Cell-Penetrating Peptide Based on Myosin Phosphatase Target Subunit Sequence Mediates Myosin Phosphatase Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Peptides and Proteins
2.2. Phosphatase Assays
2.3. Streptavidin Sepharose Affinity Chromatography
2.4. Surface Plasmon Resonance Binding Experiments
2.5. Cell Cultures, Treatments and Transfections
2.6. Immunofluorescent Staining
2.7. Pull-Down Assays
2.8. Statistical Analysis
3. Results and Discussion
3.1. Design of the PP1c-MYPT1 Disrupting Peptide
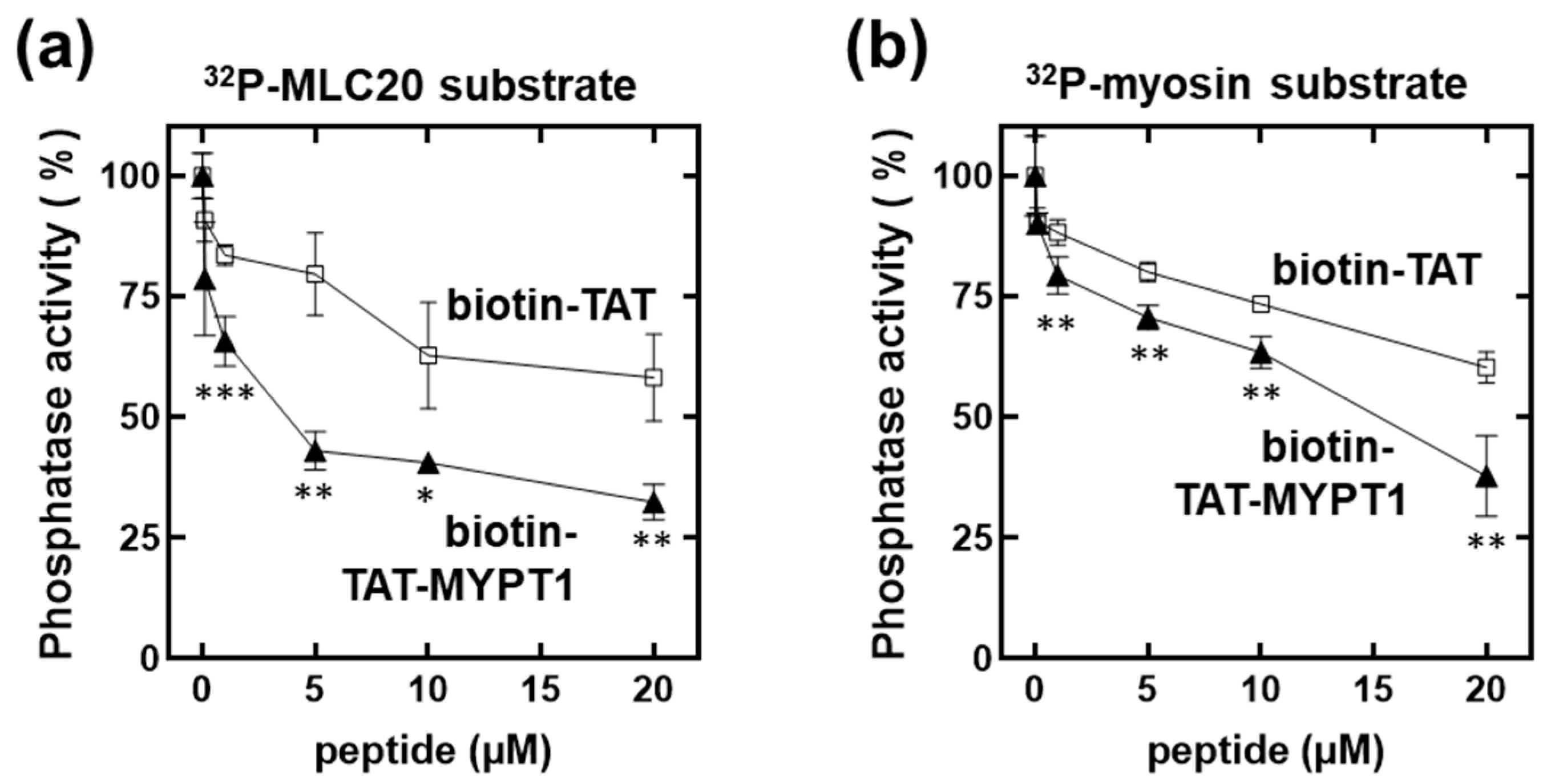
3.2. Effect of Biotin-TAT and Biotin-TAT-MYPT1 on the Activity of Myosin Phosphatase Holoenzyme
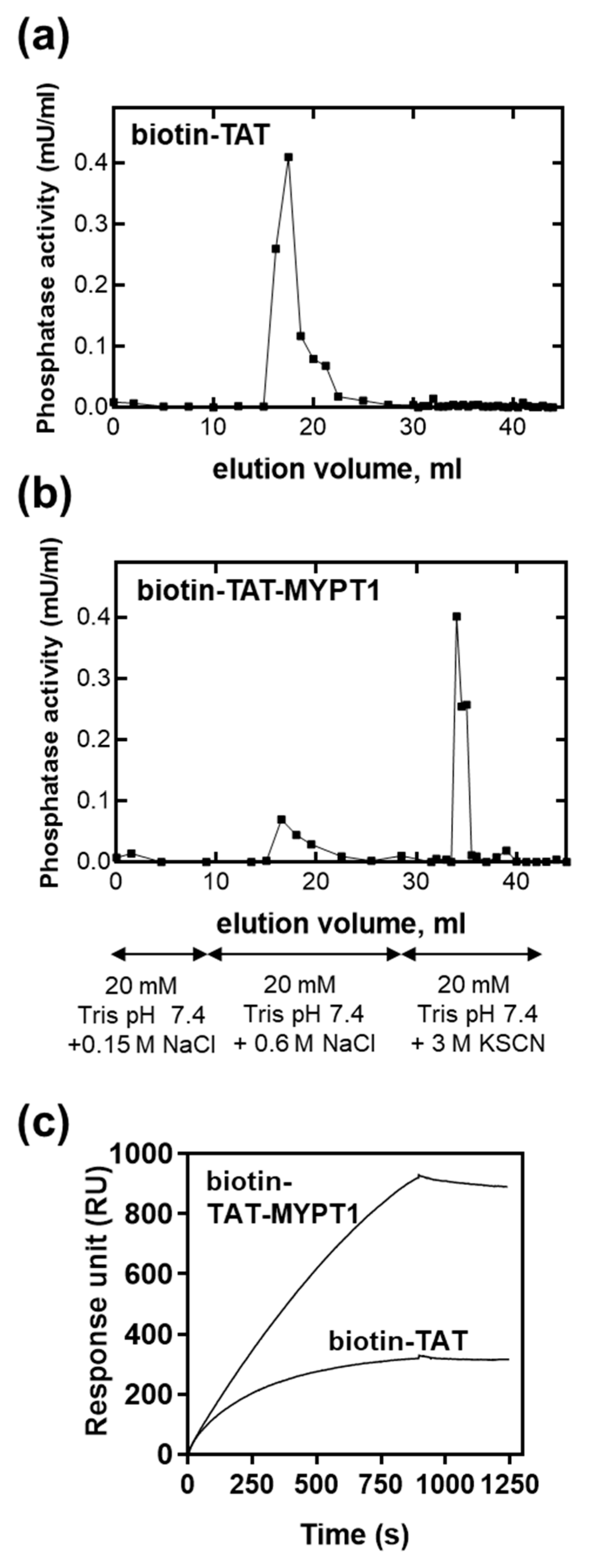
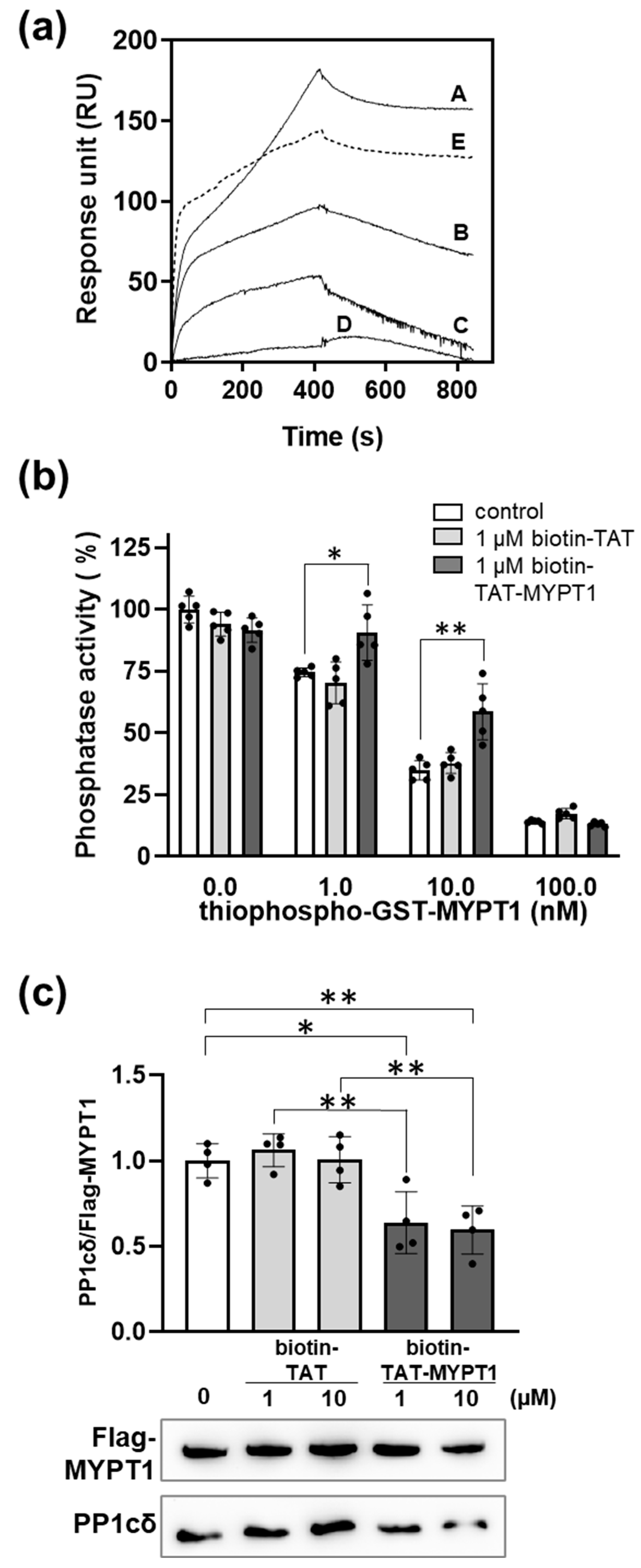
3.3. Binding of Biotin-TAT and Biotin-TAT-MYPT1 to PP1c
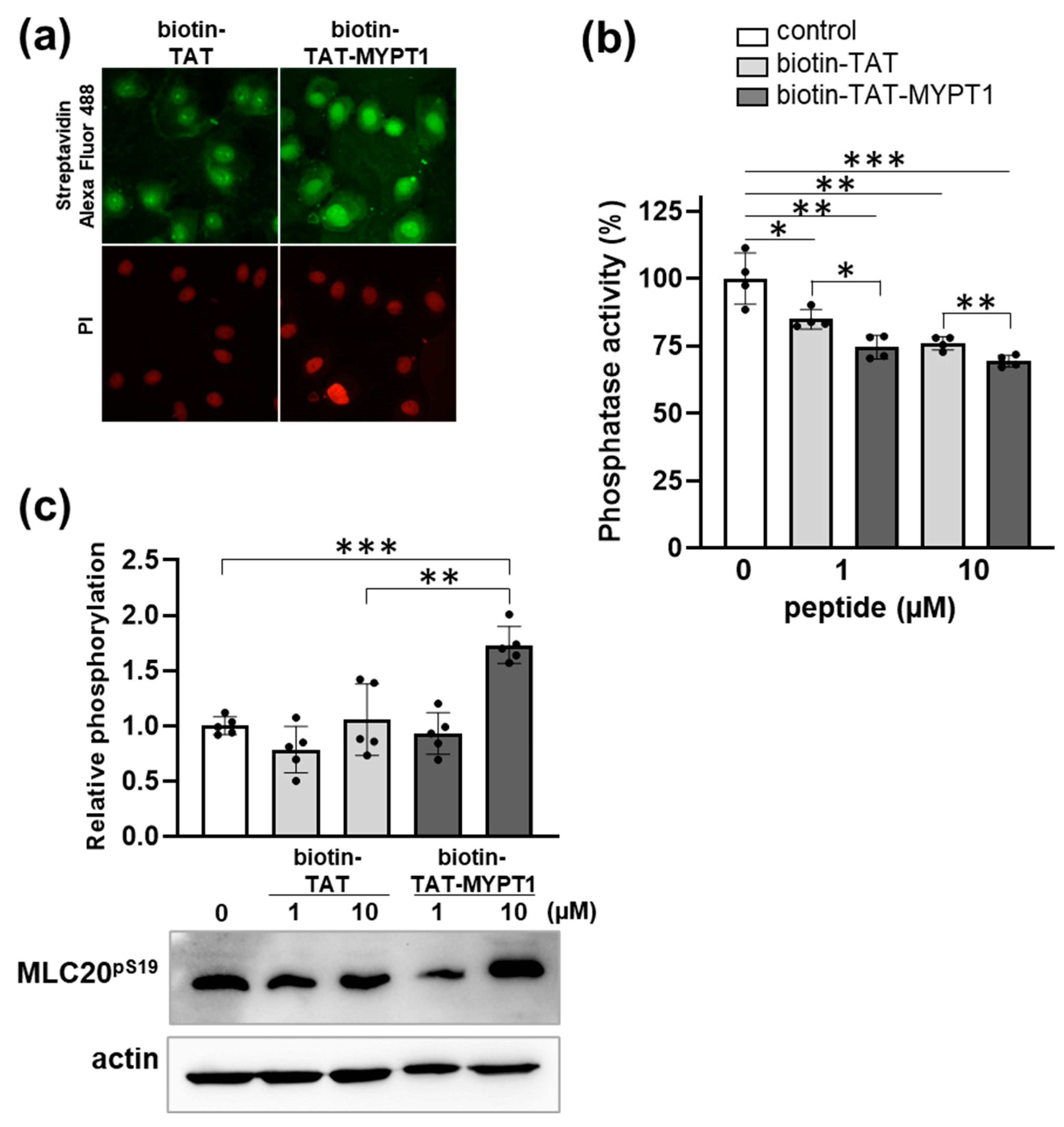
3.4. Effect of Biotin-TAT-MYPT1 on the Phosphatase Activity in A7r5 Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| biotin-TAT | biotinylated TAT peptide |
| biotin-TAT-MYPT1 | biotinylated TAT peptide coupled with MYPT132–58 fragment |
| MLC20 | 20 kDa light chain of myosin |
| MP | myosin phosphatase |
| MYPT1 | myosin phosphatase target subunit-1 |
| PP1 | protein phosphatase-1 |
| PP1c | PP1 catalytic subunit |
| SPR | surface plasmon resonance |
| TAT | HIV-derived cell-penetrating peptide (transactivating transcriptional activator) |
References
- Somlyo, A.P.; Somlyo, A.V. Ca2+ sensitivity of smooth muscle and nonmuscle myosin II: Modulated by G proteins, kinases, and myosin phosphatase. Physiol. Rev. 2003, 83, 1325–1358. [Google Scholar] [CrossRef] [PubMed]
- Takai, A.; Eto, M.; Hirano, K.; Takeya, K.; Wakimoto, T.; Watanabe, M. Protein phosphatases 1 and 2A and their naturally occurring inhibitors: Current topics in smooth muscle physiology and chemical biology. J. Physiol. Sci. 2018, 68, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Alessi, D.; MacDougall, L.K.; Sola, M.M.; Ikebe, M.; Cohen, P. The control of protein phosphatase-1 by targetting subunits. The major myosin phosphatase in avian smooth muscle is a novel form of protein phosphatase-1. Eur. J. Biochem. 1992, 210, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Hartshorne, D.J.; Ito, M.; Erdődi, F. Role of protein phosphatase type 1 in contractile functions: Myosin phosphatase. J. Biol. Chem. 2004, 279, 37211–37214. [Google Scholar] [CrossRef]
- Langsetmo, K.; Stafford, W.F., 3rd; Mabuchi, K.; Tao, T. Recombinant small subunit of smooth muscle myosin light chain phosphatase. Molecular properties and interactions with the targeting subunit. J. Biol. Chem. 2001, 276, 34318–34322. [Google Scholar] [CrossRef]
- Tóth, A.; Kiss, E.; Herberg, F.W.; Gergely, P.; Hartshorne, D.J.; Erdődi, F. Study of the subunit interactions in myosin phosphatase by surface plasmon resonance. Eur. J. Biochem. 2000, 267, 1687–1697. [Google Scholar] [CrossRef]
- Johnson, D.F.; Moorhead, G.; Caudwell, F.B.; Cohen, P.; Chen, Y.H.; Chen, M.X.; Cohen, P.T. Identification of protein-phosphatase-1-binding domains on the glycogen and myofibrillar targetting subunits. Eur. J. Biochem. 1996, 239, 317–325. [Google Scholar] [CrossRef]
- Murányi, A.; Derkach, D.; Erdődi, F.; Kiss, A.; Ito, M.; Hartshorne, D.J. Phosphorylation of Thr695 and Thr850 on the myosin phosphatase target subunit: Inhibitory effects and occurrence in A7r5 cells. FEBS Lett. 2005, 579, 6611–6615. [Google Scholar] [CrossRef]
- Eto, M.; Kitazawa, T.; Matsuzawa, F.; Aikawa, S.; Kirkbride, J.A.; Isozumi, N.; Nishimura, Y.; Brautigan, D.L.; Ohki, S.Y. Phosphorylation-induced conformational switching of CPI-17 produces a potent myosin phosphatase inhibitor. Structure 2007, 15, 1591–1602. [Google Scholar] [CrossRef]
- Bollen, M.; Peti, W.; Ragusa, M.J.; Beullens, M. The extended PP1 toolkit: Designed to create specificity. Trends Biochem. Sci. 2010, 35, 450–458. [Google Scholar] [CrossRef]
- Chatterjee, J.; Beullens, M.; Sukackaite, R.; Qian, J.; Lesage, B.; Hart, D.J.; Bollen, M.; Kohn, M. Development of a peptide that selectively activates protein phosphatase-1 in living cells. Angew. Chem. Int. Ed. Engl. 2012, 51, 10054–10059. [Google Scholar] [CrossRef] [PubMed]
- Kobrock, A.; Matos, B.; Patricio, D.; Grenho, L.; Howl, J.; Fardilha, M.; Gomes, P.S. Enhancing Dental Pulp Stem Cell Proliferation and Odontogenic Differentiation with Protein Phosphatase 1-Disrupting Peptide: An In Vitro Study. Cells 2024, 13, 1143. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.V.; Freitas, M.J.; Santiago, J.; Jones, S.; Guimaraes, S.; Vijayaraghavan, S.; Publicover, S.; Colombo, G.; Howl, J.; Fardilha, M. Disruption of protein phosphatase 1 complexes with the use of bioportides as a novel approach to target sperm motility. Fertil. Steril. 2021, 115, 348–362. [Google Scholar] [CrossRef] [PubMed]
- Hoermann, B.; Durr, E.M.; Ludwig, C.; Ercan, M.; Kohn, M. A strategy to disentangle direct and indirect effects on (de)phosphorylation by chemical modulators of the phosphatase PP1 in complex cellular contexts. Chem. Sci. 2024, 15, 2792–2804. [Google Scholar] [CrossRef]
- Terrak, M.; Kerff, F.; Langsetmo, K.; Tao, T.; Dominguez, R. Structural basis of protein phosphatase 1 regulation. Nature 2004, 429, 780–784. [Google Scholar] [CrossRef]
- Ikebe, M.; Hartshorne, D.J. Effects of Ca2+ on the conformation and enzymatic activity of smooth muscle myosin. J. Biol. Chem. 1985, 260, 13146–13153. [Google Scholar] [CrossRef]
- Shimizu, H.; Ito, M.; Miyahara, M.; Ichikawa, K.; Okubo, S.; Konishi, T.; Naka, M.; Tanaka, T.; Hirano, K.; Hartshorne, D.J.; et al. Characterization of the myosin-binding subunit of smooth muscle myosin phosphatase. J. Biol. Chem. 1994, 269, 30407–30411. [Google Scholar] [CrossRef]
- Kiss, E.; Murányi, A.; Csortos, C.; Gergely, P.; Ito, M.; Hartshorne, D.J.; Erdődi, F. Integrin-linked kinase phosphorylates the myosin phosphatase target subunit at the inhibitory site in platelet cytoskeleton. Biochem. J. 2002, 365, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Erdődi, F.; Csortos, C.; Bot, G.; Gergely, P. Effects of acidic and basic macromolecules on the activity of protein phosphatase-1. Biochim. Biophys. Acta 1985, 827, 23–29. [Google Scholar] [CrossRef]
- Pelech, S.; Cohen, P. The protein phosphatases involved in cellular regulation. 1. Modulation of protein phosphatases-1 and 2A by histone H1, protamine, polylysine and heparin. Eur. J. Biochem. 1985, 148, 245–251. [Google Scholar] [CrossRef]
- Bechara, C.; Sagan, S. Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 2013, 587, 1693–1702. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, J.; Ito, M.; Feng, J.H.; Ichikawa, K.; Hamaguchi, T.; Nakamura, M.; Hartshorne, D.J.; Nakano, T. Interaction of myosin phosphatase target subunit 1 with the catalytic subunit of type 1 protein phosphatase. Biochemistry 1998, 37, 16697–16703. [Google Scholar] [CrossRef] [PubMed]
- Taverner, A.; Almansour, K.; Gridley, K.; Marques, A.R.L.; MacKay, J.; Eggleston, I.M.; Mrsny, R.J. Structure-function analysis of tight junction-directed permeation enhancer PIP250. J. Control. Release 2023, 364, 357–370. [Google Scholar] [CrossRef]
- Kiss, A.; Erdődi, F.; Lontay, B. Myosin phosphatase: Unexpected functions of a long-known enzyme. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 2–15. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiss, A.; Mahfood, M.; Bodogán, Z.; Kónya, Z.; Bécsi, B.; Erdődi, F. Cell-Penetrating Peptide Based on Myosin Phosphatase Target Subunit Sequence Mediates Myosin Phosphatase Activity. Biomolecules 2025, 15, 705. https://doi.org/10.3390/biom15050705
Kiss A, Mahfood M, Bodogán Z, Kónya Z, Bécsi B, Erdődi F. Cell-Penetrating Peptide Based on Myosin Phosphatase Target Subunit Sequence Mediates Myosin Phosphatase Activity. Biomolecules. 2025; 15(5):705. https://doi.org/10.3390/biom15050705
Chicago/Turabian StyleKiss, Andrea, Mohamad Mahfood, Zsófia Bodogán, Zoltán Kónya, Bálint Bécsi, and Ferenc Erdődi. 2025. "Cell-Penetrating Peptide Based on Myosin Phosphatase Target Subunit Sequence Mediates Myosin Phosphatase Activity" Biomolecules 15, no. 5: 705. https://doi.org/10.3390/biom15050705
APA StyleKiss, A., Mahfood, M., Bodogán, Z., Kónya, Z., Bécsi, B., & Erdődi, F. (2025). Cell-Penetrating Peptide Based on Myosin Phosphatase Target Subunit Sequence Mediates Myosin Phosphatase Activity. Biomolecules, 15(5), 705. https://doi.org/10.3390/biom15050705





