FMR1: A Neurodevelopmental Factor Regulating Cell Metabolism in the Tumor Microenvironment
Abstract
1. Introduction
2. Methodology
3. FMR1 and FMRP: Beyond Neurodevelopment
3.1. Function of FMRP in Normal Physiology
3.2. FMRP’s Role in Cell Metabolism and Cancer Progression
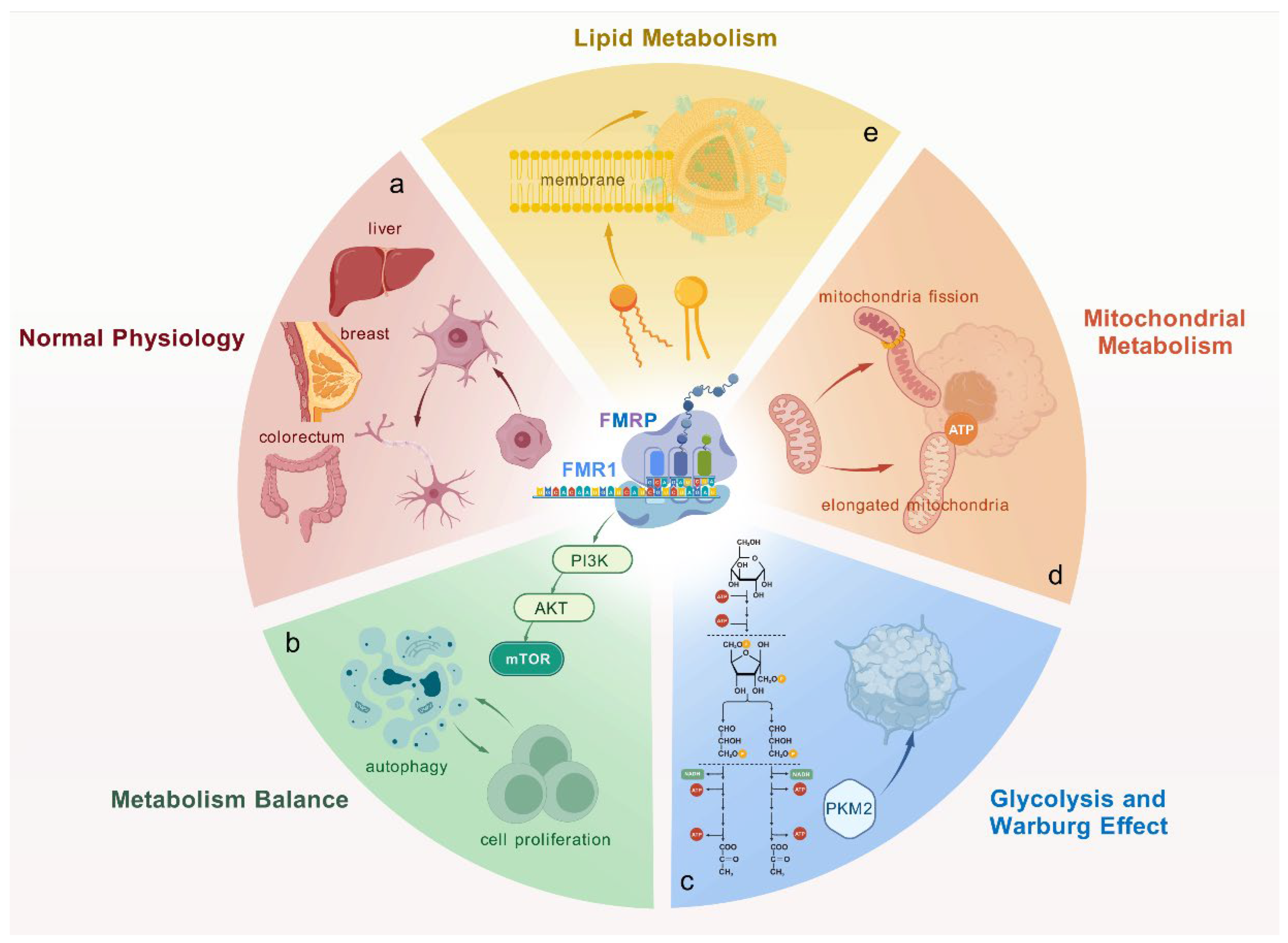
3.3. Regulation and Cell-Type-Specific Functions of FMRP
4. FMRP’s Role in the Tumor Microenvironment
4.1. FMRP and Endothelial Cells in the Tumor Microenvironment
4.2. FMRP’s Impact on Immune Cells and Tumor Immunity
4.3. Tumor Immune Evasion and FMRP’s Role
5. FMRP as a Target for Cancer Therapy
5.1. FMRP’s Role in Immunotherapy and Immune Modulation
5.2. Combination Therapies Targeting FMRP
5.3. Challenges in Targeting FMRP in Cancer Therapy
6. FMRP and Cancer Metabolism in Specific Cancer Types
6.1. Common Roles of FMRP in Cancer Metabolism
6.2. Cancer-Specific Roles of FMRP
6.2.1. Breast Cancer
6.2.2. FMRP in Glioblastoma
6.2.3. FMRP in Intrahepatic Cholangiocarcinoma
6.2.4. FMRP in Head and Neck Cancer
6.3. Controversies and Tumor-Specific Mechanisms of FMRP in Cancer Biology
7. Future Directions and Research Gaps
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hagerman, R.J.; Berry-Kravis, E.; Hazlett, H.C.; Bailey, D.B., Jr.; Moine, H.; Kooy, R.F.; Tassone, F.; Gantois, I.; Sonenberg, N.; Mandel, J.L.; et al. Fragile X syndrome. Nat. Rev. Dis. Primers 2017, 3, 17065. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.S.; Wu, H.; Krzisch, M.; Wu, X.; Graef, J.; Muffat, J.; Hnisz, D.; Li, C.H.; Yuan, B.; Xu, C.; et al. Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 2018, 172, 979–992.e976. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.D.; Zhao, X. The molecular biology of FMRP: New insights into fragile X syndrome. Nat. Rev. Neurosci. 2021, 22, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Mitsutomi, S.; Hewko, A.; Akimitsu, N.; Maquat, L.E. Integrative omics indicate FMRP sequesters mRNA from translation and deadenylation in human neuronal cells. Mol. Cell 2022, 82, 4564–4581.e4511. [Google Scholar] [CrossRef]
- Zeng, Q.; Saghafinia, S.; Chryplewicz, A.; Fournier, N.; Christe, L.; Xie, Y.Q.; Guillot, J.; Yucel, S.; Li, P.; Galván, J.A.; et al. Aberrant hyperexpression of the RNA binding protein FMRP in tumors mediates immune evasion. Science 2022, 378, eabl7207. [Google Scholar] [CrossRef]
- Yoshida, G.J. Metabolic reprogramming: The emerging concept and associated therapeutic strategies. J. Exp. Clin. Cancer Res. 2015, 34, 111. [Google Scholar] [CrossRef]
- Xia, L.; Oyang, L.; Lin, J.; Tan, S.; Han, Y.; Wu, N.; Yi, P.; Tang, L.; Pan, Q.; Rao, S.; et al. The cancer metabolic reprogramming and immune response. Mol. Cancer 2021, 20, 28. [Google Scholar] [CrossRef]
- de Esch, C.E.; Ghazvini, M.; Loos, F.; Schelling-Kazaryan, N.; Widagdo, W.; Munshi, S.T.; van der Wal, E.; Douben, H.; Gunhanlar, N.; Kushner, S.A.; et al. Epigenetic characterization of the FMR1 promoter in induced pluripotent stem cells from human fibroblasts carrying an unmethylated full mutation. Stem Cell Rep. 2014, 3, 548–555. [Google Scholar] [CrossRef]
- Elhanani, O.; Ben-Uri, R.; Keren, L. Spatial profiling technologies illuminate the tumor microenvironment. Cancer Cell 2023, 41, 404–420. [Google Scholar] [CrossRef]
- Geng, J.; Khaket, T.P.; Pan, J.; Li, W.; Zhang, Y.; Ping, Y.; Cobos Sillero, M.I.; Lu, B. Deregulation of ER-mitochondria contact formation and mitochondrial calcium homeostasis mediated by VDAC in fragile X syndrome. Dev. Cell 2023, 58, 597–615.e510. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, Y.; Wang, D.; Lin, Y.; Hou, J.; Xu, X.; Wu, J.; Zhong, L.; Zhou, Y.; Shen, J.; et al. The HNF4α-BC200-FMR1-Positive Feedback Loop Promotes Growth and Metastasis in Invasive Mucinous Lung Adenocarcinoma. Cancer Res. 2021, 81, 5904–5918. [Google Scholar] [CrossRef] [PubMed]
- Sirois, C.L.; Guo, Y.; Li, M.; Wolkoff, N.E.; Korabelnikov, T.; Sandoval, S.; Lee, J.; Shen, M.; Contractor, A.; Sousa, A.M.M.; et al. CGG repeats in the human FMR1 gene regulate mRNA localization and cellular stress in developing neurons. Cell Rep. 2024, 43, 114330. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Gao, Q.; Ma, S.; Yu, P.; Ding, S.; Yao, X.; Zhang, Z.; Lu, S.; Lu, M.; Zhang, J.; et al. FMR1 promotes the progression of colorectal cancer cell by stabilizing EGFR mRNA in an m(6)A-dependent manner. Cell Death Dis. 2022, 13, 941. [Google Scholar] [CrossRef]
- Lucá, R.; Averna, M.; Zalfa, F.; Vecchi, M.; Bianchi, F.; La Fata, G.; Del Nonno, F.; Nardacci, R.; Bianchi, M.; Nuciforo, P.; et al. The fragile X protein binds mRNAs involved in cancer progression and modulates metastasis formation. EMBO Mol. Med. 2013, 5, 1523–1536. [Google Scholar] [CrossRef]
- Taha, M.S.; Haghighi, F.; Stefanski, A.; Nakhaei-Rad, S.; Kazemein Jasemi, N.S.; Al Kabbani, M.A.; Görg, B.; Fujii, M.; Lang, P.A.; Häussinger, D.; et al. Novel FMRP interaction networks linked to cellular stress. Febs J. 2021, 288, 837–860. [Google Scholar] [CrossRef]
- Baldi, S.; Amer, B.; Alnadari, F.; Al-Mogahed, M.; Gao, Y.; Gamallat, Y. The Prognostic and Therapeutic Potential of Fragile X Mental Retardation 1 (FMR1) Gene Expression in Prostate Adenocarcinoma: Insights into Survival Outcomes and Oncogenic Pathway Modulation. Int. J. Mol. Sci. 2024, 25, 7290. [Google Scholar] [CrossRef]
- Casingal, C.R.; Kikkawa, T.; Inada, H.; Sasaki, Y.; Osumi, N. Identification of FMRP target mRNAs in the developmental brain: FMRP might coordinate Ras/MAPK, Wnt/β-catenin, and mTOR signaling during corticogenesis. Mol. Brain 2020, 13, 167. [Google Scholar] [CrossRef]
- Hua, H.; Kong, Q.; Zhang, H.; Wang, J.; Luo, T.; Jiang, Y. Targeting mTOR for cancer therapy. J. Hematol. Oncol. 2019, 12, 71. [Google Scholar] [CrossRef]
- Mossmann, D.; Park, S.; Hall, M.N. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer 2018, 18, 744–757. [Google Scholar] [CrossRef]
- Yang, H.; Wang, Y.; Xiang, Y.; Yadav, T.; Ouyang, J.; Phoon, L.; Zhu, X.; Shi, Y.; Zou, L.; Lan, L. FMRP promotes transcription-coupled homologous recombination via facilitating TET1-mediated m5C RNA modification demethylation. Proc. Natl. Acad. Sci. USA 2022, 119, e2116251119. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Leboucher, A.; Pisani, D.F.; Martinez-Gili, L.; Chilloux, J.; Bermudez-Martin, P.; Van Dijck, A.; Ganief, T.; Macek, B.; Becker, J.A.J.; Le Merrer, J.; et al. The translational regulator FMRP controls lipid and glucose metabolism in mice and humans. Mol. Metab. 2019, 21, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Tabet, R.; Vitale, N.; Moine, H. Fragile X syndrome: Are signaling lipids the missing culprits? Biochimie 2016, 130, 188–194. [Google Scholar] [CrossRef]
- Gill, K.S.; Fernandes, P.; O’Donovan, T.R.; McKenna, S.L.; Doddakula, K.K.; Power, D.G.; Soden, D.M.; Forde, P.F. Glycolysis inhibition as a cancer treatment and its role in an anti-tumour immune response. Biochim. Biophys. Acta 2016, 1866, 87–105. [Google Scholar] [CrossRef]
- Fernie, A.R.; Carrari, F.; Sweetlove, L.J. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 2004, 7, 254–261. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Vaupel, P.; Schmidberger, H.; Mayer, A. The Warburg effect: Essential part of metabolic reprogramming and central contributor to cancer progression. Int. J. Radiat. Biol. 2019, 95, 912–919. [Google Scholar] [CrossRef]
- Licznerski, P.; Park, H.A.; Rolyan, H.; Chen, R.; Mnatsakanyan, N.; Miranda, P.; Graham, M.; Wu, J.; Cruz-Reyes, N.; Mehta, N.; et al. ATP Synthase c-Subunit Leak Causes Aberrant Cellular Metabolism in Fragile X Syndrome. Cell 2020, 182, 1170–1185.e1179. [Google Scholar] [CrossRef]
- Ryu, K.W.; Fung, T.S.; Baker, D.C.; Saoi, M.; Park, J.; Febres-Aldana, C.A.; Aly, R.G.; Cui, R.; Sharma, A.; Fu, Y.; et al. Cellular ATP demand creates metabolically distinct subpopulations of mitochondria. Nature 2024, 635, 746–754. [Google Scholar] [CrossRef]
- Fenton, A.R.; Peng, R.; Bond, C.; Hugelier, S.; Lakadamyali, M.; Chang, Y.W.; Holzbaur, E.L.F.; Jongens, T.A. FMRP regulates MFF translation to locally direct mitochondrial fission in neurons. Nat. Cell Biol. 2024, 26, 2061–2074. [Google Scholar] [CrossRef]
- Sirois, C.L.; Sandoval, S.O.; Zhao, X. FMRP gains mitochondrial fission control. Nat. Cell Biol. 2024, 26, 2014–2015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Peng, X.; Du, J.X.; Boohaker, R.; Estevao, I.L.; Grajeda, B.I.; Cox, M.B.; Almeida, I.C.; Lu, W. Oncogenic KRASG12D Reprograms Lipid Metabolism by Upregulating SLC25A1 to Drive Pancreatic Tumorigenesis. Cancer Res. 2023, 83, 3739–3752. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, M.J.; Lu, W.C.; Li, Y.C.; Chang, C.J.; Yang, J.Y. Metabolic switch regulates lineage plasticity and induces synthetic lethality in triple-negative breast cancer. Cell Metab. 2024, 36, 193–208.e198. [Google Scholar] [CrossRef]
- Zhu, L.; Zhu, X.; Wu, Y. Effects of Glucose Metabolism, Lipid Metabolism, and Glutamine Metabolism on Tumor Microenvironment and Clinical Implications. Biomolecules 2022, 12, 580. [Google Scholar] [CrossRef]
- Jiang, S.; Li, H.; Zhang, L.; Mu, W.; Zhang, Y.; Chen, T.; Wu, J.; Tang, H.; Zheng, S.; Liu, Y.; et al. Generic Diagramming Platform (GDP): A comprehensive database of high-quality biomedical graphics. Nucleic Acids Res. 2024, 53, D1670–D1676. [Google Scholar] [CrossRef]
- Pieretti, M.; Zhang, F.P.; Fu, Y.H.; Warren, S.T.; Oostra, B.A.; Caskey, C.T.; Nelson, D.L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 1991, 66, 817–822. [Google Scholar] [CrossRef]
- Shah, S.; Sharp, K.J.; Raju Ponny, S.; Lee, J.; Watts, J.K.; Berry-Kravis, E.; Richter, J.D. Antisense oligonucleotide rescue of CGG expansion-dependent FMR1 mis-splicing in fragile X syndrome restores FMRP. Proc. Natl. Acad. Sci. USA 2023, 120, e2302534120. [Google Scholar] [CrossRef]
- Musco, G.; Stier, G.; Joseph, C.; Castiglione Morelli, M.A.; Nilges, M.; Gibson, T.J.; Pastore, A. Three-dimensional structure and stability of the KH domain: Molecular insights into the fragile X syndrome. Cell 1996, 85, 237–245. [Google Scholar] [CrossRef]
- Krueger, D.D.; Bear, M.F. Toward fulfilling the promise of molecular medicine in fragile X syndrome. Annu. Rev. Med. 2011, 62, 411–429. [Google Scholar] [CrossRef]
- Napoli, E.; Ross-Inta, C.; Song, G.; Wong, S.; Hagerman, R.; Gane, L.W.; Smilowitz, J.T.; Tassone, F.; Giulivi, C. Premutation in the Fragile X Mental Retardation 1 (FMR1) Gene Affects Maternal Zn-milk and Perinatal Brain Bioenergetics and Scaffolding. Front. Neurosci. 2016, 10, 159. [Google Scholar] [CrossRef]
- Bassell, G.J.; Warren, S.T. Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron 2008, 60, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Higashimori, H.; Schin, C.S.; Chiang, M.S.; Morel, L.; Shoneye, T.A.; Nelson, D.L.; Yang, Y. Selective Deletion of Astroglial FMRP Dysregulates Glutamate Transporter GLT1 and Contributes to Fragile X Syndrome Phenotypes In Vivo. J. Neurosci. 2016, 36, 7079–7094. [Google Scholar] [CrossRef] [PubMed]
- Pedini, G.; Buccarelli, M.; Bianchi, F.; Pacini, L.; Cencelli, G.; D’Alessandris, Q.G.; Martini, M.; Giannetti, S.; Sasso, F.; Melocchi, V.; et al. FMRP modulates the Wnt signalling pathway in glioblastoma. Cell Death Dis. 2022, 13, 719. [Google Scholar] [CrossRef]
- Bejarano, L.; Jordāo, M.J.C.; Joyce, J.A. Therapeutic Targeting of the Tumor Microenvironment. Cancer Discov. 2021, 11, 933–959. [Google Scholar] [CrossRef]
- Bilotta, M.T.; Antignani, A.; Fitzgerald, D.J. Managing the TME to improve the efficacy of cancer therapy. Front. Immunol. 2022, 13, 954992. [Google Scholar] [CrossRef]
- Hanahan, D.; Monje, M. Cancer hallmarks intersect with neuroscience in the tumor microenvironment. Cancer Cell 2023, 41, 573–580. [Google Scholar] [CrossRef]
- Lee, H.W.; Xu, Y.; He, L.; Choi, W.; Gonzalez, D.; Jin, S.W.; Simons, M. Role of Venous Endothelial Cells in Developmental and Pathologic Angiogenesis. Circulation 2021, 144, 1308–1322. [Google Scholar] [CrossRef]
- Leung, S.W.S.; Shi, Y. The glycolytic process in endothelial cells and its implications. Acta Pharmacol. Sin. 2022, 43, 251–259. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, Y.; Meng, C.; Fang, N. FMRP regulates endothelial cell proliferation and angiogenesis via the miR-181a-CaM-CaMKII pathway. Cell Biol. Int. 2018, 42, 1432–1444. [Google Scholar] [CrossRef]
- Ma, Y.X.; Lei, C.; Ye, T.; Wan, Q.Q.; Wang, K.Y.; Zhu, Y.N.; Li, L.; Liu, X.F.; Niu, L.Z.; Tay, F.R.; et al. Silicon Enhances Functional Mitochondrial Transfer to Improve Neurovascularization in Diabetic Bone Regeneration. Adv. Sci. 2025, 12, e2415459. [Google Scholar] [CrossRef]
- Cao, J.; Yan, Q. Cancer Epigenetics, Tumor Immunity, and Immunotherapy. Trends Cancer 2020, 6, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Saravia, J.; Raynor, J.L.; Chapman, N.M.; Lim, S.A.; Chi, H. Signaling networks in immunometabolism. Cell Res. 2020, 30, 328–342. [Google Scholar] [CrossRef] [PubMed]
- St Paul, M.; Ohashi, P.S. The Roles of CD8(+) T Cell Subsets in Antitumor Immunity. Trends Cell Biol. 2020, 30, 695–704. [Google Scholar] [CrossRef]
- Reina-Campos, M.; Scharping, N.E.; Goldrath, A.W. CD8(+) T cell metabolism in infection and cancer. Nat. Rev. Immunol. 2021, 21, 718–738. [Google Scholar] [CrossRef]
- Cao, J.; Liao, S.; Zeng, F.; Liao, Q.; Luo, G.; Zhou, Y. Effects of altered glycolysis levels on CD8(+) T cell activation and function. Cell Death Dis. 2023, 14, 407. [Google Scholar] [CrossRef]
- Geltink, R.I.K.; Kyle, R.L.; Pearce, E.L. Unraveling the Complex Interplay Between T Cell Metabolism and Function. Annu. Rev. Immunol. 2018, 36, 461–488. [Google Scholar] [CrossRef]
- Nayak, T.; Trotter, J.; Sakry, D. The Intracellular Cleavage Product of the NG2 Proteoglycan Modulates Translation and Cell-Cycle Kinetics via Effects on mTORC1/FMRP Signaling. Front. Cell Neurosci. 2018, 12, 231. [Google Scholar] [CrossRef]
- Szwed, A.; Kim, E.; Jacinto, E. Regulation and metabolic functions of mTORC1 and mTORC2. Physiol. Rev. 2021, 101, 1371–1426. [Google Scholar] [CrossRef]
- Hooshmandi, M.; Sharma, V.; Thörn Perez, C.; Sood, R.; Krimbacher, K.; Wong, C.; Lister, K.C.; Ureña Guzmán, A.; Bartley, T.D.; Rocha, C.; et al. Excitatory neuron-specific suppression of the integrated stress response contributes to autism-related phenotypes in fragile X syndrome. Neuron 2023, 111, 3028–3040.e3026. [Google Scholar] [CrossRef]
- Vitale, I.; Manic, G.; Coussens, L.M.; Kroemer, G.; Galluzzi, L. Macrophages and Metabolism in the Tumor Microenvironment. Cell Metab. 2019, 30, 36–50. [Google Scholar] [CrossRef]
- Boutilier, A.J.; Elsawa, S.F. Macrophage Polarization States in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 6995. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.S.; Wang, H.; Li, X.; Chao, T.; Teav, T.; Christen, S.; Di Conza, G.; Cheng, W.C.; Chou, C.H.; Vavakova, M.; et al. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 2017, 18, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Mehla, K.; Singh, P.K. Metabolic Regulation of Macrophage Polarization in Cancer. Trends Cancer 2019, 5, 822–834. [Google Scholar] [CrossRef]
- Su, P.; Wang, Q.; Bi, E.; Ma, X.; Liu, L.; Yang, M.; Qian, J.; Yi, Q. Enhanced Lipid Accumulation and Metabolism Are Required for the Differentiation and Activation of Tumor-Associated Macrophages. Cancer Res. 2020, 80, 1438–1450. [Google Scholar] [CrossRef]
- Wu, K.K.; Xu, X.; Wu, M.; Li, X.; Hoque, M.; Li, G.H.Y.; Lian, Q.; Long, K.; Zhou, T.; Piao, H.; et al. MDM2 induces pro-inflammatory and glycolytic responses in M1 macrophages by integrating iNOS-nitric oxide and HIF-1α pathways in mice. Nat. Commun. 2024, 15, 8624. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef]
- Haschemi, A.; Kosma, P.; Gille, L.; Evans, C.R.; Burant, C.F.; Starkl, P.; Knapp, B.; Haas, R.; Schmid, J.A.; Jandl, C.; et al. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 2012, 15, 813–826. [Google Scholar] [CrossRef]
- Beatty, G.L.; Gladney, W.L. Immune escape mechanisms as a guide for cancer immunotherapy. Clin. Cancer Res. 2015, 21, 687–692. [Google Scholar] [CrossRef]
- Kao, K.C.; Vilbois, S.; Tsai, C.H.; Ho, P.C. Metabolic communication in the tumour-immune microenvironment. Nat. Cell Biol. 2022, 24, 1574–1583. [Google Scholar] [CrossRef]
- Hatzioannou, A.; Banos, A.; Sakelaropoulos, T.; Fedonidis, C.; Vidali, M.S.; Köhne, M.; Händler, K.; Boon, L.; Henriques, A.; Koliaraki, V.; et al. An intrinsic role of IL-33 in T(reg) cell-mediated tumor immunoevasion. Nat. Immunol. 2020, 21, 75–85. [Google Scholar] [CrossRef]
- Liu, Y.; Cai, Y.; Liu, L.; Wu, Y.; Xiong, X. Crucial biological functions of CCL7 in cancer. PeerJ 2018, 6, e4928. [Google Scholar] [CrossRef] [PubMed]
- Maurin, T.; Zongaro, S.; Bardoni, B. Fragile X Syndrome: From molecular pathology to therapy. Neurosci. Biobehav. Rev. 2014, 46 Pt 2, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhang, J.; Chen, H.; Qiao, D.; Guo, F.; Hu, X.; Qin, C.; Jin, X.; Zhang, K.; Wang, C.; et al. Role of FMRP in AKT/mTOR pathway-mediated hippocampal autophagy in fragile X syndrome. Prog. Neuropsychopharmacol. Biol. Psychiatry 2024, 134, 111036. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Shi, B.; Man, X.; Wu, W.; Cao, J. [High expression of fragile X mental retardation protein inhibits ferroptosis of colorectal tumor cells by activating the RAS/MAPK signaling pathway]. Nan Fang. Yi Ke Da Xue Xue Bao 2024, 44, 885–893. [Google Scholar]
- Zhang, Y.; Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef]
- Gill, J.; Prasad, V. A reality check of the accelerated approval of immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2019, 16, 656–658. [Google Scholar] [CrossRef]
- Bagchi, S.; Yuan, R.; Engleman, E.G. Immune Checkpoint Inhibitors for the Treatment of Cancer: Clinical Impact and Mechanisms of Response and Resistance. Annu. Rev. Pathol. 2021, 16, 223–249. [Google Scholar] [CrossRef]
- Jia, Y.; Jia, R.; Chen, Y.; Lin, X.; Aishan, N.; Li, H.; Wang, L.; Zhang, X.; Ruan, J. The role of RNA binding proteins in cancer biology: A focus on FMRP. Genes Dis. 2025, 12, 101493. [Google Scholar] [CrossRef]
- Peng, R.; Huang, Q.; Wang, L.; Qiao, G.; Huang, X.; Jiang, J.; Chu, X. G-Quadruplex RNA Based PROTAC Enables Targeted Degradation of RNA Binding Protein FMRP for Tumor Immunotherapy. Angew. Chem. Int. Ed. Engl. 2024, 63, e202402715. [Google Scholar] [CrossRef]
- Szeto, G.L.; Finley, S.D. Integrative Approaches to Cancer Immunotherapy. Trends Cancer 2019, 5, 400–410. [Google Scholar] [CrossRef]
- Bader, J.E.; Voss, K.; Rathmell, J.C. Targeting Metabolism to Improve the Tumor Microenvironment for Cancer Immunotherapy. Mol. Cell 2020, 78, 1019–1033. [Google Scholar] [CrossRef] [PubMed]
- Granchi, C. ATP citrate lyase (ACLY) inhibitors: An anti-cancer strategy at the crossroads of glucose and lipid metabolism. Eur. J. Med. Chem. 2018, 157, 1276–1291. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Huang, J.; Jiang, X.; Yuan, Y.; Pang, H.; Luo, S.; Wang, N.; Yao, C.; Lin, Z.; Pu, D.; et al. Regulation of aerobic glycolysis to decelerate tumor proliferation by small molecule inhibitors targeting glucose transporters. Protein Cell 2020, 11, 446–451. [Google Scholar] [CrossRef]
- Walcher, L.; Kistenmacher, A.K.; Suo, H.; Kitte, R.; Dluczek, S.; Strauß, A.; Blaudszun, A.R.; Yevsa, T.; Fricke, S.; Kossatz-Boehlert, U. Cancer Stem Cells-Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Front. Immunol. 2020, 11, 1280. [Google Scholar] [CrossRef]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020, 5, 8. [Google Scholar] [CrossRef]
- Hagerman, P.J.; Hagerman, R. Fragile X syndrome. Curr. Biol. 2021, 31, R273–R275. [Google Scholar] [CrossRef]
- Hu, Y.; Nie, W.; Lyu, L.; Zhang, X.; Wang, W.; Zhang, Y.; He, S.; Guo, A.; Liu, F.; Wang, B.; et al. Tumor-Microenvironment-Activatable Nanoparticle Mediating Immunogene Therapy and M2 Macrophage-Targeted Inhibitor for Synergistic Cancer Immunotherapy. ACS Nano 2024, 18, 3295–3312. [Google Scholar] [CrossRef]
- Lim, Z.F.; Ma, P.C. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J. Hematol. Oncol. 2019, 12, 134. [Google Scholar] [CrossRef]
- Wei, G.; Wang, Y.; Yang, G.; Wang, Y.; Ju, R. Recent progress in nanomedicine for enhanced cancer chemotherapy. Theranostics 2021, 11, 6370–6392. [Google Scholar] [CrossRef]
- Lumaban, J.G.; Nelson, D.L. The Fragile X proteins Fmrp and Fxr2p cooperate to regulate glucose metabolism in mice. Hum. Mol. Genet. 2015, 24, 2175–2184. [Google Scholar] [CrossRef]
- Huang, M.; Liu, M.; Wang, R.; Man, Y.; Zhou, H.; Xu, Z.X.; Wang, Y. The crosstalk between glucose metabolism and telomerase regulation in cancer. Biomed. Pharmacother. 2024, 175, 116643. [Google Scholar] [CrossRef] [PubMed]
- Habbas, K.; Cakil, O.; Zámbó, B.; Tabet, R.; Riet, F.; Dembele, D.; Mandel, J.L.; Hocquemiller, M.; Laufer, R.; Piguet, F.; et al. AAV-delivered diacylglycerol kinase DGKk achieves long-term rescue of fragile X syndrome mouse model. EMBO Mol. Med. 2022, 14, e14649. [Google Scholar] [CrossRef] [PubMed]
- Zafarullah, M.; Palczewski, G.; Rivera, S.M.; Hessl, D.R.; Tassone, F. Metabolic profiling reveals dysregulated lipid metabolism and potential biomarkers associated with the development and progression of Fragile X-Associated Tremor/Ataxia Syndrome (FXTAS). Faseb J. 2020, 34, 16676–16692. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, T.; Han, J. Targeting FMRP: A new window for cancer immunotherapy. MedComm (2020) 2023, 4, e233. [Google Scholar] [CrossRef]
- Wang, N.; Shi, B.; Ding, L.; Zhang, X.; Ma, X.; Guo, S.; Qiao, X.; Wang, L.; Ma, D.; Cao, J. FMRP protects breast cancer cells from ferroptosis by promoting SLC7A11 alternative splicing through interacting with hnRNPM. Redox Biol. 2024, 77, 103382. [Google Scholar] [CrossRef]
- Zhang, D.; Xu, X.; Ye, Q. Metabolism and immunity in breast cancer. Front. Med. 2021, 15, 178–207. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, K.; Zhang, K.; Niu, M.; Chen, Q.; Liu, Y.; Wang, L.; Zhang, N.; Li, W.; Zhong, X.; et al. O-GlcNAcylation promotes topoisomerase IIα catalytic activity in breast cancer chemoresistance. EMBO Rep. 2023, 24, e56458. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X.; Yang, F.; Chen, C.; Liu, P.; Ren, Y.; Sun, P.; Wang, Z.; You, Y.; Zeng, Y.X.; et al. DHHC9-mediated GLUT1 S-palmitoylation promotes glioblastoma glycolysis and tumorigenesis. Nat. Commun. 2021, 12, 5872. [Google Scholar] [CrossRef]
- Guo, D.; Tong, Y.; Jiang, X.; Meng, Y.; Jiang, H.; Du, L.; Wu, Q.; Li, S.; Luo, S.; Li, M.; et al. Aerobic glycolysis promotes tumor immune evasion by hexokinase2-mediated phosphorylation of IκBα. Cell Metab. 2022, 34, 1312–1324.e1316. [Google Scholar] [CrossRef]
- Carotti, S.; Zingariello, M.; Francesconi, M.; D’Andrea, L.; Latasa, M.U.; Colyn, L.; Fernandez-Barrena, M.G.; Flammia, R.S.; Falchi, M.; Righi, D.; et al. Fragile X mental retardation protein in intrahepatic cholangiocarcinoma: Regulating the cancer cell behavior plasticity at the leading edge. Oncogene 2021, 40, 4033–4049. [Google Scholar] [CrossRef]
- Rahnemai-Azar, A.A.; Weisbrod, A.; Dillhoff, M.; Schmidt, C.; Pawlik, T.M. Intrahepatic cholangiocarcinoma: Molecular markers for diagnosis and prognosis. Surg. Oncol. 2017, 26, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Caredda, E.; Pedini, G.; D’Amico, F.; Scioli, M.G.; Pacini, L.; Orsaria, P.; Vanni, G.; Buonomo, O.C.; Orlandi, A.; Bagni, C.; et al. FMRP expression in primary breast tumor cells correlates with recurrence and specific site of metastasis. PLoS ONE 2023, 18, e0287062. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Liu, B.; Wu, B.; Zhou, H.; Wang, X.; Cao, J.; Jiang, M.; Zhou, Y.; Guo, F.; Xue, C.; et al. FMRP regulates STAT3 mRNA localization to cellular protrusions and local translation to promote hepatocellular carcinoma metastasis. Commun. Biol. 2021, 4, 540. [Google Scholar] [CrossRef]
- Zhu, Y.J.; Zheng, B.; Luo, G.J.; Ma, X.K.; Lu, X.Y.; Lin, X.M.; Yang, S.; Zhao, Q.; Wu, T.; Li, Z.X.; et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics 2019, 9, 3526–3540. [Google Scholar] [CrossRef]
- Zhong, C.; Long, Z.; Yang, T.; Wang, S.; Zhong, W.; Hu, F.; Teoh, J.Y.; Lu, J.; Mao, X. M6A-modified circRBM33 promotes prostate cancer progression via PDHA1-mediated mitochondrial respiration regulation and presents a potential target for ARSI therapy. Int. J. Biol. Sci. 2023, 19, 1543–1563. [Google Scholar] [CrossRef]
- Wang, X.; Xu, K.; Liao, X.; Rao, J.; Huang, K.; Gao, J.; Xu, G.; Wang, D. Construction of a survival nomogram for gastric cancer based on the cancer genome atlas of m6A-related genes. Front. Genet. 2022, 13, 936658. [Google Scholar] [CrossRef]
- Yang, J.; Wu, Z.; Wu, X.; Chen, S.; Xia, X.; Zeng, J. Constructing and validating of m6a-related genes prognostic signature for stomach adenocarcinoma and immune infiltration: Potential biomarkers for predicting the overall survival. Front. Oncol. 2022, 12, 1050288. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, R.; Lin, H.; Dou, X.; Zeng, B.; Zhao, X.; Ma, L.; Diarra, D.; Liu, B.; Deng, W.-W.; Wu, T. FMR1: A Neurodevelopmental Factor Regulating Cell Metabolism in the Tumor Microenvironment. Biomolecules 2025, 15, 779. https://doi.org/10.3390/biom15060779
Zhou R, Lin H, Dou X, Zeng B, Zhao X, Ma L, Diarra D, Liu B, Deng W-W, Wu T. FMR1: A Neurodevelopmental Factor Regulating Cell Metabolism in the Tumor Microenvironment. Biomolecules. 2025; 15(6):779. https://doi.org/10.3390/biom15060779
Chicago/Turabian StyleZhou, Renbin, Hao Lin, Xinyu Dou, Bang Zeng, Xinyi Zhao, Lei Ma, Drissa Diarra, Bing Liu, Wei-Wei Deng, and Tianfu Wu. 2025. "FMR1: A Neurodevelopmental Factor Regulating Cell Metabolism in the Tumor Microenvironment" Biomolecules 15, no. 6: 779. https://doi.org/10.3390/biom15060779
APA StyleZhou, R., Lin, H., Dou, X., Zeng, B., Zhao, X., Ma, L., Diarra, D., Liu, B., Deng, W.-W., & Wu, T. (2025). FMR1: A Neurodevelopmental Factor Regulating Cell Metabolism in the Tumor Microenvironment. Biomolecules, 15(6), 779. https://doi.org/10.3390/biom15060779






