High Bendability of Short RNA-DNA Hybrid Duplex Revealed by Single-Molecule Cyclization and Molecular Dynamics Simulations
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA and RDH Preparations
2.2. smFRET Data Acquisition
2.3. smFRET Data Analyses
2.4. DNA and RNA Sequences Used in smFRET Experiments (5′-3′)
2.5. All-Atom Molecular Dynamics Simulations
3. Results
3.1. The Experimental Design of Single-Molecule RDH Duplex Cyclization
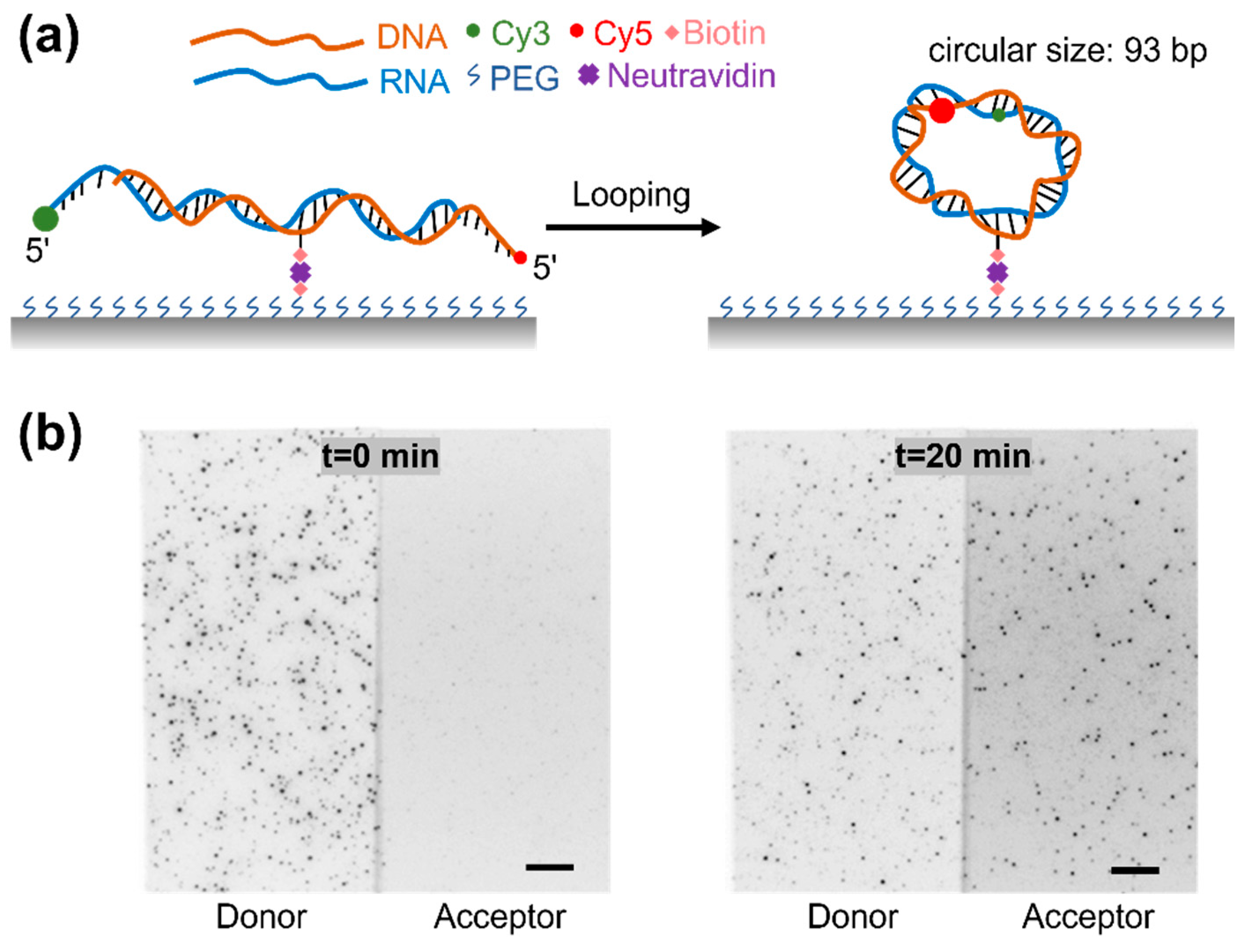
3.2. The High Bendability of Short RDH Duplex

3.3. The Curvature and Structural Fluctuations of an RDH Duplex
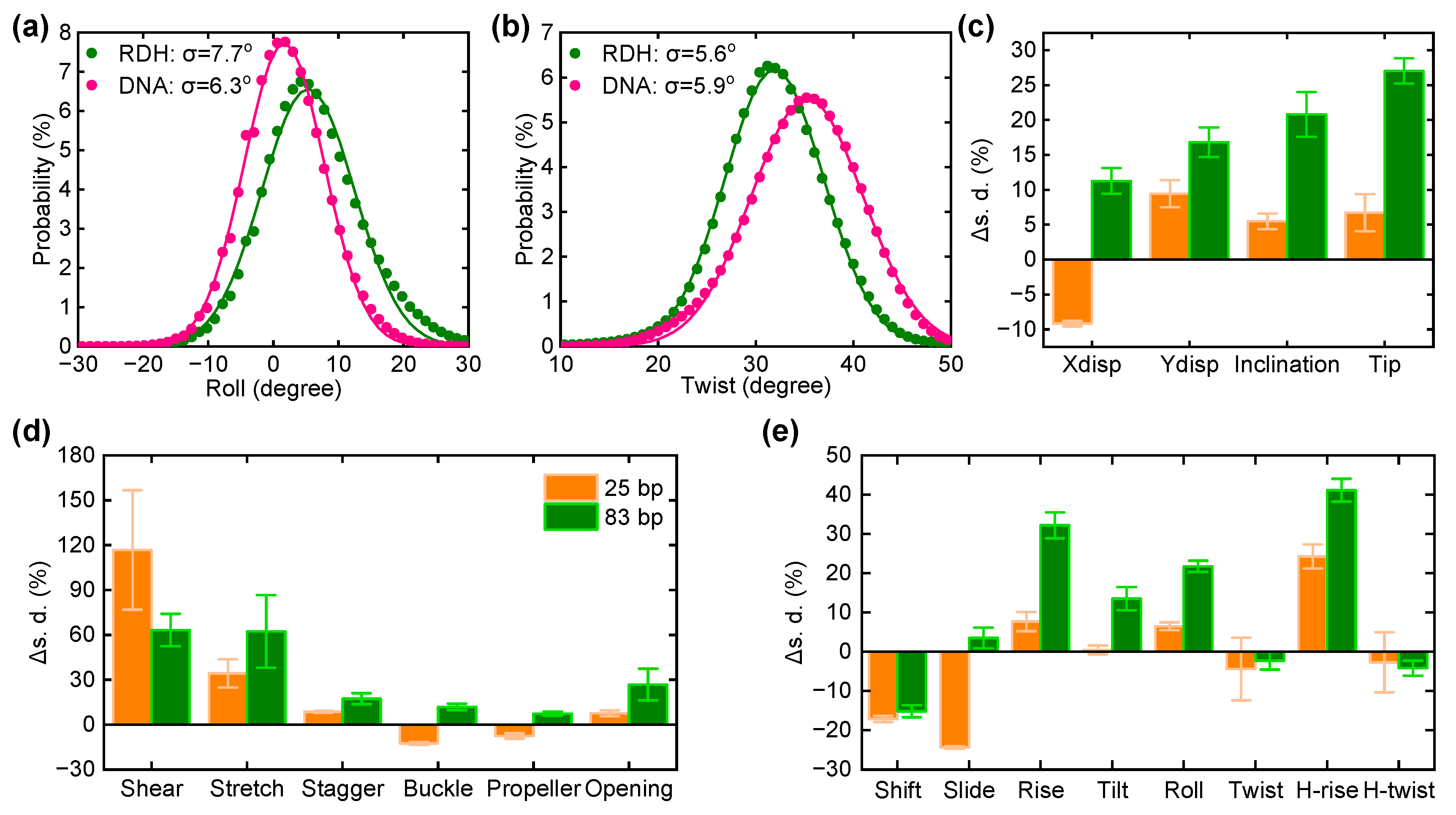
3.4. Kink Release Bending Stress in RDH Duplex

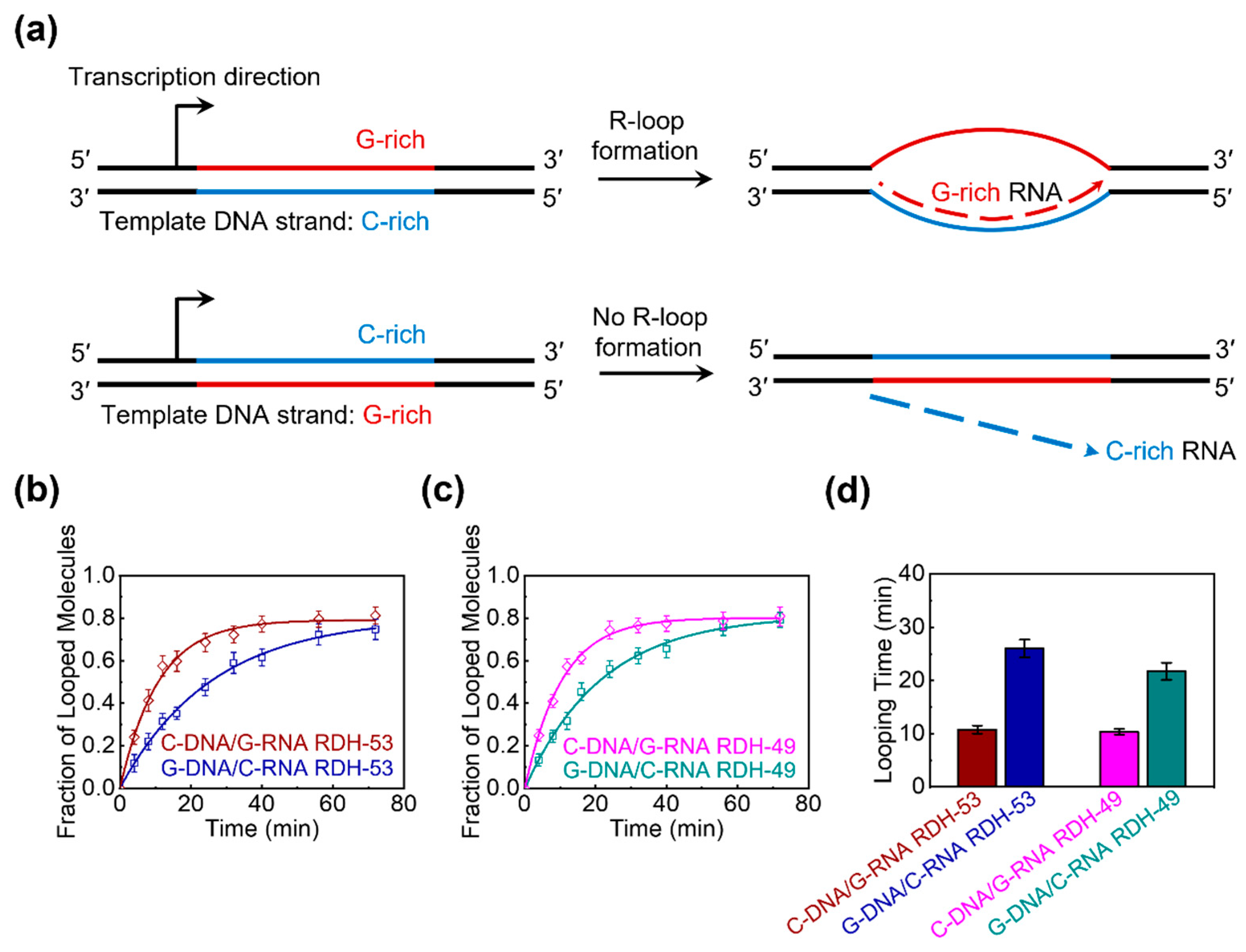
3.5. The Sequence Dependence in the Bendability of the RDH Duplex
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hamperl, S.; Cimprich, K.A. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair 2014, 19, 84–94. [Google Scholar] [CrossRef]
- Kuzminov, A. When DNA Topology Turns Deadly—RNA Polymerases Dig in Their R-Loops to Stand Their Ground: New Positive and Negative (Super) Twists in the Replication-Transcription Conflict. Trends Genet. 2018, 34, 111–120. [Google Scholar] [CrossRef]
- Toubiana, S.; Selig, S. DNA:RNA hybrids at telomeres—When it is better to be out of the (R) loop. FEBS J. 2018, 285, 2552–2566. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.W.; Csorba, T.; Skourti-Stathaki, K.; Proudfoot, N.J.; Dean, C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 2013, 340, 619–621. [Google Scholar] [CrossRef] [PubMed]
- Nakama, M.; Kawakami, K.; Kajitani, T.; Urano, T.; Murakami, Y. DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells 2012, 17, 218–233. [Google Scholar] [CrossRef] [PubMed]
- Ginno, P.A.; Lott, P.L.; Christensen, H.C.; Korf, I.; Chédin, F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 2012, 45, 814–825. [Google Scholar] [CrossRef]
- Hartono, S.R.; Korf, I.F.; Chédin, F. GC skew is a conserved property of unmethylated CpG island promoters across vertebrates. Nucleic Acids Res. 2015, 43, 9729–9741. [Google Scholar] [CrossRef]
- Sanz, L.A.; Hartono, S.R.; Lim, Y.W.; Steyaert, S.; Rajpurkar, A.; Ginno, P.A.; Xu, X.Q.; Chédin, F. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol. Cell 2016, 63, 167–178. [Google Scholar] [CrossRef]
- Wahba, L.; Costantino, L.; Tan, F.J.; Zimmer, A.; Koshland, D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016, 30, 1327–1338. [Google Scholar] [CrossRef]
- Wongsurawat, T.; Jenjaroenpun, P.; Kwoh, C.K.; Kuznetsov, V. Quantitative model of R-loop forming structures reveals a novel level of RNA-DNA interactome complexity. Nucleic Acids Res. 2012, 40, e16. [Google Scholar] [CrossRef]
- Plosky, B.S. The Good and Bad of RNA:DNA Hybrids in Double-Strand Break Repair. Mol. Cell 2016, 64, 643–644. [Google Scholar] [CrossRef]
- Costantino, L.; Koshland, D. The Yin and Yang of R-loop biology. Curr. Opin. Cell Biol. 2015, 34, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; García-Muse, T. R loops: From transcription byproducts to threats to genome stability. Mol. Cell 2012, 46, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Santos-Pereira, J.M.; Aguilera, A. R loops: New modulators of genome dynamics and function. Nat. Rev. Genet. 2015, 16, 583–597. [Google Scholar] [CrossRef]
- Fazzio, T.G. Regulation of chromatin structure and cell fate by R-loops. Transcription 2016, 7, 121–126. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, P.B.; Chen, H.V.; Acharya, D.; Rando, O.J.; Fazzio, T.G. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat. Struct. Mol. Biol. 2015, 22, 999–1007. [Google Scholar] [CrossRef]
- Ohle, C.; Tesorero, R.; Schermann, G.; Dobrev, N.; Sinning, I.; Fischer, T. Transient RNA-DNA Hybrids Are Required for Efficient Double-Strand Break Repair. Cell 2016, 167, 1001–1013. [Google Scholar] [CrossRef]
- Lu, W.T.; Hawley, B.R.; Skalka, G.L.; Baldock, R.A.; Smith, E.M.; Bader, A.S.; Malewicz, M.; Watts, F.Z.; Wilczynska, A.; Bushell, M. Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair. Nat. Commun. 2018, 9, 532. [Google Scholar] [CrossRef]
- García-Muse, T.; Aguilera, A. R Loops: From Physiological to Pathological Roles. Cell 2019, 179, 604–618. [Google Scholar] [CrossRef]
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell 2019, 73, 398–411. [Google Scholar] [CrossRef]
- Richard, P.; Manley, J.L. R Loops and Links to Human Disease. J. Mol. Biol. 2017, 429, 3168–3180. [Google Scholar] [CrossRef]
- Schmidt, M.H.M.; Pearson, C.E. Disease-associated repeat instability and mismatch repair. DNA Repair 2016, 38, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Haeusler, A.R.; Donnelly, C.J.; Rothstein, J.D. The expanding biology of the C9orf72 nucleotide repeat expansion in neurodegenerative disease. Nat. Rev. Neurosci. 2016, 17, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Groh, M.; Gromak, N. Out of balance: R-loops in human disease. PLoS Genet. 2014, 10, e1004630. [Google Scholar] [CrossRef] [PubMed]
- Perego, M.G.L.; Taiana, M.; Bresolin, N.; Comi, G.P.; Corti, S. R-Loops in Motor Neuron Diseases. Mol. Neurobiol. 2019, 56, 2579–2589. [Google Scholar] [CrossRef]
- Cui, C.; Shu, W.; Li, P. Fluorescence In situ Hybridization: Cell-Based Genetic Diagnostic and Research Applications. Front. Cell Dev. Biol. 2016, 4, 89. [Google Scholar] [CrossRef]
- Bisaria, N.; Jarmoskaite, I.; Herschlag, D. Lessons from Enzyme Kinetics Reveal Specificity Principles for RNA-Guided Nucleases in RNA Interference and CRISPR-Based Genome Editing. Cell Syst. 2017, 4, 21–29. [Google Scholar] [CrossRef]
- Wright, A.V.; Nuñez, J.K.; Doudna, J.A. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 2016, 164, 29–44. [Google Scholar] [CrossRef]
- Ko, S.H.; Su, M.; Zhang, C.A.; Ribbe, A.E.; Jiang, W.; Mao, C.D. Synergistic self-assembly of RNA and DNA molecules. Nat. Chem. 2010, 2, 1050–1055. [Google Scholar] [CrossRef]
- Janssen, H.L.A.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef]
- Xiong, Y.; Sundaralingam, M. Crystal structure of a DNA.RNA hybrid duplex with a polypurine RNA r(gaagaagag) and a complementary polypyrimidine DNA d(CTCTTCTTC). Nucleic Acids Res. 2000, 28, 2171–2176. [Google Scholar] [CrossRef] [PubMed]
- Cheatham, T.E.; Kollman, P.A. Molecular Dynamics Simulations Highlight the Structural Differences among DNA:DNA, RNA:RNA, and DNA:RNA Hybrid Duplexes. J. Am. Chem. Soc. 1997, 119, 4805–4825. [Google Scholar] [CrossRef]
- Lane, A.N.; Ebel, S.; Brown, T. NMR assignments and solution conformation of the DNA.RNA hybrid duplex d(GTGAACTT).r(AAGUUCAC). Eur. J. Biochem. 1993, 215, 297–306. [Google Scholar] [CrossRef]
- Gyi, J.I.; Lane, A.N.; Conn, G.L.; Brown, T. Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: Comparison with the homologous DNA and RNA duplexes. Biochemistry 1998, 37, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Gyi, J.I.; Conn, G.L.; Lane, A.N.; Brown, T. Comparison of the thermodynamic stabilities and solution conformations of DNA.RNA hybrids containing purine-rich and pyrimidine-rich strands with DNA and RNA duplexes. Biochemistry 1996, 35, 12538–12548. [Google Scholar] [CrossRef]
- Noy, A.; Pérez, A.; Márquez, M.; Luque, F.J.; Orozco, M. Structure, recognition properties, and flexibility of the DNA.RNA hybrid. J. Am. Chem. Soc. 2005, 127, 4910–4920. [Google Scholar] [CrossRef]
- Hantz, E.; Larue, V.; Ladam, P.; Le Moyec, L.; Gouyette, C.; Dinh, T.H. Solution conformation of an RNA--DNA hybrid duplex containing a pyrimidine RNA strand and a purine DNA strand. Int. J. Biol. Macromol. 2001, 28, 273–284. [Google Scholar] [CrossRef]
- Ratmeyer, L.; Vinayak, R.; Zhong, Y.Y.; Zon, G.; Wilson, W.D. Sequence specific thermodynamic and structural properties for DNA.RNA duplexes. Biochemistry 1994, 33, 5298–5304. [Google Scholar] [CrossRef]
- Roberts, R.W.; Crothers, D.M. Stability and properties of double and triple helices: Dramatic effects of RNA or DNA backbone composition. Science 1992, 258, 1463–1466. [Google Scholar] [CrossRef]
- Lesnik, E.A.; Freier, S.M. Relative thermodynamic stability of DNA, RNA, and DNA:RNA hybrid duplexes: Relationship with base composition and structure. Biochemistry 1995, 34, 10807–10815. [Google Scholar] [CrossRef]
- Suresh, G.; Priyakumar, U.D. DNA-RNA hybrid duplexes with decreasing pyrimidine content in the DNA strand provide structural snapshots for the A- to B-form conformational transition of nucleic acids. Phys. Chem. Chem. Phys. 2014, 16, 18148–18155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Fu, H.; Yang, Y.; Zhou, E.; Tan, Z.; You, H.; Zhang, X. The Mechanical Properties of RNA-DNA Hybrid Duplex Stretched by Magnetic Tweezers. Biophys. J. 2019, 116, 196–204. [Google Scholar] [CrossRef]
- Yang, D.; Liu, W.; Deng, X.; Xie, W.; Chen, H.; Zhong, Z.; Ma, J. GC-Content Dependence of Elastic and Overstretching Properties of DNA:RNA Hybrid Duplexes. Biophys. J. 2020, 119, 852–861. [Google Scholar] [CrossRef]
- Liu, J.; Xi, K.; Zhang, X.; Bao, L.; Zhang, X.; Tan, Z. Structural Flexibility of DNA-RNA Hybrid Duplex: Stretching and Twist-Stretch Coupling. Biophys. J. 2019, 117, 74–86. [Google Scholar] [CrossRef]
- Hansen, T.M.; Reihani, S.N.S.; Oddershede, L.B.; Sorensen, M.A. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc. Natl. Acad. Sci. USA 2007, 104, 5830–5835. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.H.; Wen, J.D.; Lancaster, L.; Noller, H.F.; Bustamante, C.; Tinoco, I. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 2011, 475, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Bercy, M.; Bockelmann, U. Hairpins under tension: RNA versus DNA. Nucleic Acids Res. 2015, 43, 9928–9936. [Google Scholar] [CrossRef]
- Wen, J.D.; Lancaster, L.; Hodges, C.; Zeri, A.C.; Yoshimura, S.H.; Noller, H.F.; Bustamante, C.; Tinoco, I. Following translation by single ribosomes one codon at a time. Nature 2008, 452, 598–603. [Google Scholar] [CrossRef]
- Jiang, F.G.; Taylor, D.W.; Chen, J.S.; Kornfeld, J.E.; Zhou, K.H.; Thompson, A.J.; Nogales, E.; Doudna, J.A. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 2016, 351, 867–871. [Google Scholar] [CrossRef]
- Davis, R.R.; Shaban, N.M.; Perrino, F.W.; Hollis, T. Crystal structure of RNA-DNA duplex provides insight into conformational changes induced by RNase H binding. Cell Cycle 2015, 14, 668–673. [Google Scholar] [CrossRef]
- Gotte, M.; Fackler, S.; Hermann, T.; Perola, E.; Cellai, L.; Gross, H.J.; Grice, S.F.L.; Heumann, H. HIV-1 reverse transcriptase-associated RNase H cleaves RNA/RNA in arrested complexes: Implications for the mechanism by which RNase H discriminates between RNA/RNA and RNA/DNA. EMBO J. 1995, 14, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Hansen, U.M.; McClure, W.R. Role of the sigma subunit of Escherichia coli RNA polymerase in initiation. II. Release of sigma from ternary complexes. J. Biol. Chem. 1980, 255, 9564–9570. [Google Scholar] [CrossRef] [PubMed]
- Dohnalová, H.; Matousková, E.; Lankas, F. Temperature-dependent elasticity of DNA, RNA, and hybrid double helices. Biophys. J. 2024, 123, 572–583. [Google Scholar] [CrossRef]
- Burkhoff, A.M.; Tullius, T.D. Structural details of an adenine tract that does not cause DNA to bend. Nature 1988, 331, 455–457. [Google Scholar] [CrossRef]
- Cherstvy, A.G. DNA cyclization: Suppression or enhancement by electrostatic repulsions? J. Phys. Chem. B 2011, 115, 4286–4294. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.J.; Olson, W.K. 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003, 31, 5108–5121. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Zgarbová, M.; Šponer, J.; Jurečka, P. Z-DNA as a Touchstone for Additive Empirical Force Fields and a Refinement of the Alpha/Gamma DNA Torsions for AMBER. J. Chem. Theory Comput. 2021, 17, 6292–6301. [Google Scholar] [CrossRef]
- Zgarbová, M.; Otyepka, M.; Sponer, J.; Mládek, A.; Banás, P.; Cheatham, T.E.; Jurecka, P. Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J. Chem. Theory Comput. 2011, 7, 2886–2902. [Google Scholar] [CrossRef]
- Joung, I.S.; Cheatham, T.E., III. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B 2008, 112, 9020–9041. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Roy, R.; Hohng, S.; Ha, T. A practical guide to single-molecule FRET. Nat. Methods 2008, 5, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Vafabakhsh, R.; Ha, T. Extreme bendability of DNA less than 100 base pairs long revealed by single-molecule cyclization. Science 2012, 337, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Ngo, T.T.M.; Yoo, J.; Dai, Q.; Zhang, Q.; He, C.; Aksimentiev, A.; Ha, T. Effects of cytosine modifications on DNA flexibility and nucleosome mechanical stability. Nat. Commun. 2016, 7, 10813. [Google Scholar] [CrossRef]
- Ngo, T.T.M.; Zhang, Q.C.; Zhou, R.B.; Yodh, J.G.; Ha, T. Asymmetric unwrapping of nucleosomes under tension directed by DNA local flexibility. Cell 2015, 160, 1135–1144. [Google Scholar] [CrossRef]
- Lavery, R.; Moakher, M.; Maddocks, J.H.; Petkeviciute, D.; Zakrzewska, K. Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res. 2009, 37, 5917–5929. [Google Scholar] [CrossRef]
- Czapla, L.; Swigon, D.; Olson, W.K. Sequence-Dependent Effects in the Cyclization of Short DNA. J. Chem. Theory Comput. 2006, 2, 685–695. [Google Scholar] [CrossRef]
- Lankaš, F.; Lavery, R.; Maddocks, J.H. Kinking occurs during molecular dynamics simulations of small DNA minicircles. Structure 2006, 14, 1527–1534. [Google Scholar] [CrossRef]
- Mitchell, J.S.; Laughton, C.A.; Harris, S.A. Atomistic simulations reveal bubbles, kinks and wrinkles in supercoiled DNA. Nucleic Acids Res. 2011, 39, 3928–3938. [Google Scholar] [CrossRef] [PubMed]
- Filip, M.R.; Giustino, F. The geometric blueprint of perovskites. Proc. Natl. Acad. Sci. USA 2018, 115, 5397–5402. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Kotlyar, A.; Vologodskii, A. Kinking the double helix by bending deformation. Nucleic Acids Res. 2008, 36, 1120–1128. [Google Scholar] [CrossRef] [PubMed]
- Curuksu, J.D. Spectral analysis of DNA superhelical dynamics from molecular minicircle simulations. J. Chem. Phys. 2023, 159, 105101. [Google Scholar] [CrossRef]
- Serrano-Pozo, A.; Das, S.; Hyman, B.T. APOE and Alzheimer’s disease: Advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 2021, 20, 68–80. [Google Scholar] [CrossRef]
- O’Neill, M.A.; Barton, J.K. 2-Aminopurine: A probe of structural dynamics and charge transfer in DNA and DNA:RNA hybrids. J. Am. Chem. Soc. 2002, 124, 13053–13066. [Google Scholar] [CrossRef]
- Priyakumar, U.D.; MacKerell, A.D. Atomic detail investigation of the structure and dynamics of DNA.RNA hybrids: A molecular dynamics study. J. Phys. Chem. B 2008, 112, 1515–1524. [Google Scholar] [CrossRef]
- Coslovich, D.; Roland, C.M. Thermodynamic scaling of diffusion in supercooled Lennard-Jones liquids. J. Phys. Chem. B 2008, 112, 1329–1332. [Google Scholar] [CrossRef]
- Zgarbová, M.; Sponer, J.; Jurecka, P. Refinement of the Sugar Puckering Torsion Potential in the AMBER DNA Force Field. J. Chem. Theory Comput. 2025, 21, 833–846. [Google Scholar] [CrossRef]
- Herrero-Galán, E.; Fuentes-Perez, M.E.; Carrasco, C.; Valpuesta, J.M.; Carrascosa, J.L.; Moreno-Herrero, F.; Arias-Gonzalez, J.R. Mechanical identities of RNA and DNA double helices unveiled at the single-molecule level. J. Am. Chem. Soc. 2013, 135, 122–131. [Google Scholar] [CrossRef]
- Bao, L.; Zhang, X.; Jin, L.; Tan, Z. Flexibility of nucleic acids: From DNA to RNA. Chinese Phys. B 2016, 25, 018703. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, B.; Tian, F.; Yang, Y.; Dai, L.; Zhang, X. High Bendability of Short RNA-DNA Hybrid Duplex Revealed by Single-Molecule Cyclization and Molecular Dynamics Simulations. Biomolecules 2025, 15, 724. https://doi.org/10.3390/biom15050724
Wu B, Tian F, Yang Y, Dai L, Zhang X. High Bendability of Short RNA-DNA Hybrid Duplex Revealed by Single-Molecule Cyclization and Molecular Dynamics Simulations. Biomolecules. 2025; 15(5):724. https://doi.org/10.3390/biom15050724
Chicago/Turabian StyleWu, Bin, Fujia Tian, Yajun Yang, Liang Dai, and Xinghua Zhang. 2025. "High Bendability of Short RNA-DNA Hybrid Duplex Revealed by Single-Molecule Cyclization and Molecular Dynamics Simulations" Biomolecules 15, no. 5: 724. https://doi.org/10.3390/biom15050724
APA StyleWu, B., Tian, F., Yang, Y., Dai, L., & Zhang, X. (2025). High Bendability of Short RNA-DNA Hybrid Duplex Revealed by Single-Molecule Cyclization and Molecular Dynamics Simulations. Biomolecules, 15(5), 724. https://doi.org/10.3390/biom15050724




