Improving the Catalytic Activity and Thermostability of FAST-PETase with a Multifunctional Short Peptide
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria, Plasmids, Media, and Reagents
2.2. Construction of Plasmids for the Expression of FAST-PETase Fused with or without S1v1 Tag
2.3. Expression of FAST-PETase Fused with or without S1v1 Tag
2.4. Purification of the Recombinant Proteins with Ni-NTA Affinity Chromatography
2.5. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.6. Western Blotting
2.7. High-Performance Liquid Chromatography (HPLC) to Analyze the Activity of PETase
2.8. Kinetic Analysis
2.9. Thermostability Assay
2.10. Melting Temperatures Assay with nanoDSF
2.11. Molecular Docking
3. Results
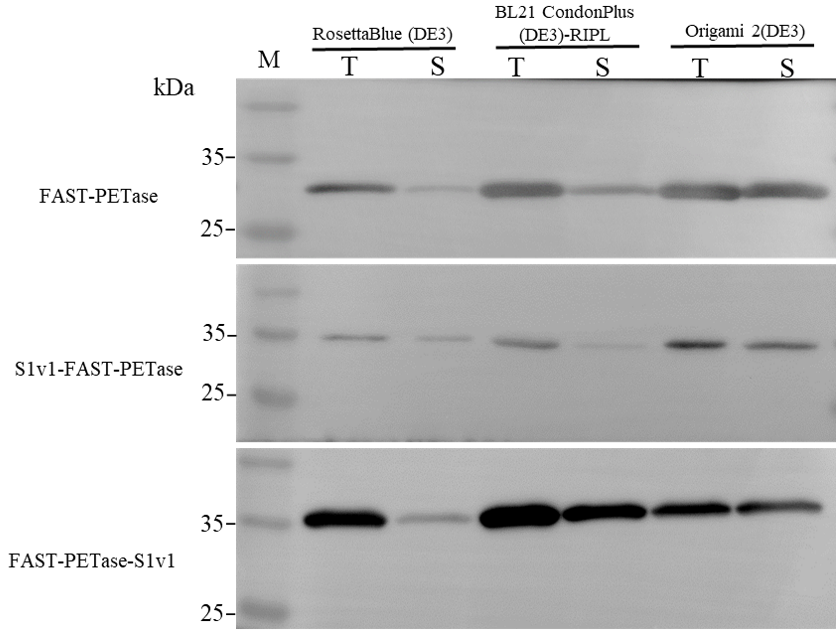
3.1. Expression of FAST-PETase Fused with or without the Multifunctional Peptide
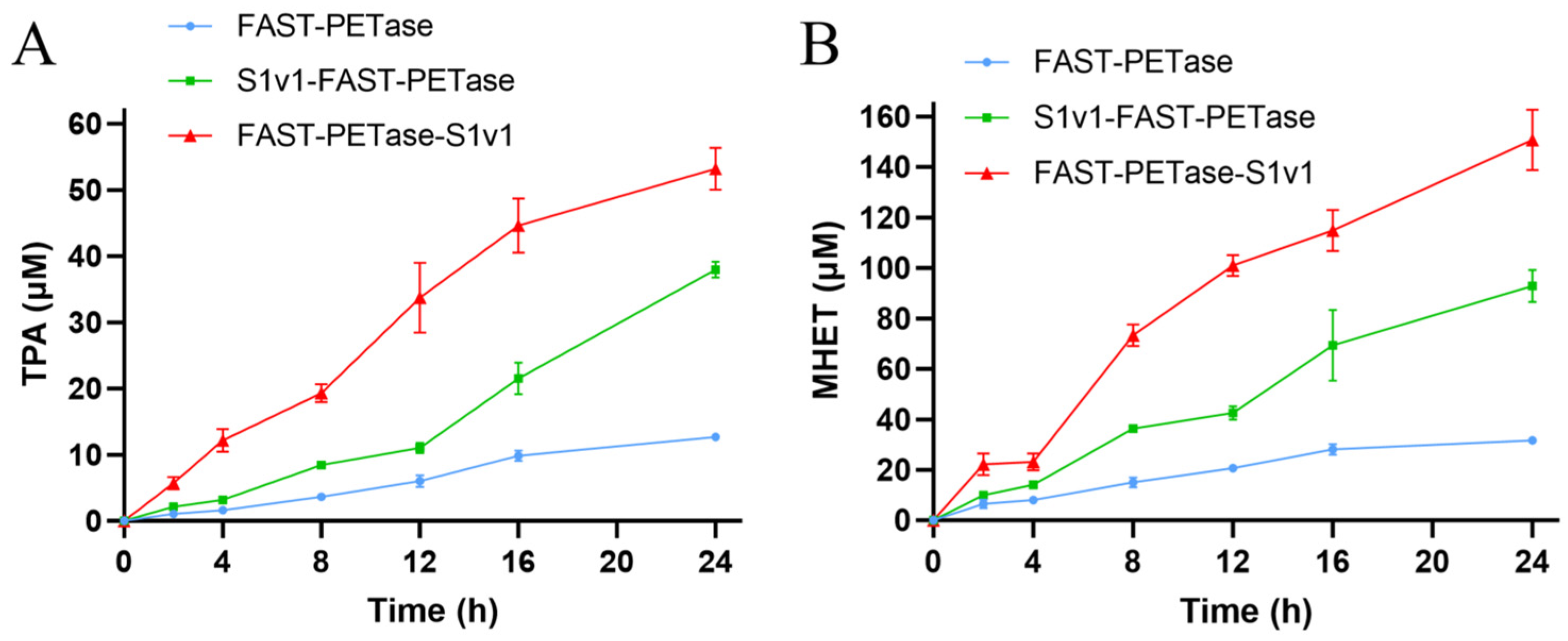
3.2. The Activities of FAST-PETase Fused with or without the Multifunctional Peptide
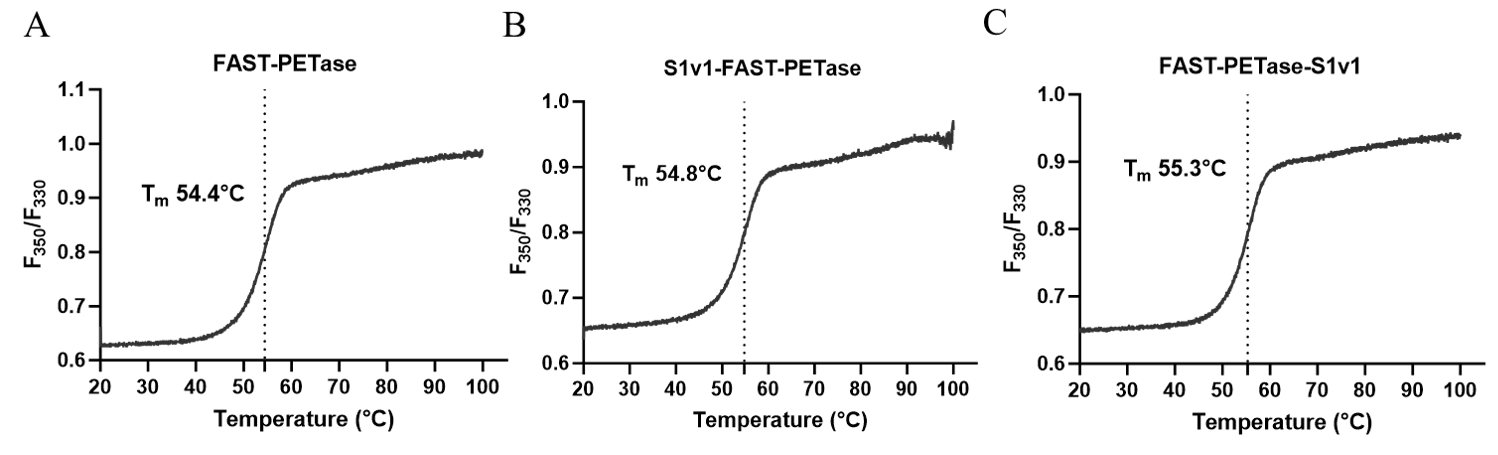
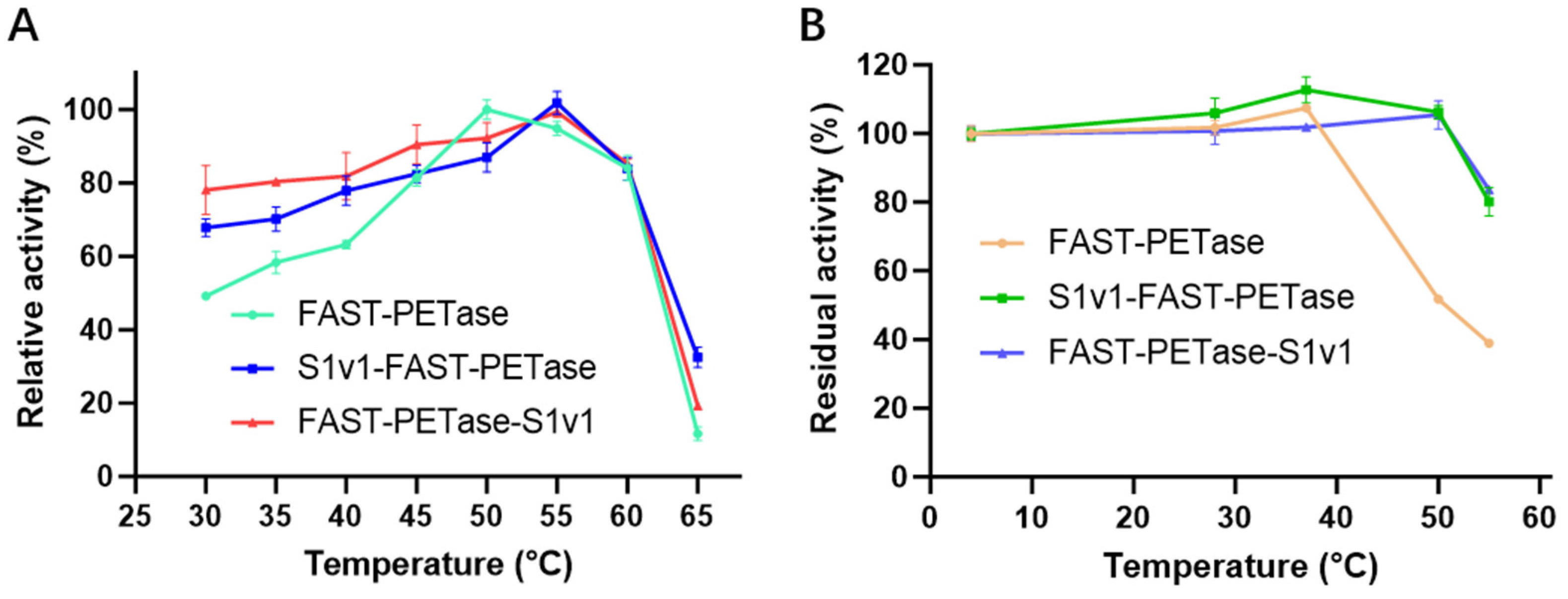
3.3. The Thermostability of FAST-PETase Fused with or without S1v1 Tag
3.4. Enzyme Kinetic of FAST-PETase Fused with or without S1v1 Tag
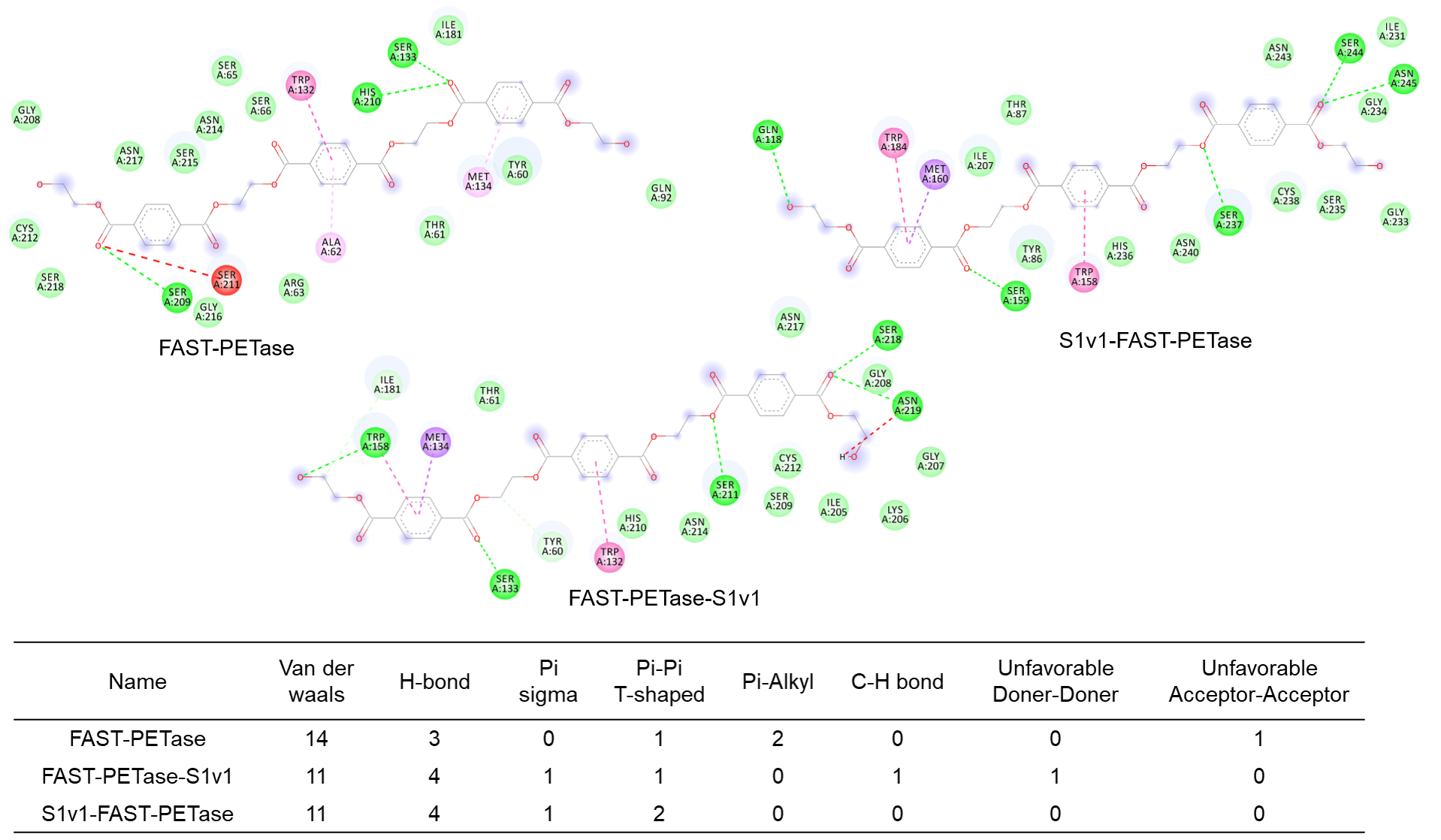
3.5. Structure Simulation of the Interactions Between Enzymes and the Substrate
4. Discussions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Magalhães, R.P.; Cunha, J.M.; Sousa, S.F. Perspectives on the Role of Enzymatic Biocatalysis for the Degradation of Plastic PET. Int. J. Mol. Sci. 2021, 22, 11257. [Google Scholar] [CrossRef]
- Chamas, A.; Moon, H.; Zheng, J.; Qiu, Y.; Suh, S. Degradation Rates of Plastics in the Environment. ACS Sustain. Chem. Eng. 2020, 8, 3494–3511. [Google Scholar] [CrossRef]
- Müller, R.J.; Schrader, H.; Profe, J.; Dresler, K.; Deckwer, W.D. Enzymatic degradation of poly (ethylene terephthalate): Rapid hydrolyse using a hydrolase from T. fusca. Macromol. Rapid Commun. 2005, 26, 1400–1405. [Google Scholar] [CrossRef]
- Danso, D.; Chow, J.; Streit, W.R. Plastics: Environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 2019, 85, e01095-19. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly (ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef]
- Han, X.; Liu, W.; Huang, J.; Ma, J.; Zheng, Y.; Ko, T.; Xu, L.; Cheng, Y.; Chen, C.; Guo, R. Structural insight into catalytic mechanism of PET hydrolase. Nat. Commun. 2017, 8, 2106. [Google Scholar] [CrossRef]
- Joo, S.; Cho, I.J.; Seo, H.; Son, H.F.; Sagong, H.-Y.; Shin, T.J.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Structural insight into molecular mechanism of poly (ethylene terephthalate) degradation. Nat. Commun. 2018, 9, 382. [Google Scholar] [CrossRef]
- Son, H.F.; Cho, I.J.; Joo, S.; Seo, H.; Sagong, H.-Y.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Rational protein engineering of thermo-stable PETase from Ideonella sakaiensis for highly efficient PET degradation. Acs Catal. 2019, 9, 3519–3526. [Google Scholar] [CrossRef]
- Cui, Y.; Chen, Y.; Liu, X.; Dong, S.; Tian, Y.e.; Qiao, Y.; Mitra, R.; Han, J.; Li, C.; Han, X. Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy. Acs Catal. 2021, 11, 1340–1350. [Google Scholar] [CrossRef]
- Lu, H.; Diaz, D.J.; Czarnecki, N.J.; Zhu, C.; Kim, W.; Shroff, R.; Acosta, D.J.; Alexander, B.R.; Cole, H.O.; Zhang, Y. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 2022, 604, 662–667. [Google Scholar] [CrossRef]
- Bell, E.L.; Smithson, R.; Kilbride, S.; Foster, J.; Hardy, F.J.; Ramachandran, S.; Tedstone, A.A.; Haigh, S.J.; Garforth, A.A.; Day, P.J. Directed evolution of an efficient and thermostable PET depolymerase. Nat. Catal. 2022, 5, 673–681. [Google Scholar] [CrossRef]
- Seo, H.; Kim, S.; Son, H.F.; Sagong, H.-Y.; Joo, S.; Kim, K.-J. Production of extracellular PETase from Ideonella sakaiensis using sec-dependent signal peptides in E. coli. Biochem. Biophys. Res. Commun. 2019, 508, 250–255. [Google Scholar] [CrossRef]
- Shi, L.; Liu, H.; Gao, S.; Weng, Y.; Zhu, L. Enhanced extracellular production of is PETase in Escherichia coli via engineering of the pelB signal peptide. J. Agric. Food Chem. 2021, 69, 2245–2252. [Google Scholar] [CrossRef]
- Huang, X.; Cao, L.; Qin, Z.; Li, S.; Kong, W.; Liu, Y. Tat-independent secretion of polyethylene terephthalate hydrolase PETase in Bacillus subtilis 168 mediated by its native signal peptide. J. Agric. Food Chem. 2018, 66, 13217–13227. [Google Scholar] [CrossRef]
- Wang, N.; Guan, F.; Lv, X.; Han, D.; Zhang, Y.; Wu, N.; Xia, X.; Tian, J. Enhancing secretion of polyethylene terephthalate hydrolase PETase in Bacillus subtilis WB600 mediated by the SPamy signal peptide. Lett. Appl. Microbiol. 2020, 71, 235–241. [Google Scholar] [CrossRef]
- Carter, L.M.; MacFarlane, C.E.; Karlock, S.P.; Sen, T.; Kaar, J.L.; Berberich, J.A.; Boock, J.T. Increased cytoplasmic expression of PETase enzymes in E. coli. Microb. Cell Factories 2024, 23, 319. [Google Scholar] [CrossRef]
- Deng, B.; Yue, Y.; Yang, J.; Yang, M.; Xing, Q.; Peng, H.; Wang, F.; Li, M.; Ma, L.; Zhai, C. Improving the activity and thermostability of PETase from Ideonella sakaiensis through modulating its post-translational glycan modification. Commun. Biol. 2023, 6, 39. [Google Scholar] [CrossRef]
- Ko, H.; Kang, M.; Kim, M.J.; Yi, J.; Kang, J.; Bae, J.H.; Sohn, J.H.; Sung, B.H. A novel protein fusion partner, carbohydrate-binding module family 66, to enhance heterologous protein expression in Escherichia coli. Microb. Cell Fact. 2021, 20, 232. [Google Scholar] [CrossRef]
- Lee, N.R.; Bowerman, C.J.; Nilsson, B.L. Effects of varied sequence pattern on the self-assembly of amphipathic peptides. Biomacromolecules 2013, 14, 3267–3277. [Google Scholar] [CrossRef]
- Zhang, S.; Lockshin, C.; Herbert, A.; Winter, E.; Rich, A. Zuotin, a putative Z-DNA binding protein in Saccharomyces cerevisiae. EMBO J. 1992, 11, 3787–3796. [Google Scholar] [CrossRef]
- Yang, H.; Lu, X.; Liu, L.; Li, J.; Shin, H.D.; Chen, R.R.; Du, G.; Chen, J. Fusion of an oligopeptide to the N terminus of an alkaline α-amylase from Alkalimonas amylolytica simultaneously improves the enzyme’s catalytic efficiency, thermal stability, and resistance to oxidation. Appl. Env. Microbiol. 2013, 79, 3049–3058. [Google Scholar] [CrossRef]
- Liu, Y.; Cui, W.; Liu, Z.; Cui, Y.; Xia, Y.; Kobayashi, M.; Zhou, Z. Enhancement of thermo-stability and product tolerance of Pseudomonas putida nitrile hydratase by fusing with self-assembling peptide. J. Biosci. Bioeng. 2014, 118, 249–252. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, L.; Du, G.; Liu, S. A multifunctional tag with the ability to benefit the expression, purification, thermostability and activity of recombinant proteins. J. Biotechnol. 2018, 283, 1–10. [Google Scholar] [CrossRef]
- Fang, J.; Sheng, L.; Ye, Y.; Gao, S.; Ji, J.; Zhang, Y.; Sun, X. Biochemical Characterization and Application of Zearalenone Lactone Hydrolase Fused with a Multifunctional Short Peptide. J. Agric. Food Chem. 2024, 72, 18146–18154. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Mannual, 3rd ed.; Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Yu, F.; Li, X.; Wang, F.; Liu, Y.; Zhai, C.; Li, W.; Ma, L.; Chen, W. TLTC, a T5 exonuclease-mediated low-temperature DNA cloning method. Front. Bioeng. Biotechnol. 2023, 11, 1167534. [Google Scholar] [CrossRef]
- Ma, Y.; Yao, M.; Li, B.; Ding, M.; He, B.; Chen, S.; Zhou, X.; Yuan, Y. Enhanced poly (ethylene terephthalate) hydrolase activity by protein engineering. Engineering 2018, 4, 888–893. [Google Scholar] [CrossRef]
- Wang, S.Z.; Fang, B.S. Microplate bioassay for determining substrate selectivity of Candida rugosa lipase. J. Chem. Educ. 2012, 89, 409–411. [Google Scholar] [CrossRef]
- Peng, Y.; Fu, S.; Liu, H.; Lucia, L.A. Accurately determining esterase activity via the isosbestic point of p-nitrophenol. BioResources 2016, 11, 10099–10111. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Wingfield, P.T. Overview of the purification of recombinant proteins. Curr. Protoc. Protein Sci. 2015, 80, 6.1.1–6.1.35. [Google Scholar] [CrossRef]
- Bell, M.R.; Engleka, M.J.; Malik, A.; Strickler, J.E. To fuse or not to fuse: What is your purpose? Protein Sci. 2013, 22, 1466–1477. [Google Scholar] [CrossRef] [PubMed]

| Enzyme | kcat (min−1) | KM (mmol·L−1) | kcat/KM (mmol−1·L·min−1) |
|---|---|---|---|
| FAST-PETase | 43.42 ± 3.19 | 0.88 ± 0.16 | 49.42 ± 6.70 |
| S1v1-FAST-PETase | 34.84 ± 2.35 | 0.86 ± 0.15 | 40.71 ± 4.33 |
| FAST-PETase-S1v1 | 81.32 ± 5.51 | 1.22 ± 0.18 | 66.82 ± 8.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Deng, B.; Liao, P.; Lin, S.; Zheng, L.; Yang, X.; Wang, F.; Zhai, C.; Ma, L. Improving the Catalytic Activity and Thermostability of FAST-PETase with a Multifunctional Short Peptide. Biomolecules 2025, 15, 888. https://doi.org/10.3390/biom15060888
Yang J, Deng B, Liao P, Lin S, Zheng L, Yang X, Wang F, Zhai C, Ma L. Improving the Catalytic Activity and Thermostability of FAST-PETase with a Multifunctional Short Peptide. Biomolecules. 2025; 15(6):888. https://doi.org/10.3390/biom15060888
Chicago/Turabian StyleYang, Jun, Binyang Deng, Pingan Liao, Siyu Lin, Liqi Zheng, Xing Yang, Fei Wang, Chao Zhai, and Lixin Ma. 2025. "Improving the Catalytic Activity and Thermostability of FAST-PETase with a Multifunctional Short Peptide" Biomolecules 15, no. 6: 888. https://doi.org/10.3390/biom15060888
APA StyleYang, J., Deng, B., Liao, P., Lin, S., Zheng, L., Yang, X., Wang, F., Zhai, C., & Ma, L. (2025). Improving the Catalytic Activity and Thermostability of FAST-PETase with a Multifunctional Short Peptide. Biomolecules, 15(6), 888. https://doi.org/10.3390/biom15060888






