Scanning Tunneling Microscope Measurement of Proteasome Conductance
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression and Purification of Proteasomes
2.2. Preparation of Unfolded Pleiotrophin
2.3. Proteasome Activity Assays
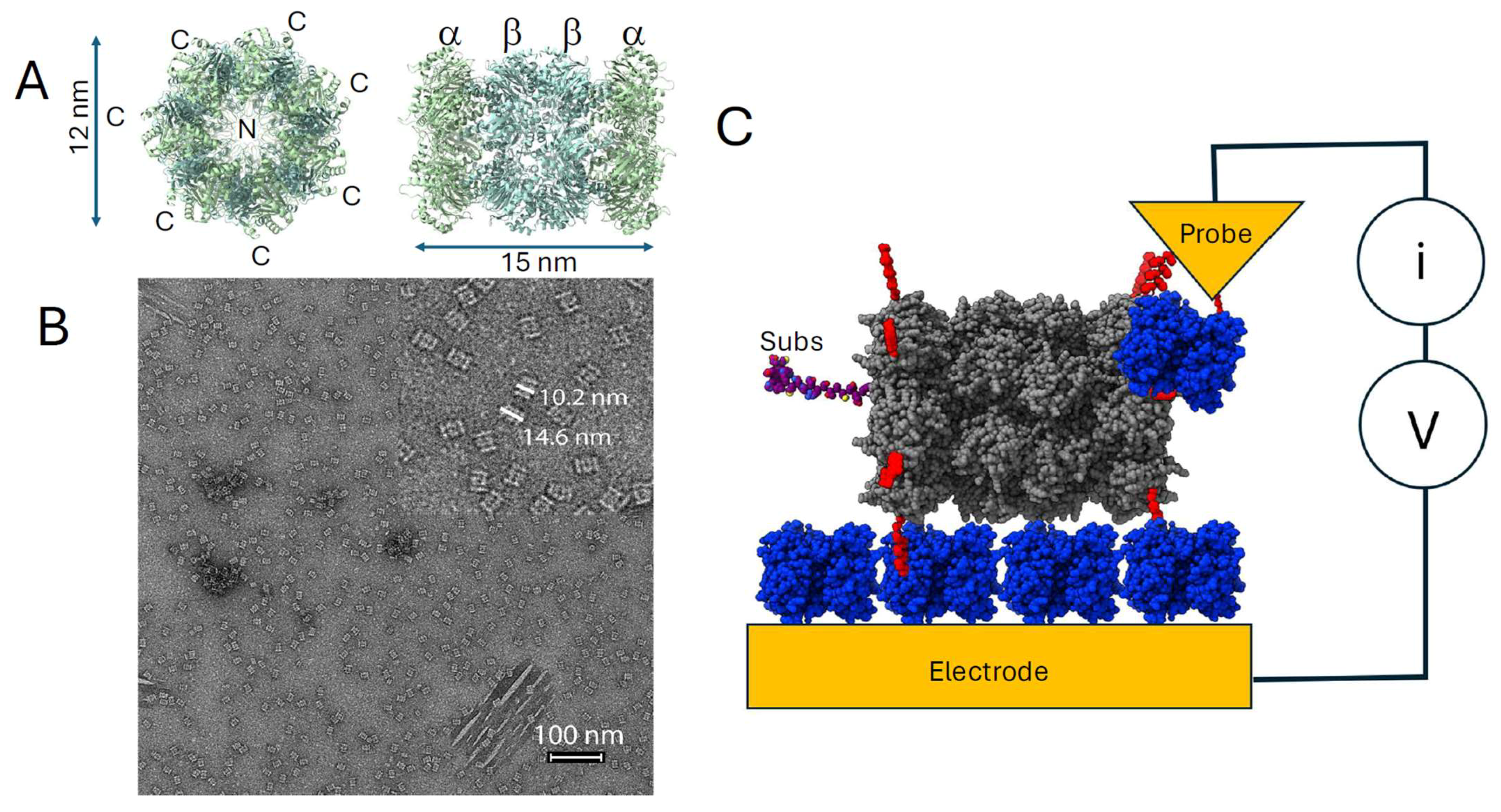
2.4. Preparation of Samples for STM
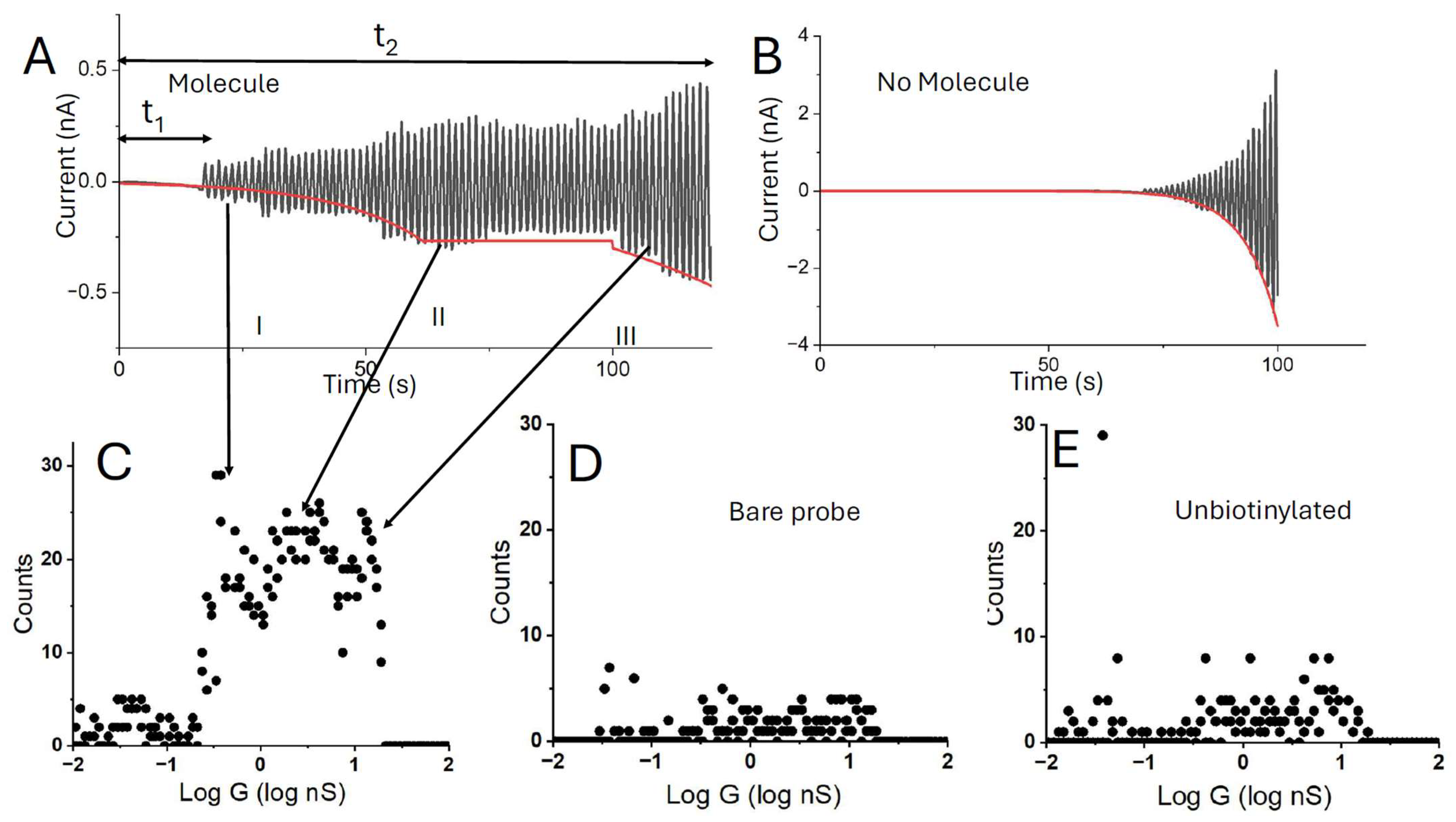
2.5. Collection of STM Data
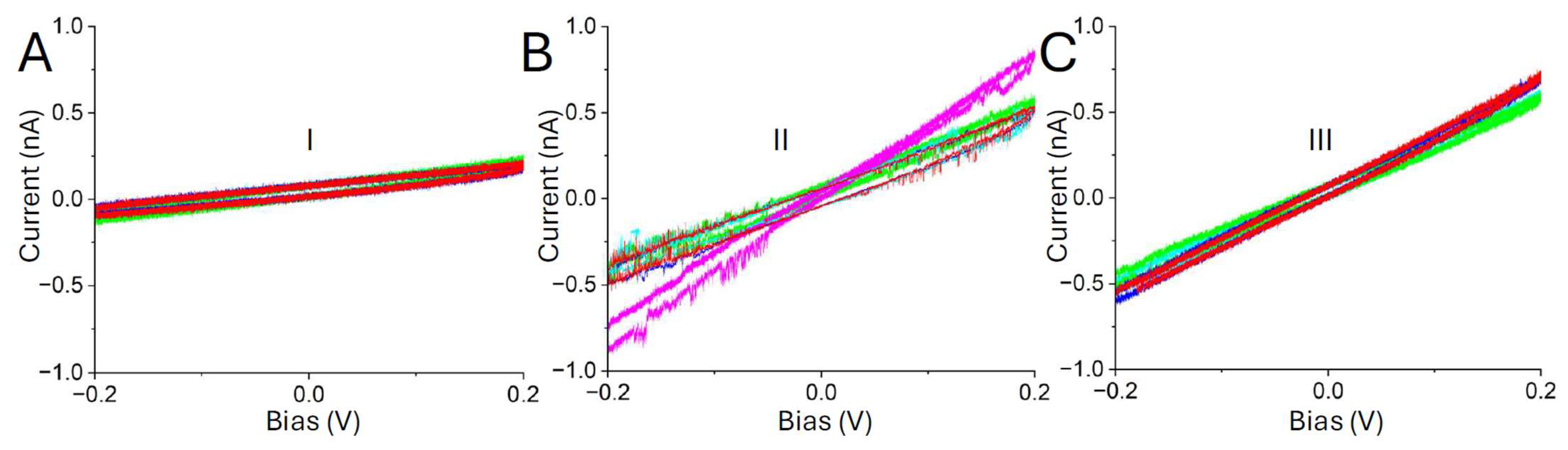
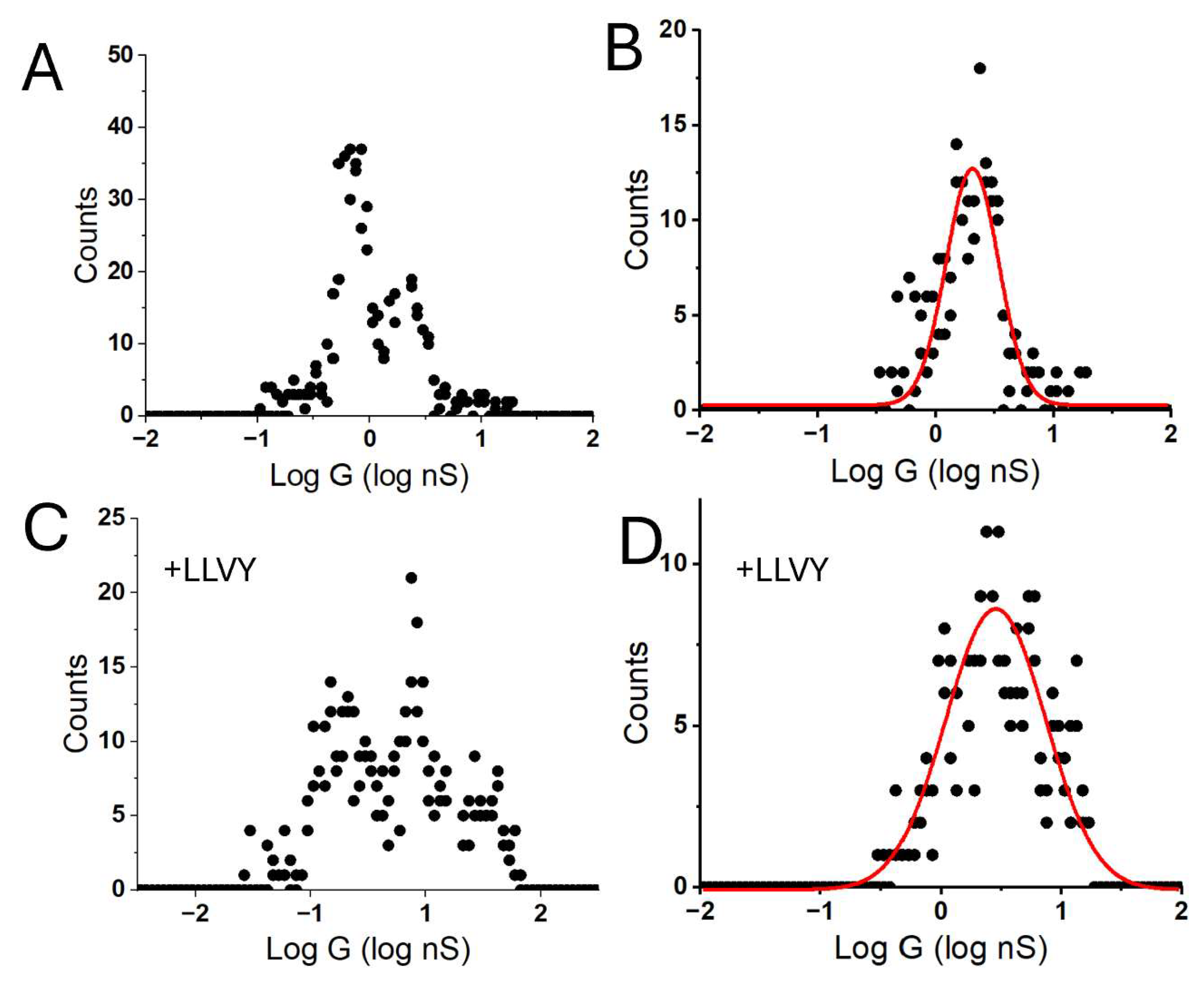
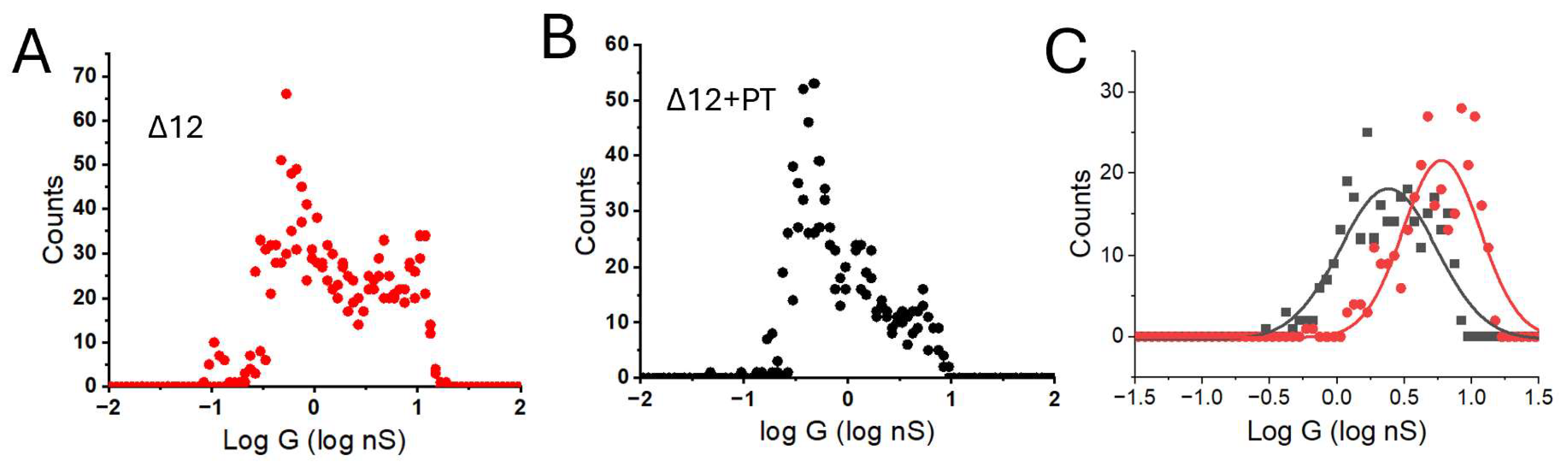
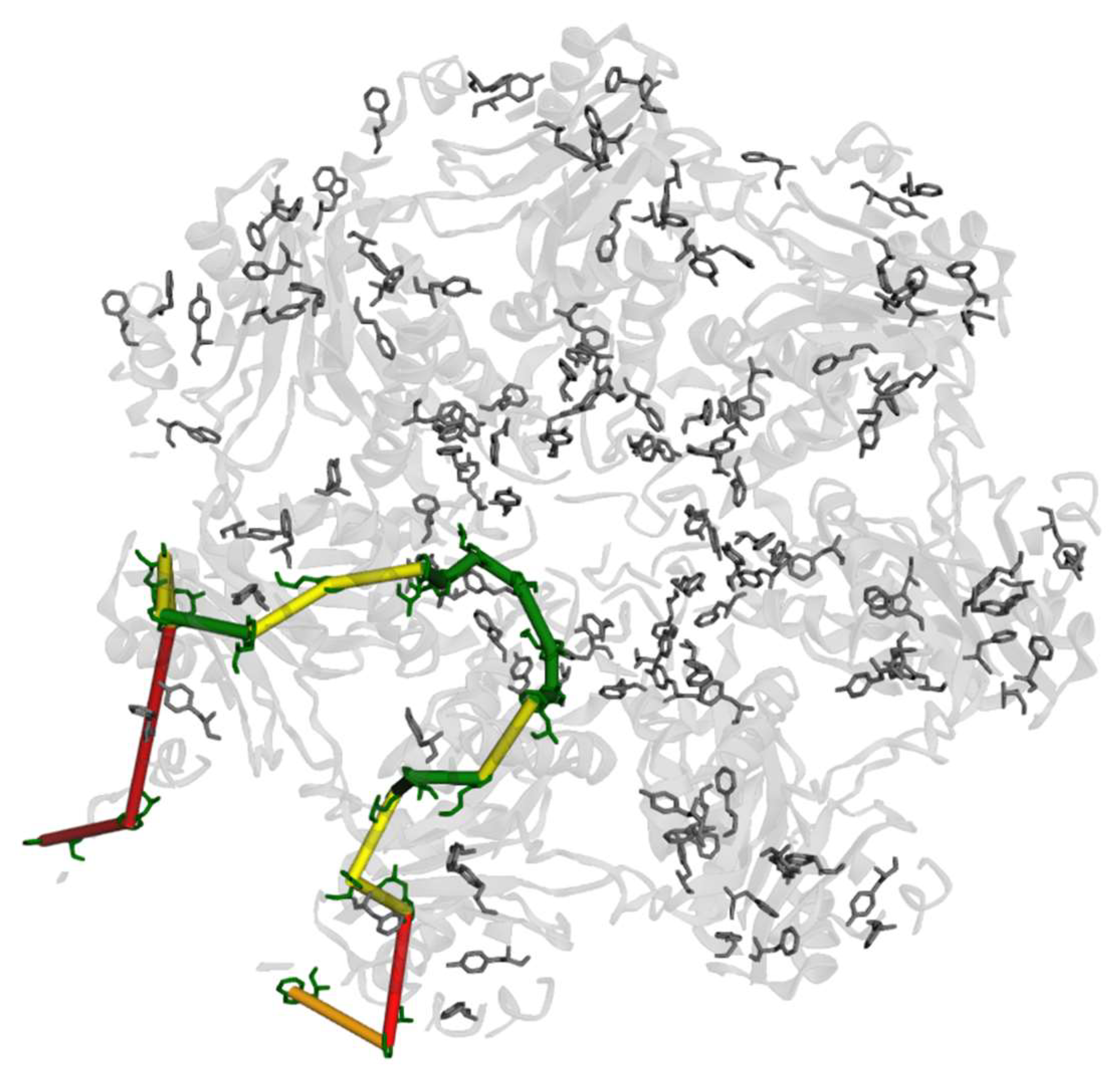
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yalcin, S.E.; Malvankar, N.S. The blind men and the filament: Understanding structures and functions of microbial nanowires. Curr. Opin. Chem. Biol. 2020, 59, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Bostick, C.D.; Mukhopadhyay, S.; Pecht, I.; Sheves, M.; Cahen, D.; Lederman, D. Protein bioelectronics: A review of what we do and do not know. Rep. Prog. Phys. 2018, 81, 026601. [Google Scholar] [CrossRef] [PubMed]
- Ing, N.L.; El-Naggar, M.Y.; Hochbaum, A.I. Going the Distance: Long-Range Conductivity in Protein and Peptide Bioelectronic Materials. J. Phys. Chem. B 2018, 122, 10403–10423. [Google Scholar] [CrossRef]
- Ha, T.Q.; Planje, I.J.; White, J.R.; Aragonès, A.C.; Díez-Pérez, I. Charge transport at the protein–electrode interface in the emerging field of BioMolecular Electronics. Curr. Opin. Electrochem. 2021, 28, 100734. [Google Scholar] [CrossRef]
- Rosenberg, B. Electrical Conductivity of Proteins. Nature 1962, 193, 364–365. [Google Scholar]
- Shipps, C.; Kelly, H.R.; Dahl, P.J.; Yi, S.M.; Vu, D.; Boyer, D.; Glynn, C.; Sawaya, M.R.; Eisenberg, D.; Batista, V.S.; et al. Intrinsic electronic conductivity of individual atomically resolved amyloid crystals reveals micrometer-long hole hopping via tyrosines. Proc. Natl. Acad. Sci. USA 2021, 118, e2014139118. [Google Scholar]
- Shapiro, D.M.; Mandava, G.; Yalcin, S.E.; Arranz-Gibert, P.; Dahl, P.J.; Shipps, C.; Gu, Y.; Srikanth, V.; Salazar-Morales, A.I.; O’Brien, J.P.; et al. Protein nanowires with tunable functionality and programmable self-assembly using sequence-controlled synthesis. Nat. Commun. 2022, 13, 829. [Google Scholar] [PubMed]
- Li, W.; Sepunaru, L.; Amdursky, N.; Cohen, S.R.; Pecht, I.; Sheves, M.; Cahen, D. Temperature and Force Dependence of Nanoscale Electron Transport via the Cu Protein Azurin. ACS Nano 2012, 6, 10816–10824. [Google Scholar] [CrossRef]
- Amdursky, N.; Marchak, D.; Sepunaru, L.; Pecht, I.; Sheves, M.; Cahen, D. Electronic Transport via Proteins. Adv. Mater. 2014, 26, 7142–7161. [Google Scholar]
- Zhang, B.; Song, W.; Pang, P.; Lai, H.; Chen, Q.; Zhang, P.; Lindsay, S. The Role of Contacts in Long-Range Protein Conductance. Proc. Natl. Acad. Sci. USA 2019, 116, 5886–5891. [Google Scholar] [CrossRef]
- Krishnan, S.; Aksimentiev, A.; Lindsay, S.; Matyushov, D. Long-Range Conductivity in Proteins Mediated by Aromatic Residues. ACS Phys. Chem. Au 2023, 3, 444–455. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.; Wang, S.X.; Sepunaru, L. What can electrochemistry tell us about individual enzymes? Curr. Opin. Electro Chem. 2021, 25, 100643. [Google Scholar]
- Zhang, B.; Deng, H.; Mukherjee, S.; Song, W.; Wang, X.; Lindsay, S. Engineering an Enzyme for Direct Electrical Monitoring of Activity. ACS Nano 2019, 14, 1360–1368. [Google Scholar] [CrossRef] [PubMed]
- Kisselev, A.F.; Akopian, T.N.; Goldberg, A.L. Range of sizes of peptide products generated during degradation of different proteins by ar-chaeal proteasomes. J. Biol. Chem. 1998, 273, 1982–1989. [Google Scholar] [CrossRef]
- Thomas, T.; Salcedo-Tacuma, D.; Smith, D.M. Structure, Function, and Allosteric Regulation of the 20S Proteasome by the 11S/PA28 Family of Proteasome Activators. Biomolecules 2023, 13, 1326. [Google Scholar] [CrossRef]
- Fei, X.; Bell, T.A.; Barkow, S.R.; Baker, T.A.; Sauer, R.T. Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates. Elife 2020, 9, e61496. [Google Scholar]
- Fairhead, M.; Howarth, M. Site-specific biotinylation of purified proteins using BirA. Methods Mol. Biol. 2015, 1266, 171–184. [Google Scholar]
- Kay, B.K.; Thai, S.; Volgina, V.V. High-throughput biotinylation of proteins. Methods Mol. Biol. 2009, 498, 185–196. [Google Scholar]
- Förster, A.; Whitby, F.G.; Hill, C.P. The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. EMBO J. 2003, 22, 4356–4364. [Google Scholar] [CrossRef]
- Yu, Z.; Yu, Y.; Wang, F.; Myasnikov, A.G.; Coffino, P.; Cheng, Y. Allosteric coupling between α-rings of the 20S proteasome. Nat. Commun. 2020, 11, 4580. [Google Scholar] [CrossRef]
- Scarff, C.A.; Fuller, M.J.G.; Thompson, R.F.; Iadanza, M.G. Variations on Negative Stain Electron Microscopy Methods: Tools for Tackling Challenging Systems. J. Vis. Exp. 2018, 132, 57199. [Google Scholar]
- Ryan, E.; Shen, D.; Wang, X. Structural studies reveal an important role for the pleiotrophin C-terminus in mediating interactions with chondroitin sulfate. FEBS J. 2016, 283, 1488–1503. [Google Scholar] [CrossRef]
- Tuchband, M.; He, J.; Huang, S.; Lindsay, S. Insulated gold scanning tunneling microscopy probes for recognition tunneling in an aqueous environment. Rev. Sci. Instrum. 2012, 83, 015102. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; He, J.; Zhang, P.; Gyarfas, B.; Lindsay, S. Gap distance and interactions in a molecular tunnel junction. J. Am. Chem. Soc. 2011, 133, 14267–14269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Lindsay, S. Electronic Decay Length in a Protein Molecule. Nano Lett. 2019, 19, 4017–4022. [Google Scholar] [CrossRef]
- Tang, L.; Yi, L.; Jiang, T.; Ren, R.; Nadappuram, B.P.; Zhang, B.; Wu, J.; Liu, X.; Lindsay, S.; Edel, J.B.; et al. Measuring conductance switching in single proteins using quantum tunneling. Sci. Adv. 2022, 8, eabm8149. [Google Scholar] [CrossRef]
- Friis, E.; Andersen, J.; Madsen, L.; Møller, P.; Ulstrup, J. In situ STM and AFM of the copper protein Pseudomonas aeruginosa azurin. J. Electroanal. Chem. 1997, 431, 35–38. [Google Scholar]
- Frascerra, V.; Calabi, F.; Maruccio, G.; Pompa, P.; Cingolani, R.; Rinaldi, R. Resonant electron tunneling through azurin in air and liquid by scanning tunnelling microscopy. IEEE Trans. Nanotechnol. 2005, 4, 637–640. [Google Scholar] [CrossRef]
- Chi, Q.; Farver, O.; Ulstrup, J. Long-range protein electron transfer observed at the single-molecule level: In situ mapping of redox-gated tunneling resonance. Proc. Natl. Acad. Sci. USA 2005, 102, 16203–16208. [Google Scholar] [CrossRef]
- Ruiz, M.P.; Aragonès, A.C.; Camarero, N.; Vilhena, J.G.; Ortega, M.; Zotti, L.A.; Pérez, R.; Cuevas, J.C.; Gorostiza, P.; Díez-Pérez, I. Bioengineering a Single-Protein Junction. J. Am. Chem. Soc. 2017, 139, 15337–15346. [Google Scholar] [CrossRef]
- Artés, J.M.; Díez-Pérez, I.; Gorostiza, P. Transistor-like behavior of single metalloprotein junctions. Nano Lett. 2011, 12, 2679–2684. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Davis, J.J.; Sansom, M.S.P.; Hung, A. Exploring the Electronic and Mechanical Properties of Protein Using Conducting Atomic Force Microscopy. J. Am. Chem. Soc. 2004, 126, 5601–5609. [Google Scholar] [CrossRef] [PubMed]
- Raccosta, S.; Baldacchini, C.; Bizzarri, A.R.; Cannistraro, S. Conductive atomic force microscopy study of single molecule electron transport through the Azurin-gold nanoparticle system. Appl. Phys. Lett. 2013, 102, 203704. [Google Scholar] [CrossRef]
- Romero-Muñiz, C.; Ortega, M.; Vilhena, J.G.; Díez-Pérez, I.; Pérez, R.; Cuevas, J.C.; Zotti, L.A. Can Electron Transport through a Blue-Copper Azurin Be Coherent? An Ab Initio Study. J. Phys. Chem. C 2021, 125, 1693–1702. [Google Scholar] [CrossRef]
- Afsari, S.; Mukherjee, S.; Halloran, N.; Ghirlanda, G.; Ryan, E.; Wang, X.; Lindsay, S. Heavy Water Reduces the Electronic Conductance of Protein Wires via Deuteron Interactions with Aromatic Residues. Nano Lett. 2023, 23, 8907–8913. [Google Scholar] [CrossRef]
- Sarhangi, S.M.; Matyushov, D.V. Anomalously Small Reorganization Energy of the Half Redox Reaction of Azurin. J. Phys. Chem. B 2022, 126, 3000–3011. [Google Scholar] [CrossRef]
- Yen, J.Y. Finding the K Shortest Loopless Paths in a Network. Manag. Sci. 1971, 17, 712–716. [Google Scholar] [CrossRef]
- Lindsay, S. Ubiquitous Electron Transport in Non-Electron Transfer Proteins. Life 2020, 10, 72. [Google Scholar] [CrossRef]
| Sample | Streptavidin | +0.5 µM CP | +1 µM CP | +1 µM Unbiotinylated CP |
|---|---|---|---|---|
| Thickness (nm) | 2.63 ± 0.13 | 3.85 ± 0.19 | 4.68 ± 0.62 | 4.79 ± 0.04 |
| Sample | Gpeak(Run 1) nS | Gpeak(Run 2) nS | Gpeak(Run 3) nS |
|---|---|---|---|
| WT proteasome | 2.69 ± 0.13 | 2.04 ± 0.1 | |
| WT + LLVY | 2.82 ± 0.14 | 2.57 ± 0.5 | |
| Δ12 | 4.68 ± 0.23 | 5.13 ± 0.26 | 6.02 ± 0.03 * |
| Δ12 + Pleiotrophin | 2.63 ± 0.13 | 1.99 ± 0.1 | 1.74 ± 0.09 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afsari, S.; Ryan, E.; Ashcroft, B.; Wang, X.; Lindsay, S. Scanning Tunneling Microscope Measurement of Proteasome Conductance. Biomolecules 2025, 15, 496. https://doi.org/10.3390/biom15040496
Afsari S, Ryan E, Ashcroft B, Wang X, Lindsay S. Scanning Tunneling Microscope Measurement of Proteasome Conductance. Biomolecules. 2025; 15(4):496. https://doi.org/10.3390/biom15040496
Chicago/Turabian StyleAfsari, Sepideh, Eathen Ryan, Brian Ashcroft, Xu Wang, and Stuart Lindsay. 2025. "Scanning Tunneling Microscope Measurement of Proteasome Conductance" Biomolecules 15, no. 4: 496. https://doi.org/10.3390/biom15040496
APA StyleAfsari, S., Ryan, E., Ashcroft, B., Wang, X., & Lindsay, S. (2025). Scanning Tunneling Microscope Measurement of Proteasome Conductance. Biomolecules, 15(4), 496. https://doi.org/10.3390/biom15040496







