The Exon-Based Transcriptomic Analysis of Parkinson’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. Workflow
2.3. Statistical Analysis
3. Results
3.1. General Outcome (Table 2)
3.2. Comparisons Between PD and CO and Between Visits (BL Versus V08)
3.2.1. At the Time of PD Diagnosis, Baseline (BL)
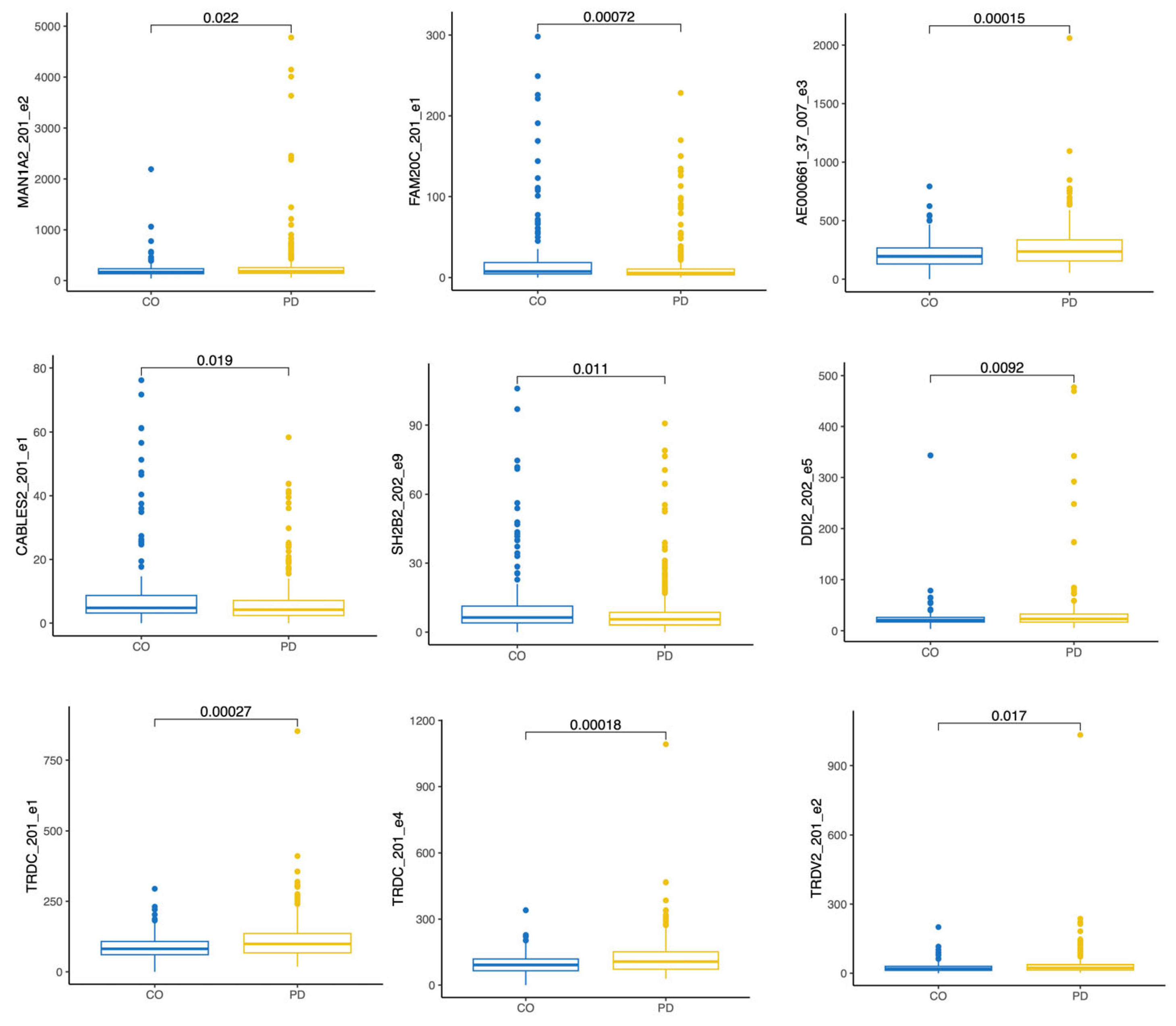
3.2.2. Three Years After the PD Diagnosis Compared to Controls, V08
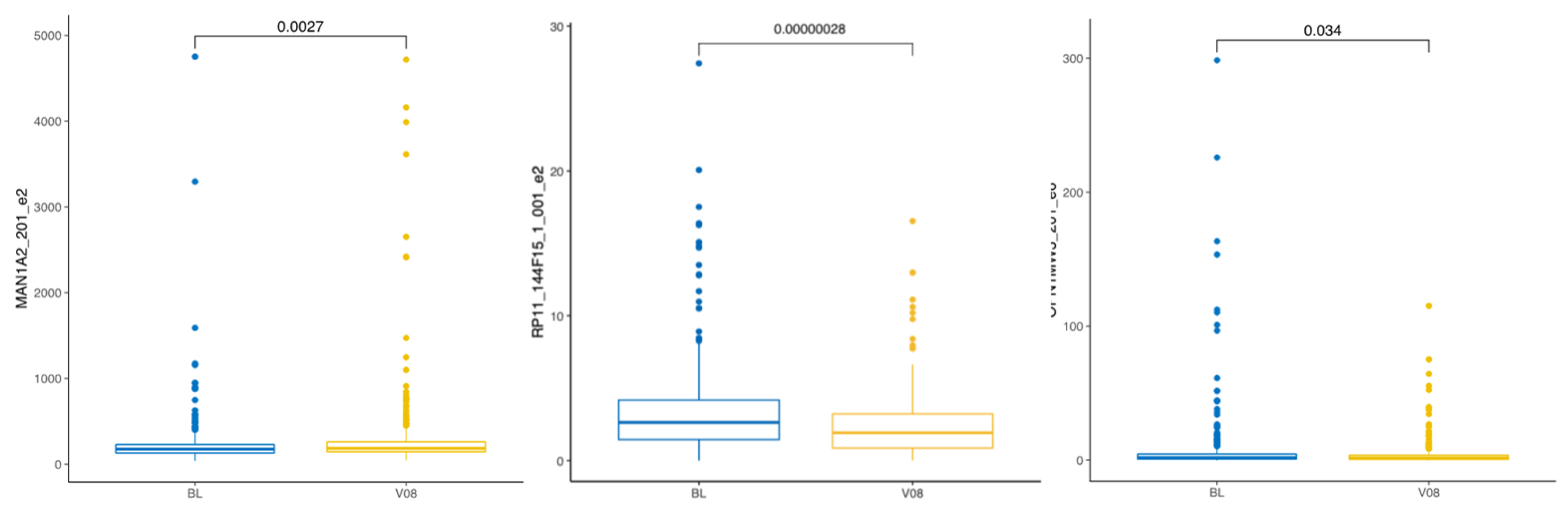
3.2.3. Longitudinal Change After the PD Diagnosis Compared to Baseline in the PD Group
3.2.4. Longitudinal Change After Three Years Compared to Baseline in the CO Group
3.3. Pairwise Comparisons for Differential Exon Expression
3.3.1. All Samples Combined Longitudinal Change
3.3.2. Change in PD Patients Three Years After Diagnosis
3.3.3. PD Progression-Related Changes Three Years After Diagnosis
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bendotti, C.; Tortarolo, M.; Suchak, S.K.; Calvaresi, N.; Carvelli, L.; Bastone, A.; Rizzi, M.; Rattray, M.; Mennini, T. Transgenic SOD1 G93A mice develop reduced GLT-1 in spinal cord without alterations in cerebrospinal fluid glutamate levels. J. Neurochem. 2001, 79, 737–746. [Google Scholar] [CrossRef]
- Koks, S.; Luuk, H.; Nelovkov, A.; Areda, T.; Vasar, E. A screen for genes induced in the amygdaloid area during cat odor exposure. Genes Brain Behav. 2004, 3, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Kingo, K.; Aunin, E.; Karelson, M.; Ratsep, R.; Silm, H.; Vasar, E.; Koks, S. Expressional changes in the intracellular melanogenesis pathways and their possible role in the pathogenesis of vitiligo. J. Dermatol. Sci. 2008, 52, 39–46. [Google Scholar] [CrossRef]
- Koks, G.; Uudelepp, M.L.; Limbach, M.; Peterson, P.; Reimann, E.; Koks, S. Smoking-induced expression of the GPR15 gene indicates its potential role in chronic inflammatory pathologies. Am. J. Pathol. 2015, 185, 2898–2906. [Google Scholar] [CrossRef]
- Butler, D.; Mozsary, C.; Meydan, C.; Foox, J.; Rosiene, J.; Shaiber, A.; Danko, D.; Afshinnekoo, E.; MacKay, M.; Sedlazeck, F.J.; et al. Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. Nat. Commun. 2021, 12, 1660. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, I.; Chung, M.; Angel, L.; Tsay, J.J.; Wu, B.G.; Yeung, S.T.; Krolikowski, K.; Li, Y.; Duerr, R.; Schluger, R.; et al. Microbial signatures in the lower airways of mechanically ventilated COVID-19 patients associated with poor clinical outcome. Nat. Microbiol. 2021, 6, 1245–1258. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef]
- Lancour, D.; Dupuis, J.; Mayeux, R.; Haines, J.L.; Pericak-Vance, M.A.; Schellenberg, G.C.; Crovella, M.; Farrer, L.A.; Kasif, S. Analysis of brain region-specific co-expression networks reveals clustering of established and novel genes associated with Alzheimer disease. Alzheimers Res. Ther. 2020, 12, 103. [Google Scholar] [CrossRef]
- Sumner, C.J.; Kolb, S.J.; Harmison, G.G.; Jeffries, N.O.; Schadt, K.; Finkel, R.S.; Dreyfuss, G.; Fischbeck, K.H. SMN mRNA and protein levels in peripheral blood: Biomarkers for SMA clinical trials. Neurology 2006, 66, 1067–1073. [Google Scholar] [CrossRef]
- Saris, C.G.; Horvath, S.; van Vught, P.W.; van Es, M.A.; Blauw, H.M.; Fuller, T.F.; Langfelder, P.; DeYoung, J.; Wokke, J.H.; Veldink, J.H.; et al. Weighted gene co-expression network analysis of the peripheral blood from Amyotrophic Lateral Sclerosis patients. BMC Genom. 2009, 10, 405. [Google Scholar] [CrossRef]
- Iwaki, H.; Blauwendraat, C.; Leonard, H.L.; Kim, J.J.; Liu, G.; Maple-Grødem, J.; Corvol, J.C.; Pihlstrøm, L.; van Nimwegen, M.; Hutten, S.J.; et al. Genomewide association study of Parkinson’s disease clinical biomarkers in 12 longitudinal patients’ cohorts. Mov. Disord. 2019, 34, 1839–1850. [Google Scholar] [CrossRef] [PubMed]
- Bandres-Ciga, S.; Ahmed, S.; Sabir, M.S.; Blauwendraat, C.; Adarmes-Gómez, A.D.; Bernal-Bernal, I.; Bonilla-Toribio, M.; Buiza-Rueda, D.; Carrillo, F.; Carrión-Claro, M.; et al. The Genetic Architecture of Parkinson Disease in Spain: Characterizing Population-Specific Risk, Differential Haplotype Structures, and Providing Etiologic Insight. Mov. Disord. 2019, 34, 1851–1863. [Google Scholar] [CrossRef] [PubMed]
- Nalls, M.A.; Blauwendraat, C.; Vallerga, C.L.; Heilbron, K.; Bandres-Ciga, S.; Chang, D.; Tan, M.; Kia, D.A.; Noyce, A.J.; Xue, A.; et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019, 18, 1091–1102. [Google Scholar] [CrossRef] [PubMed]
- Billingsley, K.J.; Barbosa, I.A.; Bandres-Ciga, S.; Quinn, J.P.; Bubb, V.J.; Deshpande, C.; Botia, J.A.; Reynolds, R.H.; Zhang, D.; Simpson, M.A.; et al. Mitochondria function associated genes contribute to Parkinson’s Disease risk and later age at onset. npj Park. Dis. 2019, 5, 8. [Google Scholar] [CrossRef]
- Kurvits, L.; Lattekivi, F.; Reimann, E.; Kadastik-Eerme, L.; Kasterpalu, K.M.; Koks, S.; Taba, P.; Planken, A. Transcriptomic profiles in Parkinson’s disease. Exp. Biol. Med. 2021, 246, 584–595. [Google Scholar] [CrossRef]
- Müller-Nedebock, A.C.; van der Westhuizen, F.H.; Kõks, S.; Bardien, S. Nuclear Genes Associated with Mitochondrial DNA Processes as Contributors to Parkinson’s Disease Risk. Mov. Disord. 2021, 36, 815–831. [Google Scholar] [CrossRef]
- Pfaff, A.L.; Bubb, V.J.; Quinn, J.P.; Koks, S. An Increased Burden of Highly Active Retrotransposition Competent L1s Is Associated with Parkinson’s Disease Risk and Progression in the PPMI Cohort. Int. J. Mol. Sci. 2020, 21, 6562. [Google Scholar] [CrossRef]
- Guelfi, S.; D’Sa, K.; Botía, J.A.; Vandrovcova, J.; Reynolds, R.H.; Zhang, D.; Trabzuni, D.; Collado-Torres, L.; Thomason, A.; Quijada Leyton, P.; et al. Regulatory sites for splicing in human basal ganglia are enriched for disease-relevant information. Nat. Commun. 2020, 11, 1041. [Google Scholar] [CrossRef]
- Reynolds, R.H.; Botía, J.; Nalls, M.A.; Hardy, J.; Gagliano Taliun, S.A.; Ryten, M. Moving beyond neurons: The role of cell type-specific gene regulation in Parkinson’s disease heritability. npj Park. Dis. 2019, 5, 6. [Google Scholar] [CrossRef]
- Kurvits, L.; Reimann, E.; Kadastik-Eerme, L.; Truu, L.; Kingo, K.; Erm, T.; Koks, S.; Taba, P.; Planken, A. Serum Amyloid Alpha Is Downregulated in Peripheral Tissues of Parkinson’s Disease Patients. Front. Neurosci. 2019, 13, 13. [Google Scholar] [CrossRef]
- Planken, A.; Kurvits, L.; Reimann, E.; Kadastik-Eerme, L.; Kingo, K.; Koks, S.; Taba, P. Looking beyond the brain to improve the pathogenic understanding of Parkinson’s disease: Implications of whole transcriptome profiling of Patients’ skin. BMC Neurol. 2017, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Koks, S.; Pfaff, A.L.; Bubb, V.J.; Quinn, J.P. Longitudinal intronic RNA-Seq analysis of Parkinson’s disease patients reveals disease-specific nascent transcription. Exp. Biol. Med. 2022, 247, 945–957. [Google Scholar] [CrossRef]
- Peng, H.; Zhang, J.; Ya, A.; Ma, W.; Villa, S.; Sukenik, S.; Ge, X. Myomegalin regulates Hedgehog pathway by controlling PDE4D at the centrosome. Mol. Biol. Cell 2021, 32, 1807–1817. [Google Scholar] [CrossRef] [PubMed]
- Potashkin, J.A.; Bottero, V.; Santiago, J.A.; Quinn, J.P. Bioinformatic Analysis Reveals Phosphodiesterase 4D-Interacting Protein as a Key Frontal Cortex Dementia Switch Gene. Int. J. Mol. Sci. 2020, 21, 3787. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.A.; Scherzer, C.R.; Potashkin, J.A. Specific splice variants are associated with Parkinson’s disease. Mov. Disord. 2013, 28, 1724–1727. [Google Scholar] [CrossRef]
- Scherzer, C.R.; Eklund, A.C.; Morse, L.J.; Liao, Z.; Locascio, J.J.; Fefer, D.; Schwarzschild, M.A.; Schlossmacher, M.G.; Hauser, M.A.; Vance, J.M.; et al. Molecular markers of early Parkinson’s disease based on gene expression in blood. Proc. Natl. Acad. Sci. USA 2007, 104, 955–960. [Google Scholar] [CrossRef]
- Calligaris, R.; Banica, M.; Roncaglia, P.; Robotti, E.; Finaurini, S.; Vlachouli, C.; Antonutti, L.; Iorio, F.; Carissimo, A.; Cattaruzza, T.; et al. Blood transcriptomics of drug-naïve sporadic Parkinson’s disease patients. BMC Genom. 2015, 16, 876. [Google Scholar] [CrossRef]
- Balducci, C.; Pierguidi, L.; Persichetti, E.; Parnetti, L.; Sbaragli, M.; Tassi, C.; Orlacchio, A.; Calabresi, P.; Beccari, T.; Rossi, A. Lysosomal hydrolases in cerebrospinal fluid from subjects with Parkinson’s disease. Mov. Disord. 2007, 22, 1481–1484. [Google Scholar] [CrossRef]
- Fiszer, U.; Mix, E.; Fredrikson, S.; Kostulas, V.; Olsson, T.; Link, H. gamma delta+ T cells are increased in patients with Parkinson’s disease. J. Neurol. Sci. 1994, 121, 39–45. [Google Scholar] [CrossRef]
- Wang, P.; Yao, L.; Luo, M.; Zhou, W.; Jin, X.; Xu, Z.; Yan, S.; Li, Y.; Xu, C.; Cheng, R.; et al. Single-cell transcriptome and TCR profiling reveal activated and expanded T cell populations in Parkinson’s disease. Cell Discov. 2021, 7, 52. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, S.; Liu, J. The role of T cells in the pathogenesis of Parkinson’s disease. Prog. Neurobiol. 2018, 169, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Ta, M.; Blauwendraat, C.; Antar, T.; Leonard, H.L.; Singleton, A.B.; Nalls, M.A.; Iwaki, H. Genome-Wide Meta-Analysis of Cerebrospinal Fluid Biomarkers in Alzheimer’s Disease and Parkinson’s Disease Cohorts. Mov. Disord. 2023, 38, 1697–1705. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Xin, J.; Le, W.; Yang, Y. Neurogranin: A Potential Biomarker of Neurological and Mental Diseases. Front. Aging Neurosci. 2020, 12, 584743. [Google Scholar] [CrossRef]
- Steiner, O.L.; de Zeeuw, J. Melanopsin retinal ganglion cell function in Alzheimer’s vs. Parkinson’s disease an exploratory meta-analysis and review of pupillometry protocols. Parkinsonism Relat. Disord. 2024, 123, 106063. [Google Scholar] [CrossRef] [PubMed]
- Gaynes, B.I.; Zaffer, A.; Yousefzai, R.; Chazaro-Cortes, M.; Colletta, K.; Kletzel, S.L.; Jost, M.B.; Park, Y.; Chawla, J.; Albert, M.V.; et al. Variable abnormality of the melanopsin-derived portion of the pupillary light reflex (PLR) in patients with Parkinson’s disease (PD) and parkinsonism features. Neurol. Sci. 2022, 43, 349–356. [Google Scholar] [CrossRef]
- Joyce, D.S.; Feigl, B.; Kerr, G.; Roeder, L.; Zele, A.J. Melanopsin-mediated pupil function is impaired in Parkinson’s disease. Sci. Rep. 2018, 8, 7796. [Google Scholar] [CrossRef]
- Bonilha, V.L.; Bell, B.A.; Rayborn, M.E.; Yang, X.; Kaul, C.; Grossman, G.H.; Samuels, I.S.; Hollyfield, J.G.; Xie, C.; Cai, H.; et al. Loss of DJ-1 elicits retinal abnormalities, visual dysfunction, and increased oxidative stress in mice. Exp. Eye Res. 2015, 139, 22–36. [Google Scholar] [CrossRef]
- Ortuño-Lizarán, I.; Sánchez-Sáez, X.; Lax, P.; Serrano, G.E.; Beach, T.G.; Adler, C.H.; Cuenca, N. Dopaminergic Retinal Cell Loss and Visual Dysfunction in Parkinson Disease. Ann. Neurol. 2020, 88, 893–906. [Google Scholar] [CrossRef]
- Ortuño-Lizarán, I.; Esquiva, G.; Beach, T.G.; Serrano, G.E.; Adler, C.H.; Lax, P.; Cuenca, N. Degeneration of human photosensitive retinal ganglion cells may explain sleep and circadian rhythms disorders in Parkinson’s disease. Acta Neuropathol. Commun. 2018, 6, 90. [Google Scholar] [CrossRef]
- Feigl, B.; Dumpala, S.; Kerr, G.K.; Zele, A.J. Melanopsin Cell Dysfunction is Involved in Sleep Disruption in Parkinson’s Disease. J. Park. Dis. 2020, 10, 1467–1476. [Google Scholar] [CrossRef]



| Study Group | Baseline (BL) | Three Years (V08) |
|---|---|---|
| CO | 189 | 157 |
| PD | 390 | 338 |
| Group | Differentially Expressed Exons | Comparison |
|---|---|---|
| BL | 2 | PD versus CO |
| V08 | 27 | PD versus CO |
| PD | 13 | V08 versus BL |
| CO | 9 | V08 versus BL |
| Exon ID | Log2 FC | FDR | Transcript and Exon |
|---|---|---|---|
| ENSE00003719556 | 1.6 | 1.8 × 10−7 | RP11-403I13.4-002 e5 |
| ENSE00003746162 | 0.5 | 0.06 | RP11-596C23.2-001 e1 |
| Exon ID | Log2 FC | FDR | Transcript and Exon |
|---|---|---|---|
| ENSE00003719556 | −2.8 | 1.03 × 10−21 | RP11-403I13.4-002 e5 |
| ENSE00001331364 | −2.0 | 1.55 × 10−16 | MST1P2-201 e2 |
| ENSE00001806909 | −0.7 | 0.007 | RNU1-4-201 e1 |
| ENSE00003742227 | −0.6 | 0.008 | RNA5-8SN2-201 e1 |
| ENSE00002197845 | −1.1 | 0.017 | RP11-304M2.3-001 e1 |
| ENSE00001635177 | 0.5 | 0.029 | MAN1A2-201 e2 |
| ENSE00002058739 | −1.0 | 0.029 | RP4-669L17.8-001 e1 |
| ENSE00001232590 | −0.8 | 0.029 | FAM20C-201 e1 |
| ENSE00003725298 | −0.6 | 0.032 | RNA5-8SN4-201 e1 |
| ENSE00001885147 | 0.3 | 0.037 | TRDC-201 e1 |
| ENSE00002496627 | 0.3 | 0.037 | TRDC-201 e4 |
| ENSE00002533311 | 0.3 | 0.037 | AE000661.37-007 e3 |
| ENSE00002663611 | −0.4 | 0.037 | RP11-498C9.3-002 e2 |
| ENSE00002724720 | −0.6 | 0.037 | ENSG110127253.1-001 e1 |
| ENSE00003734991 | −0.6 | 0.037 | RNA5-8SN5-201 e1 |
| ENSE00001820608 | −0.6 | 0.044 | CABLES2-201 e1 |
| ENSE00003746458 | −0.6 | 0.048 | 5_8S_rRNA.5-201 e1 |
| ENSE00001443289 | −0.4 | 0.052 | G0S2-201 e2 |
| ENSE00001387755 | −0.5 | 0.053 | SCAF1-201 e7 |
| ENSE00002592795 | −1.0 | 0.055 | RP11-356C4.5-001 e1 |
| ENSE00001014304 | −0.5 | 0.080 | NRGN-201 e2 |
| ENSE00001351972 | 0.4 | 0.098 | DDI2-202 e5 |
| ENSE00001242645 | −0.4 | 0.098 | HDAC4-201 e1 |
| ENSE00002480538 | −0.5 | 0.098 | SH2B2-202 e9 |
| ENSE00001508095 | 0.5 | 0.098 | TRDV2-201 e2 |
| ENSE00002222336 | −0.6 | 0.098 | RAB11FIP3-201 e1 |
| ENSE00003752463 | −0.8 | 0.098 | ENSG10010138218.1-001 e1 |
| Exon ID | Log2 FC | FDR | Transcript and Exon |
|---|---|---|---|
| ENSE00001740173 | −2.4 | 5.5 × 10−13 | RP11-403I13.4-002 e1 |
| ENSE00001806816 | 1.5 | 4.5 × 10−9 | RNU1-67P-201 e1 |
| ENSE00003763487 | 0.9 | 0.001 | CH507-513H4.6-001 e1 |
| ENSE00001267623 | 0.3 | 0.005 | SETD2-202 e6 |
| ENSE00001635177 | 0.4 | 0.015 | MAN1A2-201 e2 |
| ENSE00003760261 | −0.4 | 0.015 | RP11-144F15.1-001 e2 |
| ENSE00003762709 | 0.7 | 0.015 | CH507-513H4.4-001 e1 |
| ENSE00000830378 | −2.4 | 0.015 | CAPN6-201 e2 |
| ENSE00002089387 | 0.5 | 0.091 | Y_RNA.718-201 e1 |
| ENSE00001616358 | −0.5 | 0.099 | MST1P2-201 e11 |
| ENSE00003903060 | −0.5 | 0.099 | GDAP1-227 e2 |
| ENSE00003188750 | −0.8 | 0.099 | OPN1MW3-201 e6 |
| ENSE00001544494 | −0.5 | 0.099 | MT-TQ-201 e1 |
| Exon ID | Log2 FC | FDR | Transcript and Exon |
|---|---|---|---|
| ENSE00003719556 | 4.7 | 8.2 × 10−49 | RP11-403I13.4-002 e5 |
| ENSE00001331364 | 2.3 | 1.9 × 10−14 | MST1P2-201 e2 |
| ENSE00001838554 | 0.9 | 0.000 | IGHV3-21-201 e1 |
| ENSE00003602778 | 1.4 | 0.001 | RNVU1-31-201 e1 |
| ENSE00001808682 | 1.5 | 0.010 | RNVU1-2-201 e1 |
| ENSE00003741241 | 0.9 | 0.028 | RNVU1-2A-202 e1 |
| ENSE00001807294 | 1.1 | 0.028 | RNY1-201 e1 |
| ENSE00001806816 | 1.3 | 0.032 | RNU1-67P-201 e1 |
| ENSE00001808752 | 0.5 | 0.039 | SNORA13-201 e1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kõks, S. The Exon-Based Transcriptomic Analysis of Parkinson’s Disease. Biomolecules 2025, 15, 440. https://doi.org/10.3390/biom15030440
Kõks S. The Exon-Based Transcriptomic Analysis of Parkinson’s Disease. Biomolecules. 2025; 15(3):440. https://doi.org/10.3390/biom15030440
Chicago/Turabian StyleKõks, Sulev. 2025. "The Exon-Based Transcriptomic Analysis of Parkinson’s Disease" Biomolecules 15, no. 3: 440. https://doi.org/10.3390/biom15030440
APA StyleKõks, S. (2025). The Exon-Based Transcriptomic Analysis of Parkinson’s Disease. Biomolecules, 15(3), 440. https://doi.org/10.3390/biom15030440






