2024, 52, 8218–8240. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Mao, W.; Yang, H.; Santiago-O’Farrill, J.M.; Rask, P.J.; Mondal, J.; Chen, H.; Ivan, C.; Liu, X.; Liu, C.G.; et al. SIK2 inhibition enhances PARP inhibitor activity synergistically in ovarian and triple-negative breast cancers. J. Clin. Investig. 2022, 132, e146471. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, W.; Das, S. Temporal regulation of acetylation status determines PARP1 role in DNA damage response and metabolic homeostasis. Sci. Adv. 2024, 10, eado7720. [Google Scholar] [CrossRef] [PubMed]
- Talukdar, P.; Pal, S.; Biswas, D. Post-translational modification-dependent oligomerization switch in regulation of global transcription and DNA damage repair during genotoxic stress. Nat. Commun. 2024, 15, 4128. [Google Scholar] [CrossRef] [PubMed]
- Warnon, C.; Bouhjar, K.; Ninane, N.; Verhoyen, M.; Fattaccioli, A.; Fransolet, M.; Lambert de Rouvroit, C.; Poumay, Y.; Piel, G.; Mottet, D.; et al. HDAC2 and 7 down-regulation induces senescence in dermal fibroblasts. Aging 2021, 13, 17978–18005. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Song, M.J.; Park, J.H.; Shin, M.H.; Kim, M.K.; Hwang, D.; Lee, D.H.; Chung, J.H. Histone deacetylase 4 reverses cellular senescence via DDIT4 in dermal fibroblasts. Aging 2022, 14, 4653–4672. [Google Scholar] [CrossRef] [PubMed]
- Di Giorgio, E.; Ranzino, L.; Tolotto, V.; Dalla, E.; Burelli, M.; Gualandi, N.; Brancolini, C. Transcription of endogenous retroviruses in senescent cells contributes to the accumulation of double-stranded RNAs that trigger an anti-viral response that reinforces senescence. Cell Death Dis. 2024, 15, 157. [Google Scholar] [CrossRef] [PubMed]
- Paluvai, H.; Di Giorgio, E.; Brancolini, C. The Histone Code of Senescence. Cells 2020, 9, 466. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Qu, J.; Liu, G.H. Roles of chromatin and genome instability in cellular senescence and their relevance to ageing and related diseases. Nat. Rev. Mol. Cell Biol. 2024, 25, 979–1000. [Google Scholar] [CrossRef] [PubMed]
- Técher, H.; Kemiha, S.; Aobuli, X.; Kolinjivadi, A.M. Oncogenic RAS in Cancers from the DNA Replication Stress and Senescence Perspective. Cancers 2024, 16, 3993. [Google Scholar] [CrossRef] [PubMed]
- Jones-Weinert, C.; Mainz, L.; Karlseder, J. Telomere function and regulation from mouse models to human ageing and disease. Nat. Rev. Mol. Cell Biol. 2025, 26, 297–313. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; de Fatima Silva, F.; Liu, D.; Patel, H.U.; Xu, J.; Zhang, W.; Türk, C.; Krüger, M.; Collins, S. Salt-inducible kinase inhibition promotes the adipocyte thermogenic program and adip.ose tissue browning. Mol. Metab. 2023, 74, 101753. [Google Scholar] [CrossRef] [PubMed]
- Nam, H.; Kundu, A.; Karki, S.; Brinkley, G.J.; Chandrashekar, D.S.; Kirkman, R.L.; Liu, J.; Liberti, M.V.; Locasale, J.W.; Mitchell, T.; et al. The TGF-β/HDAC7 axis suppresses TCA cycle metabolism in renal cancer. The TGF-β/HDAC7 axis suppresses TCA cycle metabolism in renal cancer. JCI Insight 2021, 6, e148438. [Google Scholar] [CrossRef] [PubMed]
- Newton, L.M.; Fowler, V.M.; Humbert, P.O. Erythroblast enucleation at a glance. J. Cell Sci. 2024, 137, jcs261673. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, W.; Schulz, V.P.; Zhao, H.; Qu, X.; Qi, Q.; Cheng, Y.; Guo, X.; Zhang, S.; Wei, X.; et al. Impairment of human terminal erythroid differentiation by histone deacetylase 5 deficiency. Blood 2021, 138, 1615–1627. [Google Scholar] [CrossRef] [PubMed]
- Di Giorgio, E.; Dalla, E.; Franforte, E.; Paluvai, H.; Minisini, M.; Trevisanut, M.; Picco, R.; Brancolini, C. Different class IIa HDACs repressive complexes regulate specific epigenetic responses related to cell survival in leiomyosarcoma cells. Nucleic Acids Res. 2020, 48, 646–664. [Google Scholar] [CrossRef] [PubMed]
- Cutano, V.; Di Giorgio, E.; Minisini, M.; Picco, R.; Dalla, E.; Brancolini, C. HDAC7-mediated control of tumour microenvironment maintains proliferative and stemness competence of human mammary epithelial cells. Mol. Oncol. 2019, 13, 1651–1668. [Google Scholar] [CrossRef] [PubMed]
- Caslini, C.; Hong, S.; Ban, Y.J.; Chen, X.S.; Ince, T.A. HDAC7 regulates histone 3 lysine 27 acetylation and transcriptional activity at super-enhancer-associated genes in breast cancer stem cells. Oncogene 2019, 38, 6599–6614. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Shi, P.; Li, Y.; Zuo, Y.; Nie, Y.; Xu, T.; Peng, D.; An, Z.; Huang, T.; Zhang, J.; et al. Regulatory mechanisms orchestrating cellular diversity of Cd36+ olfactory sensory neurons revealed by scRNA-seq and scATAC-seq analysis. Cell Rep. 2024, 43, 114671. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Lei, I.; Tian, S.; Gao, W.; Guo, Y.; Li, Z.; Sabry, Z.; Tang, P.; Chen, Y.E.; Wang, Z. 14-3-3 binding motif phosphorylation disrupts Hdac4-organized condensates to stimulate cardiac reprogramming. Cell Rep. 2024, 43, 114054. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, H.; Park, E.J.; Chen, J.D. SMRTE inhibits MEF2C transcriptional activation by targeting HDAC4 and 5 to nuclear domains. J. Biol. Chem. 2001, 276, 24177–24185. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, J.; Moennig, T.; Hinrichs, J.H.; Weber, A.; Wagner, T.; Hemmer, T.; Schröter, R.; Weide, T.; Epting, D.; Bergmann, C.; et al. PATJ inhibits histone deacetylase 7 to control tight junction formation and cell polarity. Cell. Mol. Life Sci. 2023, 80, 333. [Google Scholar] [CrossRef] [PubMed]
- Bulgakova, N.A.; Knust, E. The Crumbs complex: From epithelial-cell polarity to retinal degeneration. J. Cell Sci. 2009, 122, 2587–2596. [Google Scholar] [CrossRef] [PubMed]
- Kasler, H.G.; Verdin, E. Histone deacetylase 7 functions as a key regulator of genes involved in both positive and negative selection of thymocytes. Mol. Cell. Biol. 2007, 27, 5184–5200. [Google Scholar] [CrossRef] [PubMed]
- Kasler, H.G.; Young, B.D.; Mottet, D.; Lim, H.W.; Collins, A.M.; Olson, E.N.; Verdin, E. Histone deacetylase 7 regulates cell survival and TCR signaling in CD4/CD8 double-positive thymocytes. J. Immunol. 2011, 186, 4782–4793. [Google Scholar] [CrossRef] [PubMed]
- Azagra, A.; Román-González, L.; Collazo, O.; Rodríguez-Ubreva, J.; de Yébenes, V.G.; Barneda-Zahonero, B.; Rodríguez, J.; Castro de Moura, M.; Grego-Bessa, J.; Fernández-Duran, I.; et al. In vivo conditional deletion of HDAC7 reveals its requirement to establish proper B lymphocyte identity and development. J. Exp. Med. 2016, 213, 2591–2601. [Google Scholar] [CrossRef] [PubMed]
- Kasler, H.G.; Lee, I.S.; Lim, H.W.; Verdin, E. Histone Deacetylase 7 mediates tissue-specific autoimmunity via control of innate effector function in invariant Natural Killer T Cells. Elife 2018, 7, e32109. [Google Scholar] [CrossRef] [PubMed]
- Azagra, A.; Meler, A.; de Barrios, O.; Tomás-Daza, L.; Collazo, O.; Monterde, B.; Obiols, M.; Rovirosa, L.; Vila-Casadesús, M.; Cabrera-Pasadas, M.; et al. The HDAC7-TET2 epigenetic axis is essential during early B lymphocyte development. Nucleic Acids Res. 2022, 50, 8471–8490. [Google Scholar] [CrossRef] [PubMed]
- Agosto, L.M.; Mallory, M.J.; Ferretti, M.B.; Blake, D.; Krick, K.S.; Gazzara, M.R.; Garcia, B.A.; Lynch, K.W. Alternative splicing of HDAC7 regulates its interaction with 14-3-3 proteins to alter histone marks and target gene expression. Cell Rep. 2023, 42, 112273. [Google Scholar] [CrossRef] [PubMed]
- Helms, R.S.; Marin-Gonzalez, A.; Patel, C.H.; Sun, I.H.; Wen, J.; Leone, R.D.; Duvall, B.; Gao, R.D.; Ha, T.; Tsukamoto, T.; et al. SIKs Regulate HDAC7 Stabilization and Cytokine Recall in Late-Stage T Cell Effector Differentiation. J. Immunol. 2023, 211, 1767–1782. [Google Scholar] [CrossRef] [PubMed]
- Walkinshaw, D.R.; Weist, R.; Kim, G.W.; You, L.; Xiao, L.; Nie, J.; Li, C.S.; Zhao, S.; Xu, M.; Yang, X.J.; et al. The tumor suppressor kinase LKB1 activates the downstream kinases SIK2 and SIK3 to stimulate nuclear export of class IIa histone deacetylases. J. Biol. Chem. 2013, 288, 9345–9362. [Google Scholar] [CrossRef] [PubMed]
- Darling, N.J.; Cohen, P. Nuts and bolts of the salt-inducible kinases (SIKs). Biochem. J. 2021, 478, 1377–1397. [Google Scholar] [CrossRef] [PubMed]
- Hsu, A.; Duan, Q.; McMahon, S.; Huang, Y.; Wood, S.A.; Gray, N.S.; Wang, B.; Bruneau, B.G.; Haldar, S.M. Salt-inducible kinase 1 maintains HDAC7 stability to promote pathologic cardiac remodeling. J. Clin. Investig. 2020, 130, 2966–2977. [Google Scholar] [CrossRef] [PubMed]
- Kanno, T.; Miyako, K.; Endo, Y. Lipid metabolism: A central modulator of RORγt-mediated Th17 cell differentiation. Int. Immunol. 2024, 36, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Jiao, J.; Wang, L.; O’Brien, S.; Newick, K.; Wang, L.C.; Falkensammer, E.; Liu, Y.; Han, R.; Kapoor, V.; et al. HDAC5 controls the functions of Foxp3(+) T-regulatory and CD8(+) T cells. Int. J. Cancer 2016, 138, 2477–2486. [Google Scholar] [CrossRef] [PubMed]
- Di Giorgio, E.; Wang, L.; Xiong, Y.; Christensen, L.M.; Akimova, T.; Han, R.; Samanta, A.; Trevisanut, M.; Brancolini, C.; Beier, U.H.; et al. A Biological Circuit Involving Mef2c, Mef2d, and Hdac9 Controls the Immunosuppressive Functions of CD4+Foxp3+ T-Regulatory Cells. Front. Immunol. 2021, 12, 703632. [Google Scholar] [CrossRef] [PubMed]
- Klarin, D.; Lynch, J.; Aragam, K.; Chaffin, M.; Assimes, T.L.; Huang, J.; Lee, K.M.; Shao, Q.; Huffman, J.E.; Natarajan, P.; et al. Genome-wide association study of peripheral artery disease in the Million Veteran Program. Nat. Med. 2019, 25, 1274–1279. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, R.; Mauer, A.C.; Lino Cardenas, C.L.; Guo, X.; Yao, J.; Zhang, X.; Wunderer, F.; Smith, A.V.; Wong, Q.; Pechlivanis, S.; et al. HDAC9 is implicated in atherosclerotic aortic calcification and affects vascular smooth muscle cell phenotype. Nat. Genet. 2019, 51, 1580–1587. [Google Scholar] [CrossRef] [PubMed]
- Asare, Y.; Campbell-James, T.A.; Bokov, Y.; Yu, L.L.; Prestel, M.; El Bounkari, O.; Roth, S.; Megens, R.T.A.; Straub, T.; Thomas, K.; et al. Histone Deacetylase 9 Activates IKK to Regulate Atherosclerotic Plaque Vulnerability. Circ. Res. 2020, 127, 811–823. [Google Scholar] [CrossRef] [PubMed]
- Lecce, L.; Xu, Y.; V’Gangula, B.; Chandel, N.; Pothula, V.; Caudrillier, A.; Santini, M.P.; d’Escamard, V.; Ceholski, D.K.; Gorski, P.A.; et al. Histone deacetylase 9 promotes endothelial-mesenchymal transition and an unfavorable atherosclerotic plaque phenotype. J. Clin. Investig. 2021, 131, e131178. [Google Scholar] [CrossRef] [PubMed]
- Asare, Y.; Yan, G.; Schlegl, C.; Prestel, M.; van der Vorst, E.P.C.; Teunissen, A.J.P.; Aronova, A.; Tosato, F.; Naser, N.; Caputo, J.; et al. A cis-regulatory element controls expression of histone deacetylase 9 to fine-tune inflammasome-dependent chronic inflammation in atherosclerosis. Immunity 2025, 58, 555–567.e9. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, I.M.; Chaves, M.M. The NLRP3 Inflammasome in inflammatory diseases: Cellular dynamics and role in granuloma formation. Cell. Immunol. 2025, 411–412, 104961. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Jin, Q.; Li, X.; Li, D.; Fu, X.; Chen, N.; Lv, Q.; Shi, Y.; He, S.; Dong, L.; et al. Crosstalk of HDAC4, PP1, and GSDMD in controlling pyroptosis. Cell Death Dis. 2024, 15, 115. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Carreira, M.B.; Cooper, Y.A.; Bobadilla, A.C.; Heinsbroek, J.A.; Koike, N.; Larson, E.B.; Balmuth, E.A.; Hughes, B.W.; Penrod, R.D.; et al. HDAC5 and Its Target Gene, Npas4, Function in the Nucleus Accumbens to Regulate Cocaine-Conditioned Behaviors. Neuron 2017, 96, 130–144.e6. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Carreira, M.B.; Smith, L.N.; Zirlin, B.C.; Neve, R.L.; Cowan, C.W. Histone deacetylase 5 limits cocaine reward through cAMP-induced nuclear import. Neuron 2012, 73, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.M.; Tsvetkov, E.; Galante, A.; DeVries, D.; McCue, L.M.; Wood, D.; Barry, S.; Berto, S.; Lavin, A.; Taniguchi, M.; et al. Epigenetic function during heroin self-administration controls future relapse-associated behavior in a cell type-specific manner. Proc. Natl. Acad. Sci. USA 2023, 120, e2210953120. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.R.; Mackay-Smith, A.; Mouri, K.; Garcia, M.F.; Dong, M.X.; Akers, J.F.; Noble, M.; Li, X.; Consortium, Z.; Lindblad-Toh, K.; et al. The functional and evolutionary impacts of human-specific deletions in conserved elements. Science 2023, 380, eabn2253. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Fu, C.; Lin, L.; Liu, S.; Su, X.; Li, A.; Wu, Q.; Jia, C.; Zhang, P.; Chen, L.; et al. miR-124 and miR-9 mediated downregulation of HDAC5 promotes neurite development through activating MEF2C-GPM6A pathway. J. Cell. Physiol. 2018, 233, 673–687. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Zhang, C.; Gan, R.; Yin, X.; Wang, M.; Shi, B.; Chen, L.; Wu, C.; Li, Q.; Liu, Q. Transcriptional regulation of daily sleep amount by TCF4-HDAC4-CREB complex in mice. Sleep 2025, 48, zsae313. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Cuffe, S.; Liang, S.; Azad, A.K.; Cheng, L.; Brhane, Y.; Qiu, X.; Cescon, D.W.; Bruce, J.; Chen, Z.; et al. BRM Promoter Polymorphisms and Survival of Advanced Non-Small Cell Lung Cancer Patients in the Princess Margaret Cohort and CCTG BR.24 Trial. Clin. Cancer Res. 2017, 23, 2460–2470. [Google Scholar] [CrossRef] [PubMed]
- Lian, B.; Pei, Y.C.; Jiang, Y.Z.; Xue, M.Z.; Li, D.Q.; Li, X.G.; Zheng, Y.Z.; Liu, X.Y.; Qiao, F.; Sun, W.L.; et al. Truncated HDAC9 identified by integrated genome-wide screen as the key modulator for paclitaxel resistance in triple-negative breast cancer. Theranostics 2020, 10, 11092–11109. [Google Scholar] [CrossRef] [PubMed]
- Di Giorgio, E.; Hancock, W.W.; Brancolini, C. MEF2 and the tumorigenic process, hic sunt leones. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Bradner, J.E.; West, N.; Grachan, M.L.; Greenberg, E.F.; Haggarty, S.J.; Warnow, T.; Mazitschek, R. Chemical phylogenetics of histone deacetylases. Nat. Chem. Biol. 2010, 6, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Meyners, C.; Mertens, M.; Wessig, P.; Meyer-Almes, F.J. A Fluorescence-Lifetime-Based Binding Assay for Class IIa Histone Deacetylases. Chemistry 2017, 23, 3107–3116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Andrade, R.; Hanna, A.A.; Pflum, M.K.H. Evidence that HDAC7 acts as an epigenetic “reader” of AR acetylation through NCoR-HDAC3 dissociation. Cell Chem. Biol. 2022, 29, 1162–1173.e5. [Google Scholar] [CrossRef] [PubMed]
- Hohl, M.; Wagner, M.; Reil, J.C.; Müller, S.A.; Tauchnitz, M.; Zimmer, A.M.; Lehmann, L.H.; Thiel, G.; Böhm, M.; Backs, J.; et al. HDAC4 controls histone methylation in response to elevated cardiac load. J. Clin. Investig. 2013, 123, 1359–1370. [Google Scholar] [CrossRef] [PubMed]
- Finke, D.; Schanze, L.M.; Schreiter, F.; Kreußer, M.M.; Katus, H.A.; Backs, J.; Lehmann, L.H. Histone deacetylase 4 deletion broadly affects cardiac epigenetic repression and regulates transcriptional susceptibility via H3K9 methylation. J. Mol. Cell. Cardiol. 2022, 162, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Margariti, A.; Zampetaki, A.; Xiao, Q.; Zhou, B.; Karamariti, E.; Martin, D.; Yin, X.; Mayr, M.; Li, H.; Zhang, Z.; et al. Histone deacetylase 7 controls endothelial cell growth through modulation of beta-catenin. Circ. Res. 2010, 106, 1202–1211. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).