Conformational Analyses of the AHD1-UBAN Region of TNIP1 Highlight Key Amino Acids for Interaction with Ubiquitin
Abstract
1. Introduction

2. Materials and Methods
2.1. In Silico Analyses of Disorder and Disorder-to-Order (D-O) Variant Design
2.2. Production of Variants
Protein Purification and Quantification
2.3. Far-UV Circular Dichroism
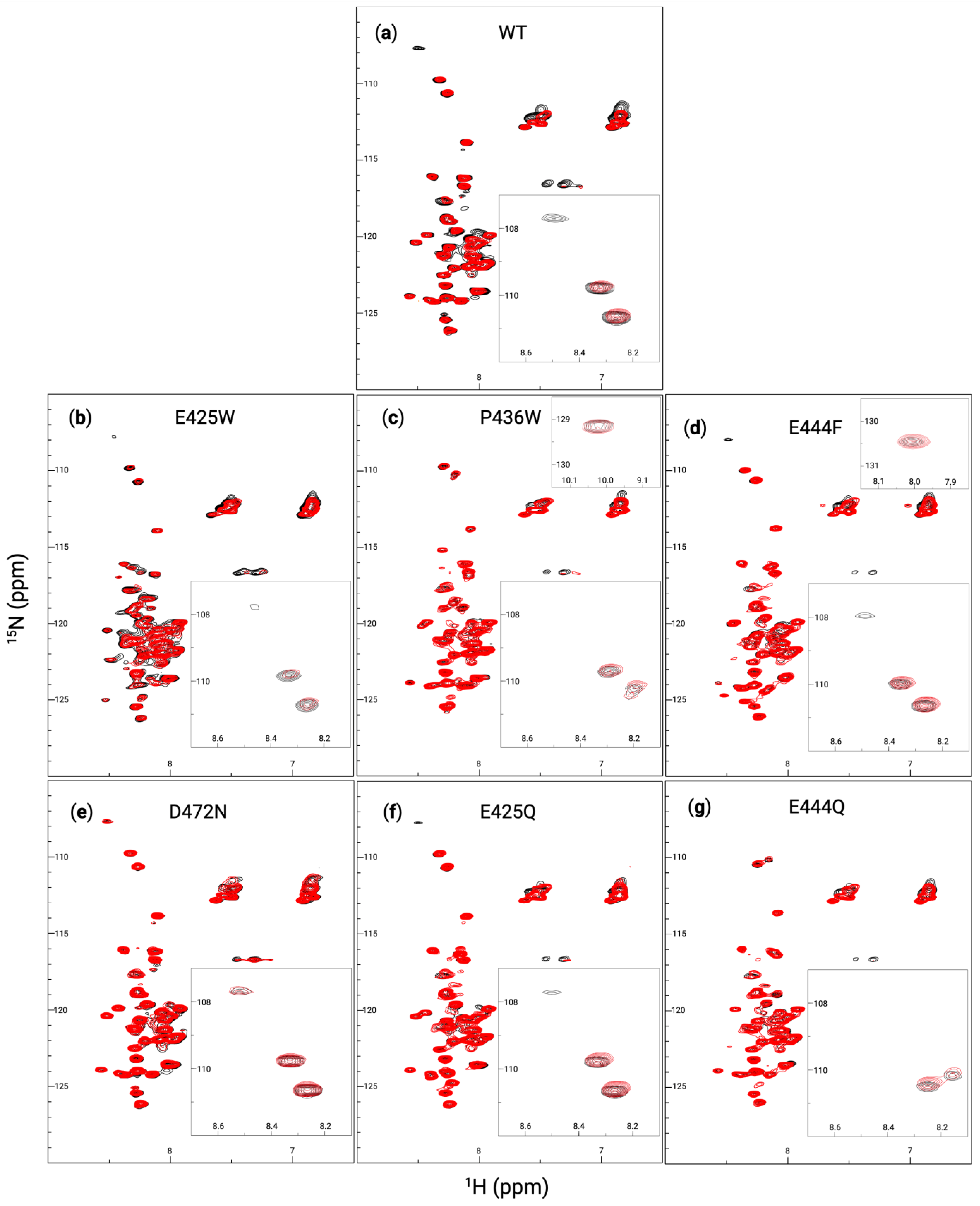
2.4. Heteronuclear Single Quantum Coherence-Nuclear Magnetic Resonance
2.5. Microscale Thermophoresis
3. Results
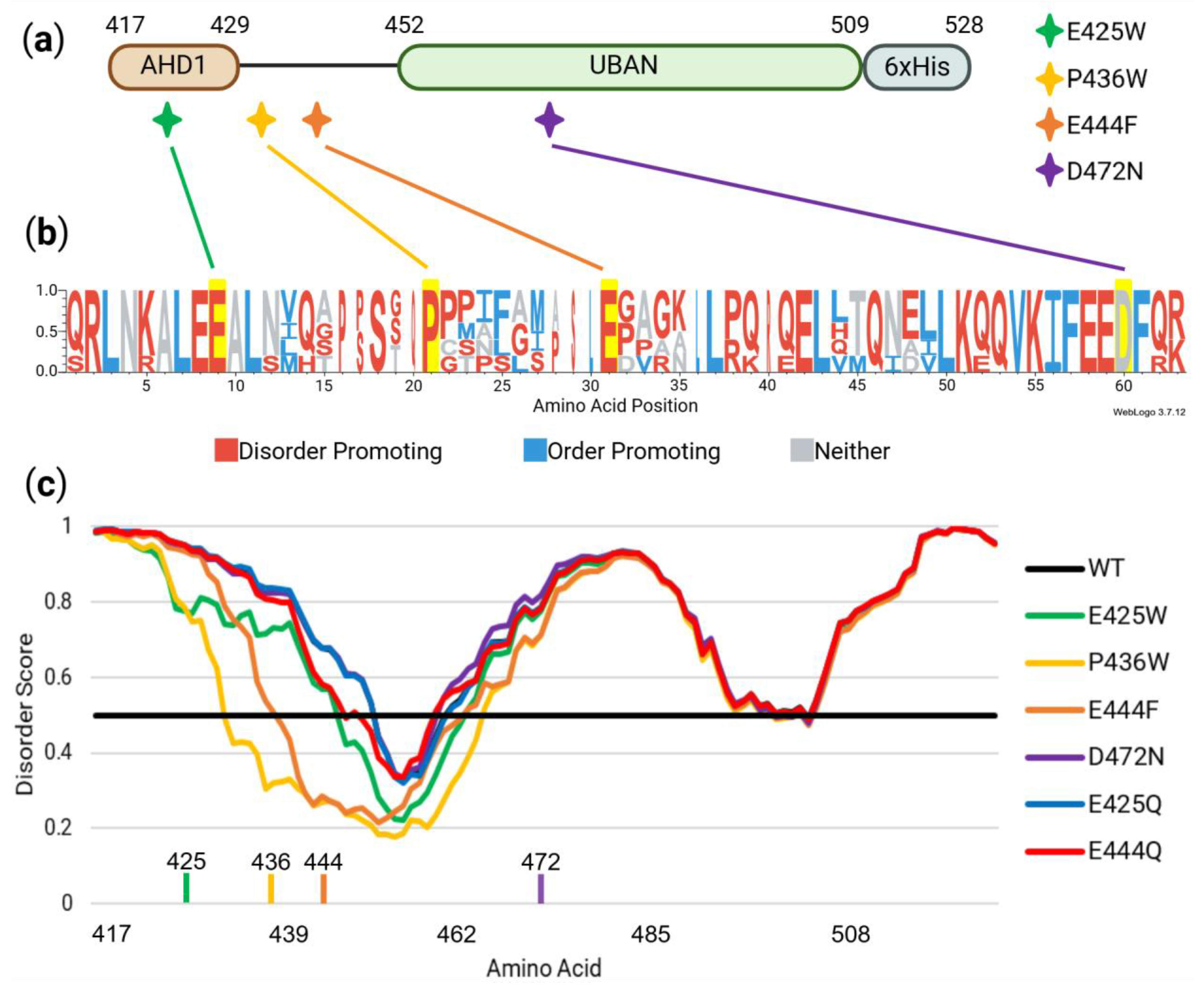
3.1. Selection of Residues and Their Predicted Gain-of-Order Following Amino Acid Mutation
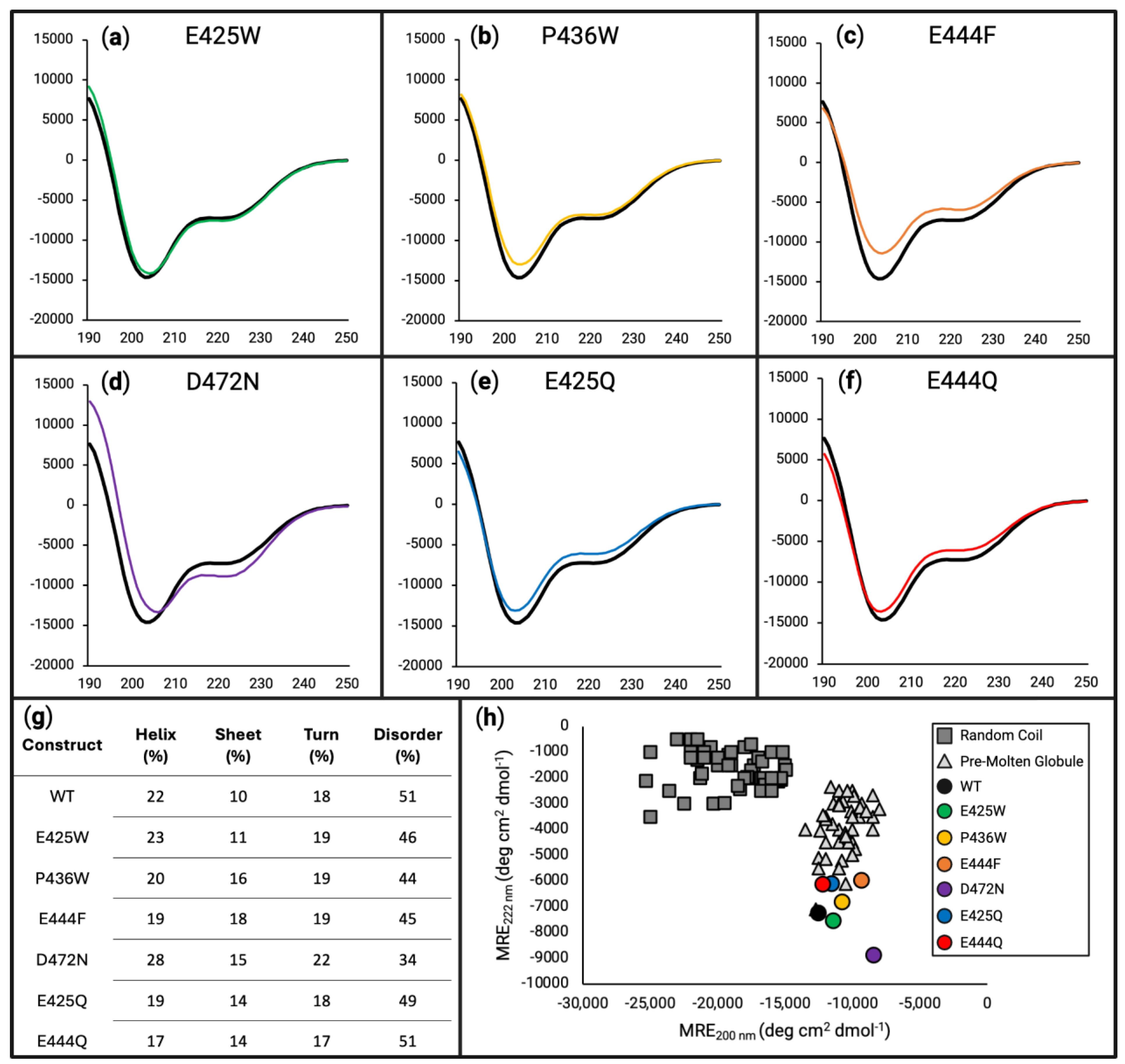
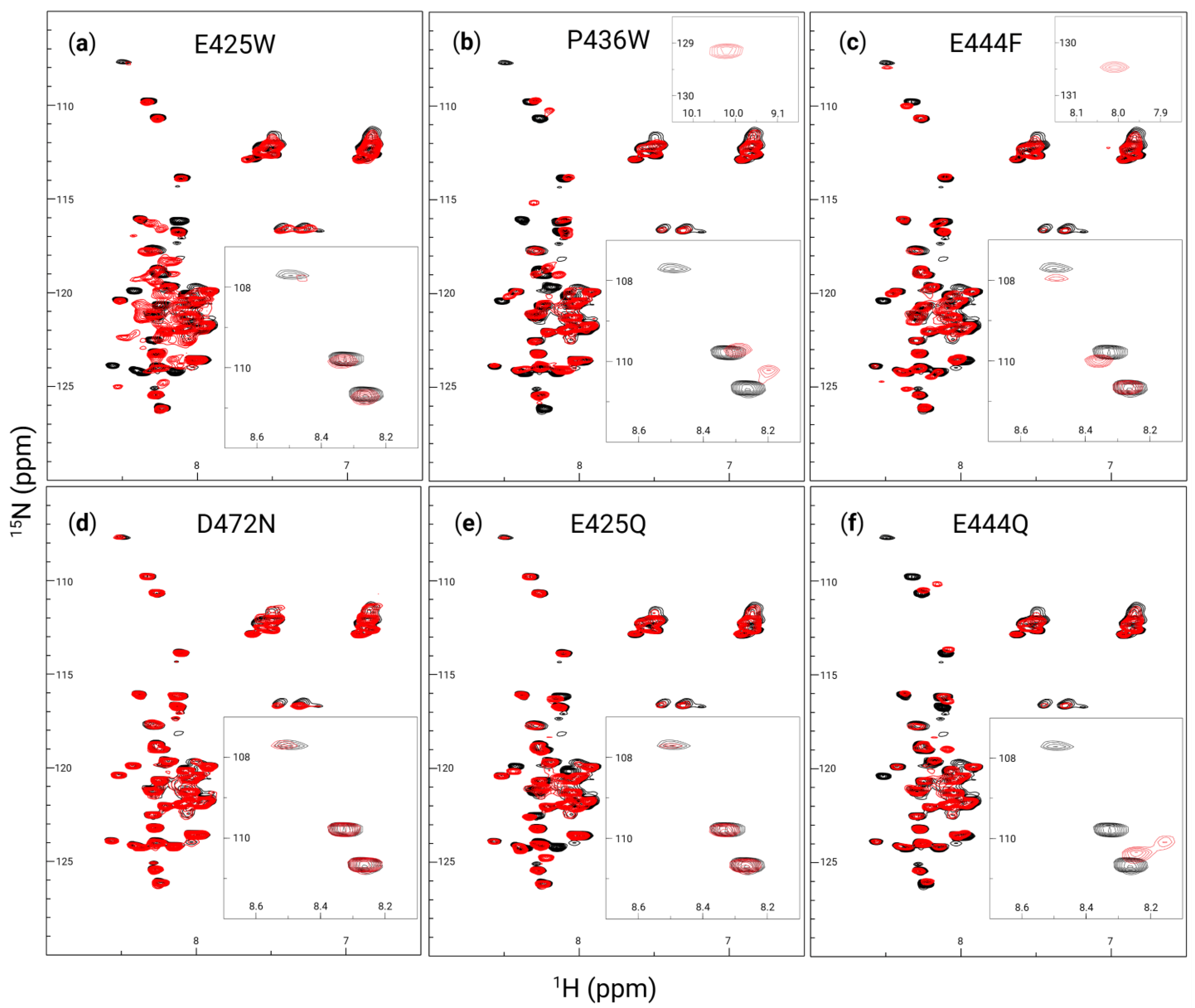
3.2. In Vitro Analysis of Disorder-to-Order Variants via Circular Dichroism and HSQC-NMR
3.3. Maximum Propensity Towards Secondary Structure Is Reduced by Variants Determined by Far-UV Circular Dichroism
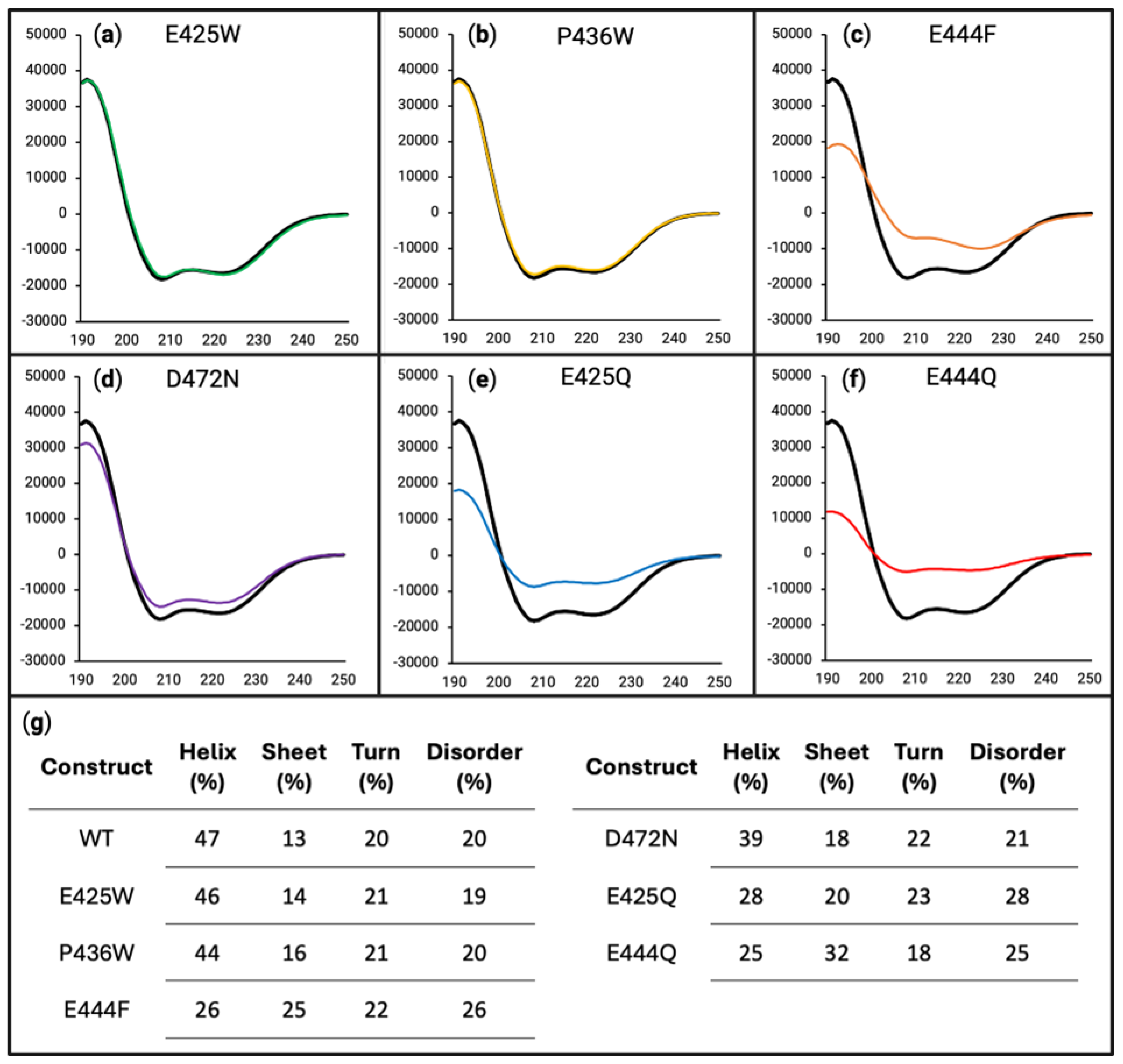
3.4. Partner-Induced Secondary Structure Varies Between AHD1-UBAN Variants as Determined by Far-UV Circular Dichroism and HSQC-NMR
3.5. AHD1-UBAN Variants Show Differences in Recognition of Partner Protein M1-Linked Triubiquitin as Determined with Microscale Thermophoresis
4. Discussion
4.1. In Silico Analysis Reveals Conservation of Disorder-Contributing Residues
4.2. In Vitro Characterization of Secondary Structure Demonstrates Ordering Effect
4.3. Partner-Induced Structural Effect Versus Loss of Conformational Flexibility
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IDP | Intrinsically disordered protein |
| IDR | Intrinsically disordered region |
| WT | Wildtype |
| D-O | Disorder-to-order |
| CD | Circular dichroism |
| HSQC | Heteronuclear single quantum coherence |
| NMR | Nuclear magnetic resonance |
| MST | Microscale thermophoresis |
| M1-triUb | Methionine-1-linked triubiquitin |
| MRE | Molar residue ellipticity |
| ME | Molar extinction |
| TFE | 2,2,2-trifluoroethanol |
| IMAC | Immobilized metal affinity chromatography |
| SEC | Size exclusion chromatography |
References
- Flores, A.M.; Gurevich, I.; Zhang, C.; Ramirez, V.P.; Devens, T.R.; Aneskievich, B.J. TNIP1 is a corepressor of agonist-bound PPARs. Arch. Biochem. Biophys. 2011, 516, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, V.P.; Gurevich, I.; Aneskievich, B.J. Emerging roles for TNIP1 in regulating post-receptor signaling. Cytokine Growth Factor Rev. 2012, 23, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Rudraiah, S.; Shamilov, R.; Aneskievich, B.J. TNIP1 reduction sensitizes keratinocytes to post-receptor signalling following exposure to TLR agonists. Cell. Signal. 2018, 45, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Heyninck, K.; Kreike, M.M.; Beyaert, R. Structure-function analysis of the A20-binding inhibitor of NF-kappa B activation, ABIN-1. FEBS Lett. 2003, 536, 135–140. [Google Scholar] [CrossRef]
- Shamilov, R.; Vinogradova, O.; Aneskievich, B.J. The Anti-Inflammatory Protein TNIP1 Is Intrinsically Disordered with Structural Flexibility Contributed by Its AHD1-UBAN Domain. Biomolecules 2020, 10, 1531. [Google Scholar] [CrossRef]
- Vandereyken, K.; Van Leene, J.; De Coninck, B.; Cammue, B.P.A. Hub Protein Controversy: Taking a Closer Look at Plant Stress Response Hubs. Front. Plant Sci. 2018, 9, 694. [Google Scholar] [CrossRef]
- Rödig, H. Regulation of Inflammation and Autophagy: Role of CD14 in Biglycan-Mediated Sterile Inflammation and Impact of ABIN-1 on Selective Autophagy. Ph.D. Thesis, Goethe University Frankfurt, Frankfurt, Germany, 2020. [Google Scholar]
- Samulevich, M.L.; Shamilov, R.; Aneskievich, B.J. Thermostable Proteins from HaCaT Keratinocytes Identify a Wide Breadth of Intrinsically Disordered Proteins and Candidates for Liquid-Liquid Phase Separation. Int. J. Mol. Sci. 2022, 23, 14323. [Google Scholar] [CrossRef]
- Galea, C.A.; High, A.A.; Obenauer, J.C.; Mishra, A.; Park, C.G.; Punta, M.; Schlessinger, A.; Ma, J.; Rost, B.; Slaughter, C.A.; et al. Large-scale analysis of thermostable, mammalian proteins provides insights into the intrinsically disordered proteome. J. Proteome Res. 2009, 8, 211–226. [Google Scholar] [CrossRef]
- Haynes, C.; Oldfield, C.J.; Ji, F.; Klitgord, N.; Cusick, M.E.; Radivojac, P.; Uversky, V.N.; Vidal, M.; Iakoucheva, L.M. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2006, 2, e100. [Google Scholar] [CrossRef]
- Lindsay, R.J.; Mansbach, R.A.; Gnanakaran, S.; Shen, T. Effects of pH on an IDP conformational ensemble explored by molecular dynamics simulation. Biophys. Chem. 2021, 271, 106552. [Google Scholar] [CrossRef]
- Beveridge, R.; Calabrese, A.N. Structural Proteomics Methods to Interrogate the Conformations and Dynamics of Intrinsically Disordered Proteins. Front. Chem. 2021, 9, 603639. [Google Scholar] [CrossRef]
- Shamilov, R.; Ackley, T.W.; Aneskievich, B.J. Enhanced Wound Healing- and Inflammasome-Associated Gene Expression in TNFAIP3-Interacting Protein 1- (TNIP1-) Deficient HaCaT Keratinocytes Parallels Reduced Reepithelialization. Mediat. Inflamm. 2020, 2020, 5919150. [Google Scholar] [CrossRef]
- Erdei, L.; Bolla, B.S.; Bozó, R.; Tax, G.; Urbán, E.; Kemény, L.; Szabó, K. TNIP1 Regulates Cutibacterium acnes-Induced Innate Immune Functions in Epidermal Keratinocytes. Front. Immunol. 2018, 9, 2155. [Google Scholar] [CrossRef]
- Kattah, M.G.; Shao, L.; Rosli, Y.Y.; Shimizu, H.; Whang, M.I.; Advincula, R.; Achacoso, P.; Shah, S.; Duong, B.H.; Onizawa, M.; et al. A20 and ABIN-1 synergistically preserve intestinal epithelial cell survival. J. Exp. Med. 2018, 215, 1839–1852. [Google Scholar] [CrossRef] [PubMed]
- Indhumathi, S.; Rajappa, M.; Chandrashekar, L.; Ananthanarayanan, P.H.; Thappa, D.M.; Negi, V.S. TNFAIP3 and TNIP1 polymorphisms confer psoriasis risk in South Indian Tamils. Br. J. Biomed. Sci. 2015, 72, 168–173. [Google Scholar] [CrossRef]
- Ippagunta, S.K.; Gangwar, R.; Finkelstein, D.; Vogel, P.; Pelletier, S.; Gingras, S.; Redecke, V.; Häcker, H. Keratinocytes contribute intrinsically to psoriasis upon loss of Tnip1 function. Proc. Natl. Acad. Sci. USA 2016, 113, E6162–E6171. [Google Scholar] [CrossRef]
- Bowes, J.; Orozco, G.; Flynn, E.; Ho, P.; Brier, R.; Marzo-Ortega, H.; Coates, L.; McManus, R.; Ryan, A.W.; Kane, D.; et al. Confirmation of TNIP1 and IL23A as susceptibility loci for psoriatic arthritis. Ann. Rheum. Dis. 2011, 70, 1641–1644. [Google Scholar] [CrossRef]
- Han, J.W.; Zheng, H.F.; Cui, Y.; Sun, L.D.; Ye, D.Q.; Hu, Z.; Xu, J.H.; Cai, Z.M.; Huang, W.; Zhao, G.P.; et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 2009, 41, 1234–1237. [Google Scholar] [CrossRef]
- Shamilov, R.; Aneskievich, B.J. TNIP1 in Autoimmune Diseases: Regulation of Toll-like Receptor Signaling. J. Immunol. Res. 2018, 2018, 3491269. [Google Scholar] [CrossRef]
- Chen, Y.; Yan, H.; Song, Z.; Chen, F.; Wang, H.; Niu, J.; Shi, X.; Zhang, D.; Zhang, N.; Zhai, Z.; et al. Downregulation of TNIP1 Expression Leads to Increased Proliferation of Human Keratinocytes and Severer Psoriasis-Like Conditions in an Imiquimod-Induced Mouse Model of Dermatitis. PLoS ONE 2015, 10, e0127957. [Google Scholar] [CrossRef]
- Nanda, S.K.; Venigalla, R.K.; Ordureau, A.; Patterson-Kane, J.C.; Powell, D.W.; Toth, R.; Arthur, J.S.; Cohen, P. Polyubiquitin binding to ABIN1 is required to prevent autoimmunity. J. Exp. Med. 2011, 208, 1215–1228. [Google Scholar] [CrossRef] [PubMed]
- Akbar, N.; Nanda, S.; Belch, J.; Cohen, P.; Khan, F. An important role for A20-binding inhibitor of nuclear factor-kB-1 (ABIN1) in inflammation-mediated endothelial dysfunction: An in vivo study in ABIN1 (D485N) mice. Arthritis Res. Ther. 2015, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Allanore, Y.; Saad, M.; Dieudé, P.; Avouac, J.; Distler, J.H.; Amouyel, P.; Matucci-Cerinic, M.; Riemekasten, G.; Airo, P.; Melchers, I.; et al. Genome-wide scan identifies TNIP1, PSORS1C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet. 2011, 7, e1002091. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.A.; Childs, E.E.; Amatya, N.; Garg, A.V.; Beyaert, R.; Kane, L.P.; Aneskievich, B.J.; Ma, A.; Gaffen, S.L. Interleukin-17 signaling triggers degradation of the constitutive NF-κB inhibitor ABIN-1. Immunohorizons 2017, 1, 133–141. [Google Scholar] [CrossRef]
- Samulevich, M.L.; Carman, L.E.; Aneskievich, B.J. Investigating Protein-Protein Interactions of Autophagy-Involved TNIP1. In Autophagy in Development and Disease; Methods in Molecular Biology; Humana: New York, NY, USA, 2024. [Google Scholar] [CrossRef]
- Zhou, J.; Rasmussen, N.L.; Olsvik, H.L.; Akimov, V.; Hu, Z.; Evjen, G.; Kaeser-Pebernard, S.; Sankar, D.S.; Roubaty, C.; Verlhac, P.; et al. TBK1 phosphorylation activates LIR-dependent degradation of the inflammation repressor TNIP1. J. Cell Biol. 2023, 222, e202108144. [Google Scholar] [CrossRef]
- Medhavy, A.; Athanasopoulos, V.; Bassett, K.; He, Y.; Stanley, M.; Enosi Tuipulotu, D.; Cappello, J.; Brown, G.J.; Gonzalez-Figueroa, P.; Turnbull, C.; et al. A TNIP1-driven systemic autoimmune disorder with elevated IgG4. Nat. Immunol. 2024, 25, 1678–1691. [Google Scholar] [CrossRef]
- G’Sell, R.T.; Gaffney, P.M.; Powell, D.W. A20-Binding Inhibitor of NF-κB Activation 1 is a Physiologic Inhibitor of NF-κB: A Molecular Switch for Inflammation and Autoimmunity. Arthritis. Rheumatol. 2015, 67, 2292–2302. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, J.; Kim, M.S.; Kwon, Y.; Shin, S.; Yi, H.; Kim, H.; Chang, M.J.; Chang, C.B.; Kang, S.B.; et al. Coordinate regulation of the senescent state by selective autophagy. Dev. Cell 2021, 56, 1512–1525.e7. [Google Scholar] [CrossRef]
- Mauro, C.; Pacifico, F.; Lavorgna, A.; Mellone, S.; Iannetti, A.; Acquaviva, R.; Formisano, S.; Vito, P.; Leonardi, A. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J. Biol. Chem. 2006, 281, 18482–18488. [Google Scholar] [CrossRef]
- Le Guerroué, F.; Bunker, E.N.; Rosencrans, W.M.; Nguyen, J.T.; Basar, M.A.; Werner, A.; Chou, T.F.; Wang, C.; Youle, R.J. TNIP1 inhibits selective autophagy via bipartite interaction with LC3/GABARAP and TAX1BP1. Mol. Cell 2023, 83, 927–941.e8. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Jamecna, D.; Antonny, B. Intrinsically disordered protein regions at membrane contact sites. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 159020. [Google Scholar] [CrossRef]
- Lazar, T.; Tantos, A.; Tompa, P.; Schad, E. Intrinsic protein disorder uncouples affinity from binding specificity. Protein Sci. 2022, 31, e4455. [Google Scholar] [CrossRef] [PubMed]
- Fennell, L.M.; Rahighi, S.; Ikeda, F. Linear ubiquitin chain-binding domains. FEBS J. 2018, 285, 2746–2761. [Google Scholar] [CrossRef]
- Hong, J.Y.; Lin, S.C.; Kuo, B.J.; Lo, Y.C. Structural and Biochemical Basis for Higher-Order Assembly between A20-Binding Inhibitor of NF-κB 1 (ABIN1) and M1-Linked Ubiquitins. J. Mol. Biol. 2021, 433, 167116. [Google Scholar] [CrossRef]
- Verstrepen, L.; Carpentier, I.; Verhelst, K.; Beyaert, R. ABINs: A20 binding inhibitors of NF-kappa B and apoptosis signaling. Biochem. Pharmacol. 2009, 78, 105–114. [Google Scholar] [CrossRef]
- Samulevich, M.L.; Carman, L.E.; Aneskievich, B.J. Critical Analysis of Cytoplasmic Progression of Inflammatory Signaling Suggests Potential Pharmacologic Targets for Wound Healing and Fibrotic Disorders. Biomedicines 2024, 12, 2723. [Google Scholar] [CrossRef]
- Carman, L.E.; Samulevich, M.L.; Aneskievich, B.J. Repressive Control of Keratinocyte Cytoplasmic Inflammatory Signaling. Int. J. Mol. Sci. 2023, 24, 11943. [Google Scholar] [CrossRef]
- Kim, D.H.; Han, K.H. PreSMo Target-Binding Signatures in Intrinsically Disordered Proteins. Mol. Cells 2018, 41, 889–899. [Google Scholar] [CrossRef]
- van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Campen, A.; Williams, R.M.; Brown, C.J.; Meng, J.; Uversky, V.N.; Dunker, A.K. TOP-IDP-scale: A new amino acid scale measuring propensity for intrinsic disorder. Protein Pept. Lett. 2008, 15, 956–963. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Brown, C.J.; Uversky, V.N.; Dunker, A.K. Comparing and combining predictors of mostly disordered proteins. Biochemistry 2005, 44, 1989–2000. [Google Scholar] [CrossRef] [PubMed]
- Lieutaud, P.; Ferron, F.; Uversky, A.V.; Kurgan, L.; Uversky, V.N.; Longhi, S. How disordered is my protein and what is its disorder for? A guide through the “dark side” of the protein universe. Intrinsically Disord Proteins 2016, 4, e1259708. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Uversky, V.N.; Kurgan, L. Comprehensive review of methods for prediction of intrinsic disorder and its molecular functions. Cell. Mol. Life Sci. 2017, 74, 3069–3090. [Google Scholar] [CrossRef]
- Nielsen, J.T.; Mulder, F.A.A. Quality and bias of protein disorder predictors. Sci. Rep. 2019, 9, 5137. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Dembinski, H.; Wismer, K.; Balasubramaniam, D.; Gonzalez, H.A.; Alverdi, V.; Iakoucheva, L.M.; Komives, E.A. Predicted disorder-to-order transition mutations in IκBα disrupt function. Phys. Chem. Chem. Phys. 2014, 16, 6480–6485. [Google Scholar] [CrossRef]
- Nagibina, G.S.; Melnik, T.N.; Glukhova, K.A.; Uversky, V.N.; Melnik, B.S. Intrinsic disorder-based design of stable globular proteins. Prog. Mol. Biol. Transl. Sci. 2020, 174, 157–186. [Google Scholar] [CrossRef]
- Mohan, A.; Oldfield, C.J.; Radivojac, P.; Vacic, V.; Cortese, M.S.; Dunker, A.K.; Uversky, V.N. Analysis of molecular recognition features (MoRFs). J. Mol. Biol. 2006, 362, 1043–1059. [Google Scholar] [CrossRef]
- Daughdrill, G.W.; Borcherds, W.M.; Wu, H. Disorder predictors also predict backbone dynamics for a family of disordered proteins. PLoS ONE 2011, 6, e29207. [Google Scholar] [CrossRef] [PubMed]
- Chemes, L.B.; Alonso, L.G.; Noval, M.G.; de Prat-Gay, G. Circular dichroism techniques for the analysis of intrinsically disordered proteins and domains. Methods Mol. Biol. 2012, 895, 387–404. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.J.; Wallace, B.A. CDtoolX, a downloadable software package for processing and analyses of circular dichroism spectroscopic data. Protein Sci. 2018, 27, 1717–1722. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef]
- Miles, A.J.; Drew, E.D.; Wallace, B.A. DichroIDP: A method for analyses of intrinsically disordered proteins using circular dichroism spectroscopy. Commun. Biol. 2023, 6, 823. [Google Scholar] [CrossRef]
- Nepravishta, R.; Ferrentino, F.; Mandaliti, W.; Mattioni, A.; Weber, J.; Polo, S.; Castagnoli, L.; Cesareni, G.; Paci, M.; Santonico, E. CoCUN, a Novel Ubiquitin Binding Domain Identified in N4BP1. Biomolecules 2019, 9, 284. [Google Scholar] [CrossRef]
- Jiao, L.; Shubbar, M.; Yang, X.; Zhang, Q.; Chen, S.; Wu, Q.; Chen, Z.; Rizo, J.; Liu, X. A partially disordered region connects gene repression and activation functions of EZH2. Proc. Natl. Acad. Sci. USA 2020, 117, 16992–17002. [Google Scholar] [CrossRef]
- Schramm, A.; Bignon, C.; Brocca, S.; Grandori, R.; Santambrogio, C.; Longhi, S. An arsenal of methods for the experimental characterization of intrinsically disordered proteins—How to choose and combine them? Arch. Biochem. Biophys. 2019, 676, 108055. [Google Scholar] [CrossRef]
- Maciejewski, M.W.; Schuyler, A.D.; Gryk, M.R.; Moraru, I.I.; Romero, P.R.; Ulrich, E.L.; Eghbalnia, H.R.; Livny, M.; Delaglio, F.; Hoch, J.C. NMRbox: A Resource for Biomolecular NMR Computation. Biophys. J. 2017, 112, 1529–1534. [Google Scholar] [CrossRef]
- Shaffer, R.; DeMaria, A.M.; Kagermazova, L.; Liu, Y.; Babaei, M.; Caban-Penix, S.; Cervantes, A.; Jehle, S.; Makowski, L.; Gilmore, T.D.; et al. A Central Region of NF-kappaB Essential Modulator Is Required for IKKbeta-Induced Conformational Change and for Signal Propagation. Biochemistry 2019, 58, 2906–2920. [Google Scholar] [CrossRef]
- Schuster, B.S.; Dignon, G.L.; Tang, W.S.; Kelley, F.M.; Ranganath, A.K.; Jahnke, C.N.; Simpkins, A.G.; Regy, R.M.; Hammer, D.A.; Good, M.C.; et al. Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Proc. Natl. Acad. Sci. USA 2020, 117, 11421–11431. [Google Scholar] [CrossRef] [PubMed]
- Ustinova, K.; Novakova, Z.; Saito, M.; Meleshin, M.; Mikesova, J.; Kutil, Z.; Baranova, P.; Havlinova, B.; Schutkowski, M.; Matthias, P.; et al. The disordered N-terminus of HDAC6 is a microtubule-binding domain critical for efficient tubulin deacetylation. J. Biol. Chem. 2020, 295, 2614–2628. [Google Scholar] [CrossRef] [PubMed]
- Schnatwinkel, J.; Herrmann, C. The interaction strength of an intrinsically disordered protein domain with its binding partner is little affected by very different cosolutes. Phys. Chem. Chem. Phys. 2020, 22, 27903–27911. [Google Scholar] [CrossRef] [PubMed]
- Ostendorp, A.; Ostendorp, S.; Zhou, Y.; Chaudron, Z.; Wolffram, L.; Rombi, K.; von Pein, L.; Falke, S.; Jeffries, C.M.; Svergun, D.I.; et al. Intrinsically disordered plant protein PARCL colocalizes with RNA in phase-separated condensates whose formation can be regulated by mutating the PLD. J. Biol. Chem. 2022, 298, 102631. [Google Scholar] [CrossRef]
- Müller-Späth, S.; Soranno, A.; Hirschfeld, V.; Hofmann, H.; Rüegger, S.; Reymond, L.; Nettels, D.; Schuler, B. From the Cover: Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 14609–14614. [Google Scholar] [CrossRef]
- Herhaus, L.; van den Bedem, H.; Tang, S.; Maslennikov, I.; Wakatsuki, S.; Dikic, I.; Rahighi, S. Molecular Recognition of M1-Linked Ubiquitin Chains by Native and Phosphorylated UBAN Domains. J. Mol. Biol. 2019, 431, 3146–3156. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef]
- Schiavina, M.; Bracaglia, L.; Bolognesi, T.; Rodella, M.A.; Tagliaferro, G.; Tino, A.S.; Pierattelli, R.; Felli, I.C. Intrinsically disordered proteins studied by NMR spectroscopy. J. Magn. Reson. 2024, 18, 100143. [Google Scholar] [CrossRef]
- Konrat, R. NMR contributions to structural dynamics studies of intrinsically disordered proteins. J. Magn. Reson. 2014, 241, 74–85. [Google Scholar] [CrossRef]
- Wiegand, T.; Malär, A.A.; Cadalbert, R.; Ernst, M.; Böckmann, A.; Meier, B.H. Asparagine and Glutamine Side-Chains and Ladders in HET-s(218-289) Amyloid Fibrils Studied by Fast Magic-Angle Spinning NMR. Front. Mol. Biosci. 2020, 7, 582033. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef] [PubMed]
- Buck, M. Trifluoroethanol and colleagues: Cosolvents come of age. Recent studies with peptides and proteins. Q. Rev. Biophys. 1998, 31, 297–355. [Google Scholar] [CrossRef]
- Elkjær, S.; Due, A.D.; Christensen, L.F.; Theisen, F.F.; Staby, L.; Kragelund, B.B.; Skriver, K. Evolutionary fine-tuning of residual helix structure in disordered proteins manifests in complex structure and lifetime. Commun. Biol. 2023, 6, 63. [Google Scholar] [CrossRef]
- Morris, O.M.; Torpey, J.H.; Isaacson, R.L. Intrinsically disordered proteins: Modes of binding with emphasis on disordered domains. Open Biol. 2021, 11, 210222. [Google Scholar] [CrossRef]
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta 2010, 1804, 1231–1264. [Google Scholar] [CrossRef]
- Uesugi, M.; Nyanguile, O.; Lu, H.; Levine, A.J.; Verdine, G.L. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science 1997, 277, 1310–1313. [Google Scholar] [CrossRef]
- Wagner, S.; Carpentier, I.; Rogov, V.; Kreike, M.; Ikeda, F.; Löhr, F.; Wu, C.J.; Ashwell, J.D.; Dötsch, V.; Dikic, I.; et al. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene 2008, 27, 3739–3745. [Google Scholar] [CrossRef]
- Bondos, S.E.; Dunker, A.K.; Uversky, V.N. Intrinsically disordered proteins play diverse roles in cell signaling. Cell Commun. Signal. 2022, 20, 20. [Google Scholar] [CrossRef]
- Holehouse, A.S.; Kragelund, B.B. The molecular basis for cellular function of intrinsically disordered protein regions. Nat. Rev. Mol. Cell Biol. 2024, 25, 187–211. [Google Scholar] [CrossRef]
- Li, S.C.; Goto, N.K.; Williams, K.A.; Deber, C.M. Alpha-helical, but not beta-sheet, propensity of proline is determined by peptide environment. Proc. Natl. Acad. Sci. USA 1996, 93, 6676–6681. [Google Scholar] [CrossRef] [PubMed]
- Vacic, V.; Uversky, V.N.; Dunker, A.K.; Lonardi, S. Composition Profiler: A tool for discovery and visualization of amino acid composition differences. BMC Bioinform. 2007, 8, 211. [Google Scholar] [CrossRef]
- Ahrens, J.; Dos Santos, H.G.; Siltberg-Liberles, J. The Nuanced Interplay of Intrinsic Disorder and Other Structural Properties Driving Protein Evolution. Mol. Biol. Evol. 2016, 33, 2248–2256. [Google Scholar] [CrossRef] [PubMed]
- Pajkos, M.; Dosztanyi, Z. Functions of intrinsically disordered proteins through evolutionary lenses. Prog. Mol. Biol. Transl. Sci. 2021, 183, 45–74. [Google Scholar] [CrossRef]
- Zhou, J.; Oldfield, C.J.; Yan, W.; Shen, B.; Dunker, A.K. Intrinsically disordered domains: Sequence ➔ disorder ➔ function relationships. Protein Sci. 2019, 28, 1652–1663. [Google Scholar] [CrossRef]
- Riley, A.C.; Ashlock, D.A.; Graether, S.P. The difficulty of aligning intrinsically disordered protein sequences as assessed by conservation and phylogeny. PLoS ONE 2023, 18, e0288388. [Google Scholar] [CrossRef]
- Ali, H.; Urolagin, S.; Gurarslan, O.; Vihinen, M. Performance of protein disorder prediction programs on amino acid substitutions. Hum. Mutat. 2014, 35, 794–804. [Google Scholar] [CrossRef]
- Atkins, J.D.; Boateng, S.Y.; Sorensen, T.; McGuffin, L.J. Disorder Prediction Methods, Their Applicability to Different Protein Targets and Their Usefulness for Guiding Experimental Studies. Int. J. Mol. Sci. 2015, 16, 19040–19054. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samulevich, M.L.; Carman, L.E.; Shamilov, R.; Aneskievich, B.J. Conformational Analyses of the AHD1-UBAN Region of TNIP1 Highlight Key Amino Acids for Interaction with Ubiquitin. Biomolecules 2025, 15, 453. https://doi.org/10.3390/biom15030453
Samulevich ML, Carman LE, Shamilov R, Aneskievich BJ. Conformational Analyses of the AHD1-UBAN Region of TNIP1 Highlight Key Amino Acids for Interaction with Ubiquitin. Biomolecules. 2025; 15(3):453. https://doi.org/10.3390/biom15030453
Chicago/Turabian StyleSamulevich, Michael L., Liam E. Carman, Rambon Shamilov, and Brian J. Aneskievich. 2025. "Conformational Analyses of the AHD1-UBAN Region of TNIP1 Highlight Key Amino Acids for Interaction with Ubiquitin" Biomolecules 15, no. 3: 453. https://doi.org/10.3390/biom15030453
APA StyleSamulevich, M. L., Carman, L. E., Shamilov, R., & Aneskievich, B. J. (2025). Conformational Analyses of the AHD1-UBAN Region of TNIP1 Highlight Key Amino Acids for Interaction with Ubiquitin. Biomolecules, 15(3), 453. https://doi.org/10.3390/biom15030453





