Discovery of a Potent Antimicrobial Peptide Through Rational Design: A New Frontier in Pathogen Control
Abstract
1. Introduction
2. Materials and Methods
2.1. Synthesis and In Silico Design of RKW
2.2. Circular Dichroism Spectroscopy
2.3. Fluorescence Spectroscopy
2.4. Bacterial Strains
2.5. Antimicrobial Assays
2.6. Antimicrobial Resistance of ESKAPE Bacteria
2.7. Antimicrobial and Antibiofilm Activity Against ESKAPE Bacteria
2.8. In Vitro Cytotoxicity Assays
2.9. In Vitro Evaluation of the Stability of RKW in Newborn Calf Serum
2.10. In Vitro Evaluation of Bactericidal and Fungicidal Activity of RKW According to European Standard Guidelines
2.11. Statistical Analysis
3. Results and Discussion
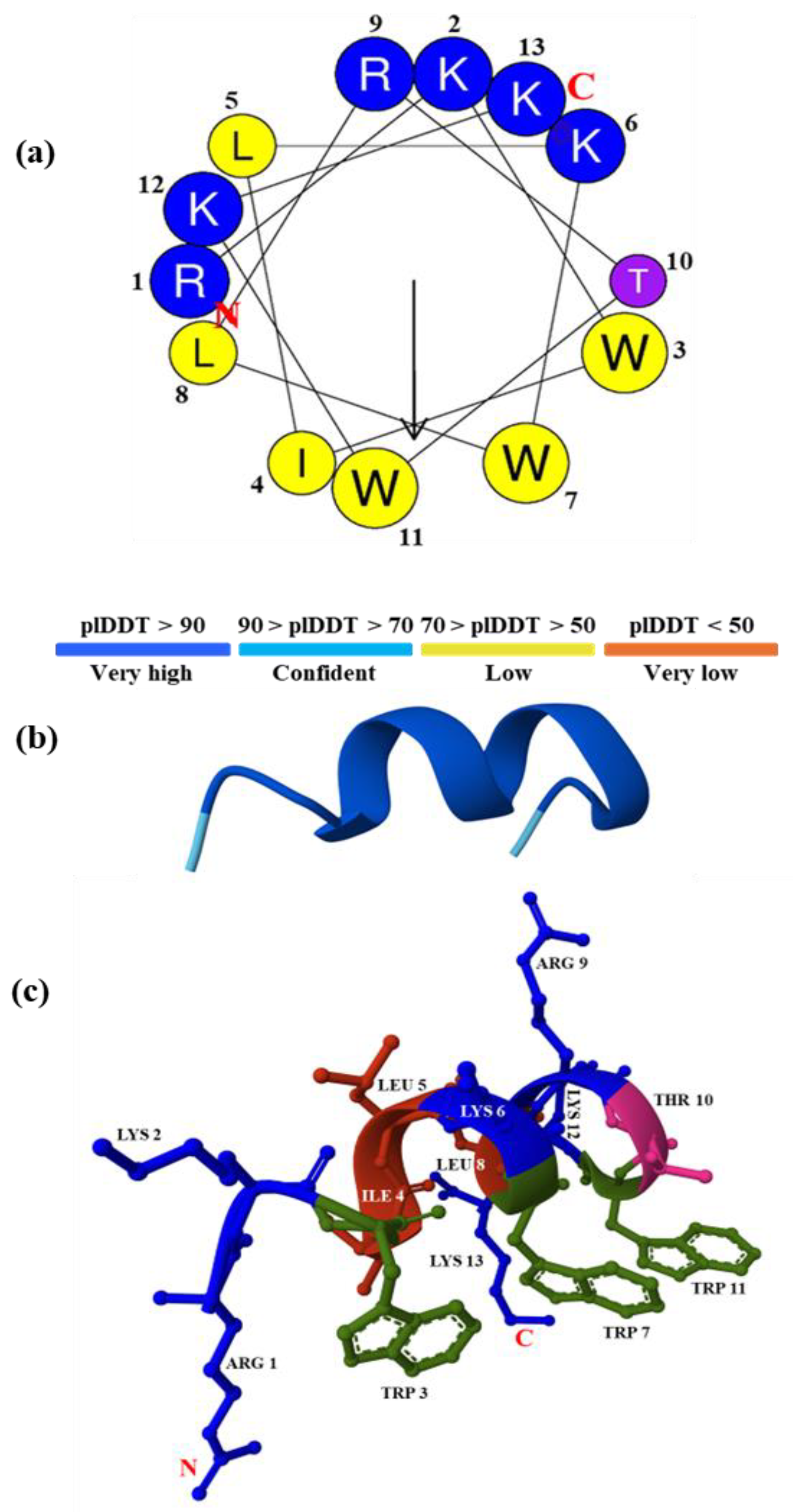
3.1. Design of a New Lysine-Tryptophan-Rich Peptide
3.2. Peptide Molecular Modeling
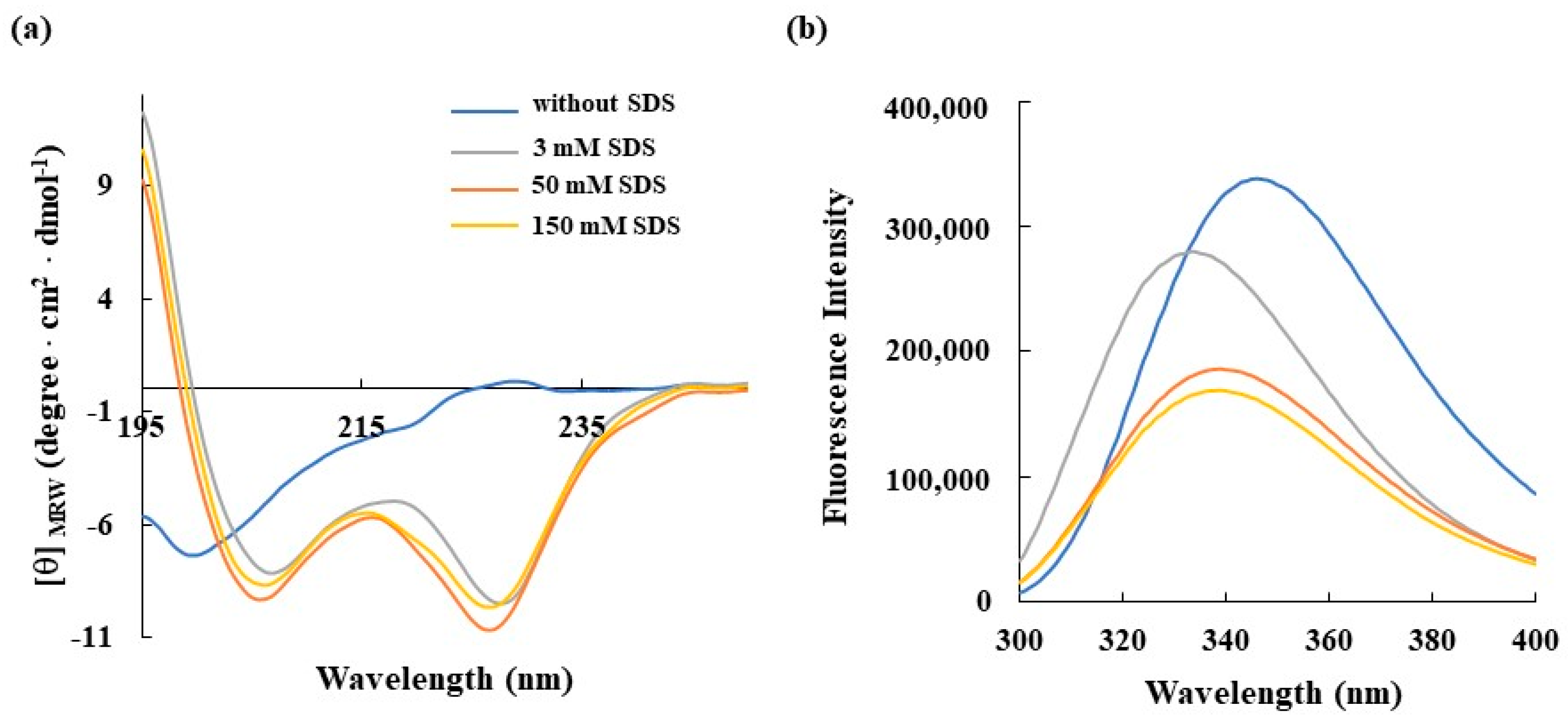
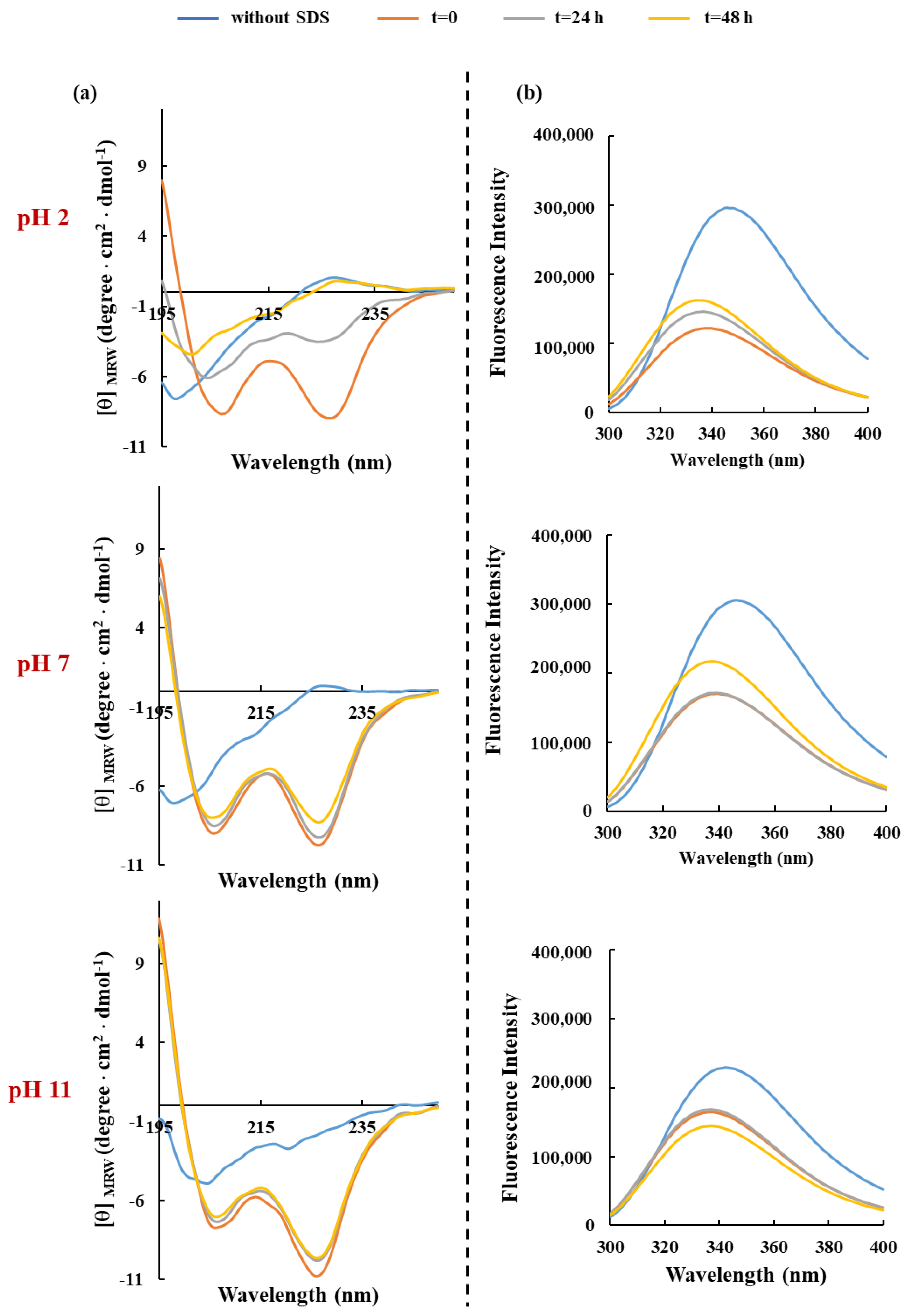
3.3. Spectroscopic Characterization
3.4. In Vitro Evaluation of Antibacterial Activity of RKW
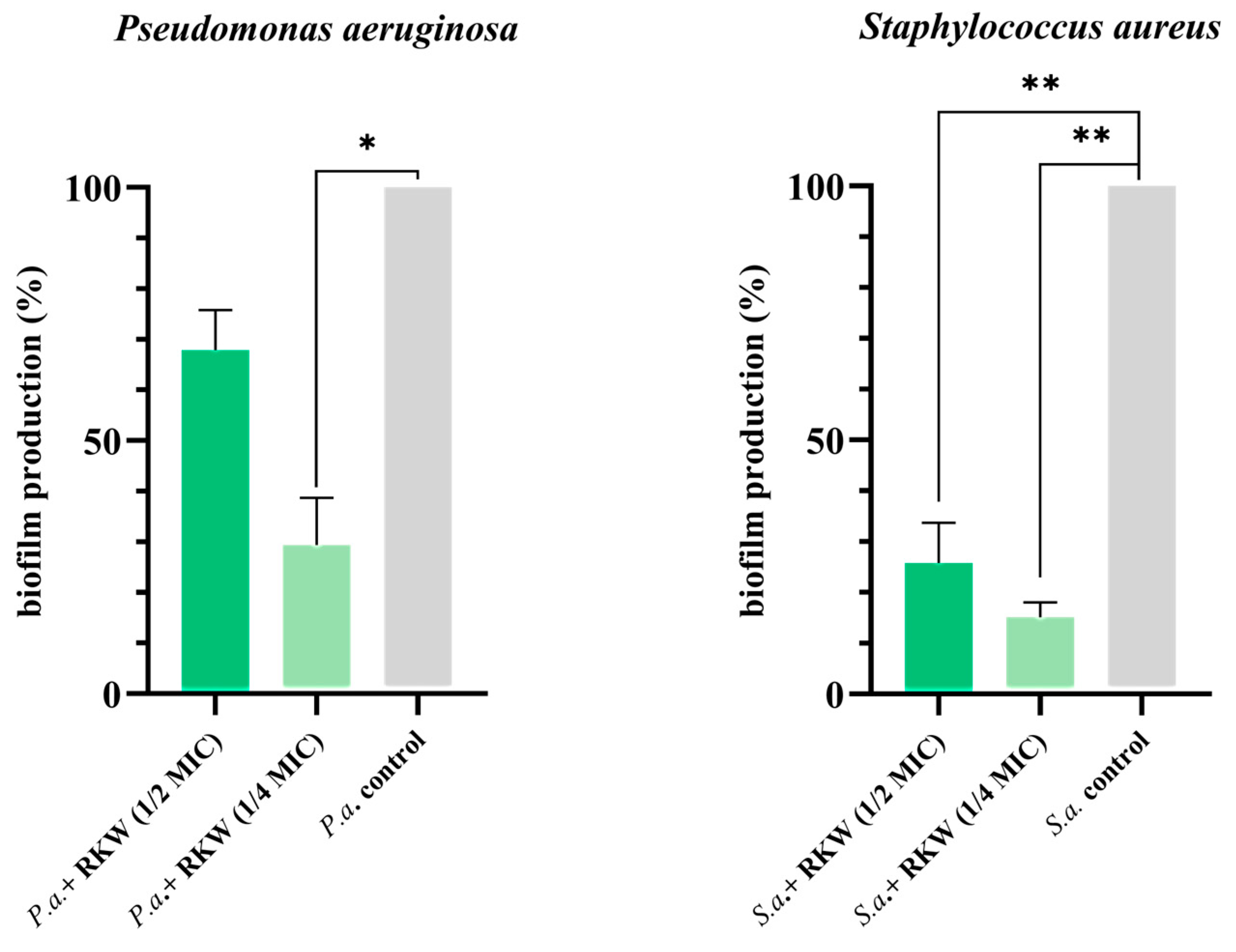
3.5. Peptide Investigation to Address Antimicrobial Resistance and Tolerance of ESKAPE Bacteria
3.6. In Vitro Cytotoxicity of RKW on Mammalian Cells
3.7. In Vitro Evaluation of Bactericidal and Fungicidal Activity of RKW According to European Standard Guidelines
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cizman, M.; Plankar Srovin, T. Antibiotic consumption and resistance of gram-negative pathogens (collateral damage). GMS Infect. Dis. 2018, 6, Doc05. [Google Scholar] [CrossRef]
- Woolhouse, M.; Waugh, C.; Perry, M.R.; Nair, H. Global disease burden due to antibiotic resistance—State of the evidence. J. Glob. Health 2016, 6, 010306. [Google Scholar] [CrossRef]
- Zhen, X.; Lundborg, C.S.; Sun, X.; Hu, X.; Dong, H. Economic burden of antibiotic resistance in ESKAPE organisms: A systematic review. Antimicrob. Resit. Infect. Control 2019, 8, 137. [Google Scholar] [CrossRef]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Marturano, J.E.; Lowery, T.J. ESKAPE Pathogens in bloodstream infections are associated with higher cost and mortality but can be predicted using diagnoses upon Admission. Open Forum Infect. Dis. 2019, 6, ofz03. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.; Panteli, D.; van Kessel, R.; Ljungqvist, G.; Colombo, F.; Mossialos, E. Challenges and opportunities for incentivising antibiotic research and development in Europe. Lancet Reg. Health-Eur. 2023, 33, 100705. [Google Scholar] [CrossRef] [PubMed]
- Buchmann, K. Evolution of Innate Immunity: Clues from Invertebrates via Fish to Mammals. Front. Immunol. 2014, 5, 459. [Google Scholar] [CrossRef]
- Flajnik, M.F.; Du Pasquier, L. Evolution of innate and adaptive immunity: Can we draw a line? Trends Immunol. 2005, 25, 640–644. [Google Scholar] [CrossRef]
- Silva, R.C.M.C.; Gomes, F.M. Evolution of the Major Components of Innate Immunity in Animals. J. Mol. Evol. 2024, 92, 3–20. [Google Scholar] [CrossRef]
- Su, Y.; Zhang, K.; Schluesener, H.J. Antimicrobial peptides in the brain. Arch. Immunol. Ther. Exp. 2010, 58, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Buda De Cesare, G.; Cristy, S.A.; Garsin, D.A.; Lorenz, M.C. Antimicrobial Peptides: A New Frontier in Antifungal Therapy. mBio 2020, 11, e02123-20. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Niu, J.; Wang, X.; Niu, M.; Liao, C. The Contribution of Antimicrobial Peptides to Immune Cell Function: A Review of Recent Advances. Pharmaceutics 2023, 15, 2278. [Google Scholar] [CrossRef]
- D’Aquila, P.; De Rose, E.; Sena, G.; Scorza, A.; Cretella, B.; Passarino, G.; Bellizzi, D. Quorum Quenching Approaches against Bacterial-Biofilm-Induced Antibiotic Resistance. Antibiotics 2024, 13, 619. [Google Scholar] [CrossRef]
- Leistikow, K.R.; May, D.S.; Suh, W.S.; Vargas Asensio, G.; Schaenzer, A.J.; Currie, C.R.; Hristova, K.R. Bacillus subtilis-derived peptides disrupt quorum sensing and biofilm assembly in multidrug-resistant Staphylococcus aureus. mSystems 2024, 9, e00712–e00724. [Google Scholar] [CrossRef]
- Bin Hafeez, A.; Jiang, X.; Bergen, P.J.; Zhu, Y. Antimicrobial Peptides: An Update on Classifications and Databases. Int. J. Mol. Sci. 2021, 22, 11691. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse the trend? Nat. Rev. Microbiol. 2011, 9, 260–271. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Liang, Q.; Liu, Z.; Liang, Z.; Zhu, C.; Li, D.; Kong, Q.; Mou, H. Development strategies and application of antimicrobial peptides as future alternatives to in-feed antibiotics. Sci. Total Environ. 2024, 927, 172150. [Google Scholar] [CrossRef]
- Epand, R.M.; Vogel, H.J. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta 1999, 1462, 11–28. [Google Scholar] [CrossRef]
- Mihaylova-Garnizova, R.; Davidova, S.; Hodzhev, Y.; Satchanska, G. Antimicrobial Peptides Derived from Bacteria: Classification, Sources, and Mechanism of Action against Multidrug-Resistant Bacteria. Int. J. Mol. Sci. 2024, 25, 10788. [Google Scholar] [CrossRef] [PubMed]
- Hollmann, A.; Martínez, M.; Noguera, M.E.; Augusto, M.T.; Disalvo, A.; Santos, N.C.; Semorile, L.; Maffía, P.C. Role of amphipathicity and hydrophobicity in the balance between hemolysis and peptide-membrane interactions of three related antimicrobial peptides. Colloids Surf. B Biointerfaces 2016, 141, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Mba, I.E.; Nweze, E.I. Antimicrobial Peptides Therapy: An Emerging Alternative for Treating Drug-Resistant Bacteria. Yale J. Biol. Med. 2022, 95, 445–463. [Google Scholar] [PubMed]
- Xuan, J.; Feng, W.; Wang, J.; Wang, R.; Zhang, B.; Bo, L.; Chen, Z.S.; Yang, H.; Sun, L. Antimicrobial peptides for combating drug-resistant bacterial infections. Drug Resist. Updat. 2023, 68, 100954. [Google Scholar] [CrossRef]
- Kang, S.J.; Nam, S.H.; Lee, B.J. Engineering Approaches for the Development of Antimicrobial Peptide-Based Antibiotics. Antibiotics 2022, 11, 1338. [Google Scholar] [CrossRef]
- Palmieri, G.; Balestrieri, M.; Capuano, F.; Proroga, Y.T.R.; Pomilio, F.; Centorame, P.; Riccio, A.; Marrone, R.; Anastasio, A. Bactericidal and antibiofilm activity of bactenecin-derivative peptides against the food-pathogen Listeria monocytogenes: New perspectives for food processing industry. Int. J. Food Microbiol. 2018, 279, 33–42. [Google Scholar] [CrossRef]
- Palmieri, G.; Balestrieri, M.; Proroga, Y.T.; Falcigno, L.; Facchiano, A.; Riccio, A.; Capuano, F.; Marrone, R.; Neglia, G.; Anastasio, A. New antimicrobial peptides against foodborne pathogens: From in silico design to experimental evidence. Food Chem. 2016, 211, 546–554. [Google Scholar] [CrossRef]
- Agrillo, B.; Proroga, Y.T.R.; Gogliettino, M.; Balestrieri, M.; Tatè, R.; Nicolais, L.; Palmieri, G. A Safe and Multitasking Antimicrobial Decapeptide: The Road from De Novo Design to Structural and Functional Characterization. Int. J. Mol. Sci. 2020, 21, 6952. [Google Scholar] [CrossRef]
- Agrillo, B.; Porritiello, A.; Gratino, L.; Balestrieri, M.; Proroga, Y.T.; Mancusi, A.; Cozzi, L.; Vicenza, T.; Dardano, P.; Miranda, B.; et al. Antimicrobial activity, membrane interaction and structural features of short arginine-rich antimicrobial peptides. Front. Microbiol. 2023, 14, 1244325. [Google Scholar] [CrossRef] [PubMed]
- Galatola, E.; Agrillo, B.; Gogliettino, M.; Palmieri, G.; Maccaroni, S.; Vicenza, T.; Proroga, Y.T.R.; Mancusi, A.; Di Pasquale, S.; Suffredini, E.; et al. A Reliable Multifaceted Solution against Foodborne Viral Infections: The Case of RiLK1 Decapeptide. Molecules 2024, 29, 2305. [Google Scholar] [CrossRef] [PubMed]
- Falcigno, L.; Palmieri, G.; Balestrieri, M.; Proroga, Y.T.; Facchiano, A.; Riccio, A.; Capuano, F.; Marrone, R.; Campanile, G.; Anastasio, A. NMR and computational data of two novel antimicrobial peptides. Data Brief 2016, 8, 562–569. [Google Scholar] [CrossRef][Green Version]
- Festa, R.; Ambrosio, R.L.; Lamas, A.; Gratino, L.; Palmieri, G.; Franco, C.M.; Cepeda, A.; Anastasio, A. A Study on the Antimicrobial and Antibiofilm Peptide 1018-K6 as Potential Alternative to Antibiotics against Food Pathogen Salmonella enterica. Foods 2021, 10, 1372. [Google Scholar] [CrossRef]
- Mwangi, J.; Kamau, P.M.; Thuku, R.C.; Lai, R. Design methods for antimicrobial peptides with improved performance. Zool. Res. 2023, 44, 1095–1114. [Google Scholar] [CrossRef]
- Verma, D.P.; Tripathi, A.K.; Thakur, A.K. Innovative Strategies and Methodologies in Antimicrobial Peptide Design. J. Funct. Biomater. 2024, 15, 320. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Walker, J.M., Ed.; Protein Identification and Analysis Tools on the Expasy Server. In The Proteomics Protocols Handbook. Springer Protocols Handbooks; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Mathur, D.; Singh, S.; Mehta, A.; Agrawal, P.; Raghava, G.P.S. In silico approaches for predicting the half-life of natural and modified peptides in blood. PLoS ONE 2018, 13, e0196829. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific α-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, version 15.0; The European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2025. [Google Scholar]
- Gratino, L.; Gogliettino, M.; Balestrieri, M.; Porritiello, A.; Dardano, P.; Miranda, B.; Ambrosio, R.L.; Ambrosio, M.; Nicolais, L.; Palmieri, G. Functional interplay between short antimicrobial peptides and model lipid membranes. Bioorg. Chem. 2024, 153, 107939. [Google Scholar] [CrossRef]
- Saubenova, M.; Rapoport, A.; Yermekbay, Z.; Oleinikova, Y. Antimicrobial Peptides, Their Production, and Potential in the Fight Against Antibiotic-Resistant Pathogens. Fermentation 2025, 11, 36. [Google Scholar] [CrossRef]
- Mabrouk, D.M. Antimicrobial peptides: Features, applications and the potential use against COVID-19. Mol. Biol. Rep. 2022, 49, 10039–10050. [Google Scholar] [CrossRef]
- Oliveira, M.; Antunes, W.; Mota, S.; Madureira-Carvalho, Á.; Dinis-Oliveira, R.J.; Dias da Silva, D. An Overview of the Recent Advances in Antimicrobial Resistance. Microorganisms 2024, 12, 1920. [Google Scholar] [CrossRef] [PubMed]
- Hetta, H.F.; Sirag, N.; Alsharif, S.M.; Alharbi, A.A.; Alkindy, T.T.; Alkhamali, A.; Albalawi, A.S.; Ramadan, Y.N.; Rashed, Z.I.; Alanazi, F.E. Antimicrobial Peptides: The Game-Changer in the Epic Battle Against Multidrug-Resistant Bacteria. Pharmaceuticals 2024, 17, 1555. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Fang, H.; Fang, L.; Liu, D.; Liu, J.; Su, M.; Fang, Z.; Ren, W.; Jiao, H. The Modification and Design of Antimicrobial Peptide. Curr. Pharm. Des. 2018, 24, 904–910. [Google Scholar] [CrossRef]
- Wang, Y.; Song, M.; Chang, W. Antimicrobial peptides and proteins against drug-resistant pathogens. Cell Surf. 2024, 12, 100135. [Google Scholar] [CrossRef]
- Kim, H.; Jang, J.H.; Kim, S.C.; Cho, J.H. De novo generation of short antimicrobial peptides with enhanced stability and cell specificity. J. Antimicrob. Chemother. 2014, 69, 121–132. [Google Scholar] [CrossRef]
- Falcigno, L.; D’Auria, G.; Palmieri, G.; Gogliettino, M.; Agrillo, B.; Tatè, R.; Dardano, P.; Nicolais, L.; Balestrieri, M. Key Physicochemical Determinants in the Antimicrobial Peptide RiLK1 Promote Amphipathic Structures. Int. J. Mol. Sci. 2021, 22, 10011. [Google Scholar] [CrossRef]
- Saravanan, R.; Li, X.; Lim, K.; Mohanram, H.; Peng, L.; Mishra, B.; Basu, A.; Lee, J.M.; Bhattacharjya, S.; Leong, S.S. Design of short membrane selective antimicrobial peptides containing tryptophan and arginine residues for improved activity, salt-resistance, and biocompatibility. Biotechnol. Bioeng. 2014, 111, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Yau, W.M.; Wimley, W.C.; Gawrisch, K.; White, S.H. The preference of tryptophan for membrane interfaces. Biochemistry 1998, 37, 14713–14718. [Google Scholar] [CrossRef]
- Feng, X.; Jin, S.; Wang, M.; Pang, Q.; Liu, C.; Liu, R.; Wang, Y.; Yang, H.; Liu, F.; Liu, Y. The Critical Role of Tryptophan in the Antimicrobial Activity and Cell Toxicity of the Duck Antimicrobial Peptide DCATH. Front. Microbiol. 2020, 11, 1146. [Google Scholar] [CrossRef] [PubMed]
- Barreto-Santamaría, A.; Arévalo-Pinzón, G.; Patarroyo, M.A.; Patarroyo, M.E. How to Combat Gram-Negative Bacteria Using Antimicrobial Peptides: A Challenge or an Unattainable Goal? Antibiotics 2021, 10, 1499. [Google Scholar] [CrossRef]
- Dombach, J.L.; Quintana, J.L.J.; Nagy, T.A.; Wan, C.; Crooks, A.L.; Yu, H.; Su, C.C.; Yu, E.W.; Shen, J.; Detweiler, C.S. A small molecule that mitigates bacterial infection disrupts Gram-negative cell membranes and is inhibited by cholesterol and neutral lipids. PLoS Pathog. 2020, 16, e1009119. [Google Scholar] [CrossRef]
- Nishino, K.; Nikaido, E.; Yamaguchi, A. Regulation and physiological function of multidrug efflux pumps in Escherichia coli and Salmonella. Biochim. Biophys. Acta 2009, 1794, 834–843. [Google Scholar] [CrossRef]
- Darnell, R.L.; Paxie, O.; Rose, F.O.T.; Morris, S.; Krause, A.L.; Monk, I.R.; Smith, M.J.B.; Stinear, T.P.; Cook, G.M.; Gebhard, S. Antimicrobial tolerance and its role in the development of resistance: Lessons from enterococci. Adv. Microb. Physiol. 2022, 81, 25–65. [Google Scholar] [CrossRef]
- World Health Organisation (WHO). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. Available online: http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (accessed on 16 June 2021).
- WHO. Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2024.
- Zhang, Y.; Du, M.; Chang, Y.; Chen, L.A.; Zhang, Q. Incidence, clinical characteristics, and outcomes of nosocomial Enterococcus spp. bloodstream infections in a tertiary-care hospital in Beijing, China: A four-year retrospective study. Antimicrob. Resist. Infect. Control. 2017, 6, 73. [Google Scholar] [CrossRef]
- Bonjean, M.; Hodille, E.; Dumitrescu, O.; Dupieux, C.; Nkoud Mongo, C.; Allam, C.; Beghin, M.; Paris, M.; Borrel, O.; Chardon, H.; et al. Disk diffusion testing for detection of methicillin-resistant staphylococci: Does Moxalactam improve upon Cefoxitin? J. Clin. Microbiol. 2016, 54, 2905–2909. [Google Scholar] [CrossRef] [PubMed]
- Gimza, B.D.; Cassat, J.E. Mechanisms of antibiotic failure during Staphylococcus aureus osteomyelitis. Front. Immunol. 2021, 12, 638085. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). 10 Threats to Global Health in 2018. 9 February 2018. Available online: https://medium.com/@who/10-threats-to-global-health-in-2018-232daf0bbef3 (accessed on 15 April 2019).
- Denissen, J.; Reyneke, B.; Waso-Reyneke, M.; Havenga, B.; Barnard, T.; Khan, S.; Khan, W. Prevalence of ESKAPE pathogens in the environment: Antibiotic resistance status, community-acquired infection and risk to human health. J. Hyg. Environ. Health 2022, 244, 114006. [Google Scholar] [CrossRef]
- Ciofu, O.; Moser, C.; Jensen, P.Ø.; Høiby, N. Tolerance and resistance of microbial biofilms. Nat. Rev. Microbiol. 2022, 20, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Campoccia, D.; Bottau, G.; De Donno, A.; Bua, G.; Ravaioli, S.; Capponi, E.; Sotgiu, G.; Bellotti, C.; Costantini, S.; Arciola, C.R. Assessing Cytotoxicity, Proteolytic Stability, and Selectivity of Antimicrobial Peptides: Implications for Orthopedic Applications. Int. J. Mol. Sci. 2024, 25, 13241. [Google Scholar] [CrossRef]
- Llor, C.; Bjerrum, L. Antimicrobial resistance: Risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 2014, 5, 229–241. [Google Scholar] [CrossRef]
- McDonnell, G.; Russell, A.D. Antiseptics and disinfectants: Activity, action, and resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. [Google Scholar] [CrossRef]
- Duarte-Mata, D.I.; Salinas-Carmona, M.C. Antimicrobial peptides immune modulation role in intracellular bacterial infection. Front. Immunol. 2023, 14, 1119574. [Google Scholar] [CrossRef]
- Petkovic, M.; Mouritzen, M.V.; Mojsoska, B.; Jenssen, H. Immunomodulatory Properties of Host Defence Peptides in Skin Wound Healing. Biomolecules 2021, 11, 952. [Google Scholar] [CrossRef]
- Li, S.; Wang, Y.; Xue, Z.; Jia, Y.; Li, R.; He, C.; Chen, H. The structure-mechanism relationship and mode of actions of antimicrobial peptides: A review. Trends Food Sci. Technol. 2021, 109, 103–115. [Google Scholar] [CrossRef]



| Parameters | RKW RKWILKWLRTWKK |
|---|---|
| Net Charge | +6 |
| Mol Weight | 1842.31 |
| Half-life (s) | 911.91 |
| Hydrophobicity | 0.479 |
| Hydrophobic moment | 0.753 |
| Total Hydrophobic Ratio (%) | 46 |
| Amphipathicity | 1.51 |
| GRAVY | −1.22 |
| Wimley-White whole-residue hydrophobicity (kcal/mol) | −1.26 |
| Hydrophilicity | 0.15 |
| Instability Index | 30.26 ** |
| Aliphatic index | 90 |
| Boman index (kcal/mol) | 2.52 |
| Total Trp ratio (%) | 23.07 |
| Strain | MBC (µM) |
|---|---|
| Escherichia coli | 10 |
| Salmonella Typhimurium | 10 |
| Listeria monocytogenes | 10 |
| Staphylococcus aureus | 15 |
| Pseudomonas aeruginosa | 5 |
| Campylobacter jejuni | 80 |
| Monophasic Salmonella Typhimurium | 20 |
| Salmonella Napoli | 20 |
| Strain | MBC (µM) |
|---|---|
| Escherichia coli | 10.0 |
| Salmonella Typhimurium | 10.0 |
| Listeria monocytogenes | 10.0 |
| Staphylococcus aureus | 20.0 |
| Pseudomonas aeruginosa | 5.0 |
| Bacteria | Antibiotic Class | Antibiotic | Disc Content (μg) | R * < (mm) | Inhibition Zone Diameter (mm) | Results |
|---|---|---|---|---|---|---|
| Acinetobacter baumannii | Aminoglycosides | Amikacin | 30 | 19 | - | R |
| Gentamicin | 10 | 17 | - | R | ||
| Tobramycin | 10 | 17 | - | R | ||
| Carbapenems | Imipenem | 10 | 21 | 16 | R | |
| Meropenem | 10 | 21 | 10 | R | ||
| Fluoroquinolones | Ciprofloxacin | 5 | 21 | - | R | |
| Levofloxacin | 5 | 20 | - | R | ||
| Miscellaneous agents | Trimethoprim-sulfamethoxazole | 1.25–23.75 | 11 | - | R | |
| Enterococcus faecium | Carbapenems | Imipenem | 10 | 21 | 50 | S |
| Fluoroquinolones | Ciprofloxacin | 5 | 15 | - | R | |
| Levofloxacin | 5 | 15 | - | R | ||
| Glycopeptide | Vancomycin | 5 | 12 | - | R | |
| Penicillins | Ampicillin | 2 | 10 | - | R | |
| Staphylococcus aureus (MRSA) | Aminoglycosides | Amikacin | 30 | 15 | 25 | S |
| Gentamicin | 10 | 18 | 28 | S | ||
| Tobramycin | 10 | 18 | 29 | S | ||
| Cephalosporins | Cefoxitin | 30 | 17 | 15 | R | |
| Fluoroquinolones | Ciprofloxacin | 5 | 17 | - | R | |
| Levofloxacin | 5 | 22 | - | R | ||
| Macrolides | Erythromycin | 15 | 21 | - | R | |
| Tetracyclines | Tetracycline | 30 | 22 | 32 | S | |
| Miscellaneous agents | Trimethoprim-sulfamethoxazole | 1.25–23.75 | 14 | 30 | S |
| Bacterial Inoculum Concentration | 5 × 103 CFU/mL | 5 × 105 CFU/mL | ||
|---|---|---|---|---|
| MIC | MBC | MIC | MBC | |
| Acinetobacter baumannii (CRAB) | 50 μM | 50 μM | 75 μM | >75 μM |
| Enterococcus faecium (VREfm) | 20 μM | 20 μM | 20 μM | 50 μM |
| Pseudomonas aeruginosa | 20 μM | 20 μM | 75 μM | >75 μM |
| Staphylococcus aureus (MRSA) | 50 μM | 50 μM | 50 μM | 50 μM |
| Bacteria | Test Suspension | Peptide Concentration (µM) | ||||
|---|---|---|---|---|---|---|
| 100 | 50 | 20 | 10 | 5 | ||
| Staphylococcus aureus ATCC 6538 | N: 2.00 × 108 N0: 2.00 × 107 log N0: 7.30 | Na = 1.47 × 104 log Na = 4.20 log R = 3.13 | Na = 1.47 × 104 log Na = 4.20 log R = 3.13 | Na= 1.47 × 104 log Na = 4.20 log R = 3.13 Not active | Na = 5.10 × 104 log Na = 4.71 log R = 2.59 Not active | Na = 1.11 × 105 log Na = 5.10 log R = 2.25 Not active |
| Pseudomonas aeruginosa ATCC 15442 | N: 2.50 × 108 N0: 2.50 × 107 log N0: 7.40 | Na < 1.40 × 102 log Na < 2.15 log R > 5.25 Active | Na < 1.40 × 102 log Na < 2.15 log R > 5.25 Active | Na < 1.40 × 102 log Na < 2.15 log R > 5.25 Active | Na < 1.40 × 102 log Na < 2.15 log R > 5.25 Active | Na < 1.40 × 102 log Na < 2.15 log R > 5.25 Active |
| Staphylococcus epidermidis ATCC 12228 | N: 1.90 × 108 N0: 1.90 × 107 log N0: 7.28 | Na = 1.60 × 102 log Na = 2.20 log R = 5.08 Active | Na = 1.60 × 102 log Na = 2.20 log R = 5.08 Active | Na = 1.60 × 102 log Na = 2.20 log R = 5.08 Active | Na = 2.40 × 102 log Na = 2.38 log R = 4.90 Not active | Na = 3.70 × 102 log Na = 2.57 log R = 4.71 Not active |
| Fungi | ||||||
| Candida albicans ATCC 10231 | N: 3.95 × 107 N0: 3.95 × 106 log N0: 6.60 | Na = 2.75 × 102 log Na = 2.44 log R = 4.16 Active | Na = 2.75 × 102 log Na = 2.44 log R = 4.16 Active | Na = 2.75 × 102 log Na = 2.44 log R = 4.16 Active | Na = 1.08 × 103 log Na = 3.03 log R = 3.57 Not Active | Na = 7.70 × 104 log Na = 4.89 log R = 1.71 Not Active |
| Aspergillus brasiliensis ATCC 16404 | N: 3.95 × 107 N0: 3.95 × 106 log N0: 6.60 | Na = 2.20 × 106 log Na = 6.34 log R = −0.09 Not Active | Na = 2.20 × 106 log Na = 6.34 log R = −0.09 Not Active | Na = 2.20 × 106 log Na = 6.34 log R = −0.09 Not Active | Na = 3.30 × 106 log Na = 6.52 log R = −0.29 Not Active | Na = 3.40 × 106 log Na = 6.53 log R = −0.28 Not Active |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agrillo, B.; Ambrosio, M.; Ambrosio, R.L.; Gogliettino, M.; Balestrieri, M.; Porritiello, A.; Peruzy, M.F.; Mancusi, A.; Nicolais, L.; Palmieri, G. Discovery of a Potent Antimicrobial Peptide Through Rational Design: A New Frontier in Pathogen Control. Biomolecules 2025, 15, 989. https://doi.org/10.3390/biom15070989
Agrillo B, Ambrosio M, Ambrosio RL, Gogliettino M, Balestrieri M, Porritiello A, Peruzy MF, Mancusi A, Nicolais L, Palmieri G. Discovery of a Potent Antimicrobial Peptide Through Rational Design: A New Frontier in Pathogen Control. Biomolecules. 2025; 15(7):989. https://doi.org/10.3390/biom15070989
Chicago/Turabian StyleAgrillo, Bruna, Monica Ambrosio, Rosa Luisa Ambrosio, Marta Gogliettino, Marco Balestrieri, Alessandra Porritiello, Maria Francesca Peruzy, Andrea Mancusi, Luigi Nicolais, and Gianna Palmieri. 2025. "Discovery of a Potent Antimicrobial Peptide Through Rational Design: A New Frontier in Pathogen Control" Biomolecules 15, no. 7: 989. https://doi.org/10.3390/biom15070989
APA StyleAgrillo, B., Ambrosio, M., Ambrosio, R. L., Gogliettino, M., Balestrieri, M., Porritiello, A., Peruzy, M. F., Mancusi, A., Nicolais, L., & Palmieri, G. (2025). Discovery of a Potent Antimicrobial Peptide Through Rational Design: A New Frontier in Pathogen Control. Biomolecules, 15(7), 989. https://doi.org/10.3390/biom15070989








