Artificial Intelligence: A New Tool for Structure-Based G Protein-Coupled Receptor Drug Discovery
Abstract
1. Introduction
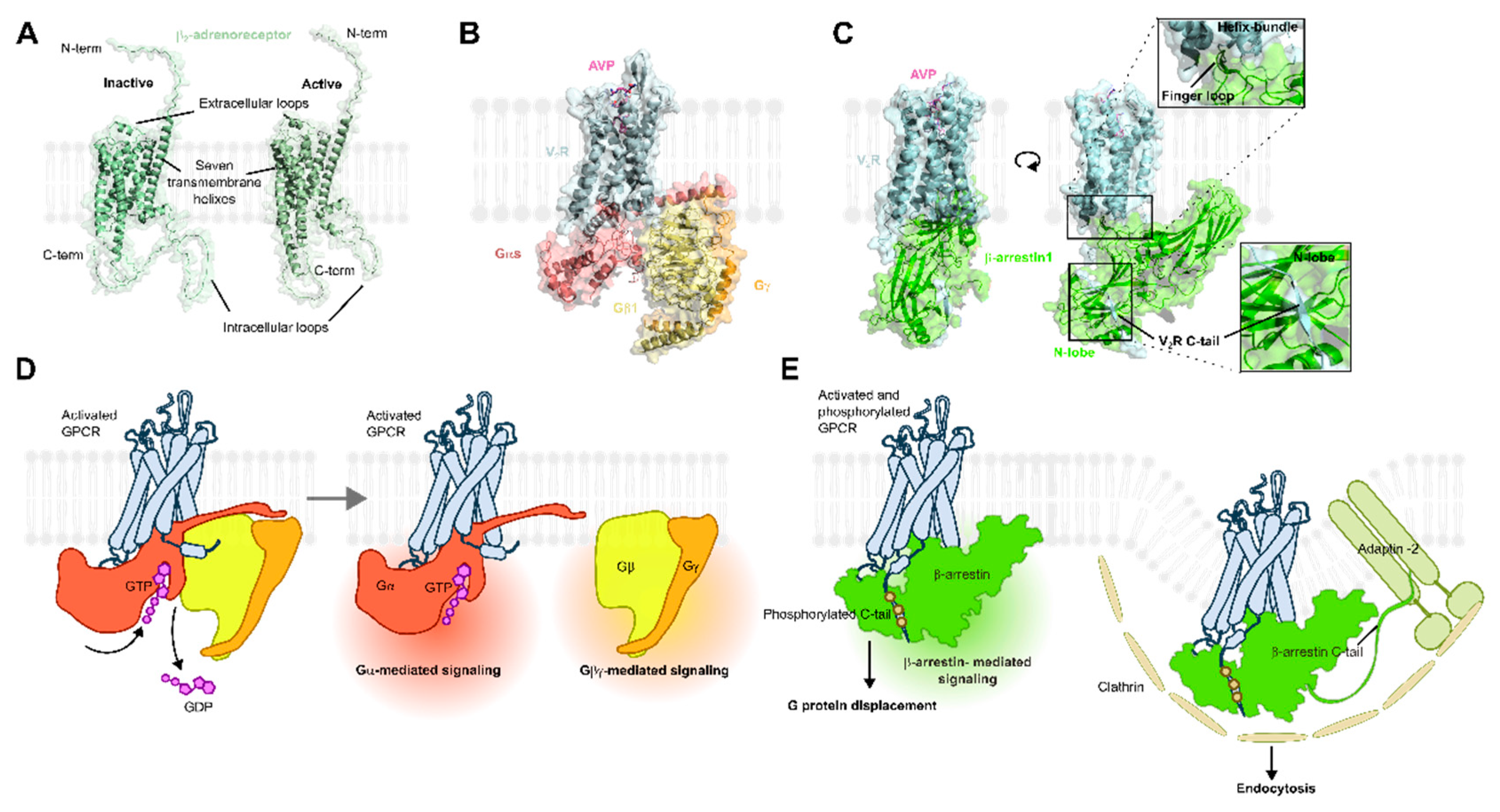
2. GPCR Structure and Function
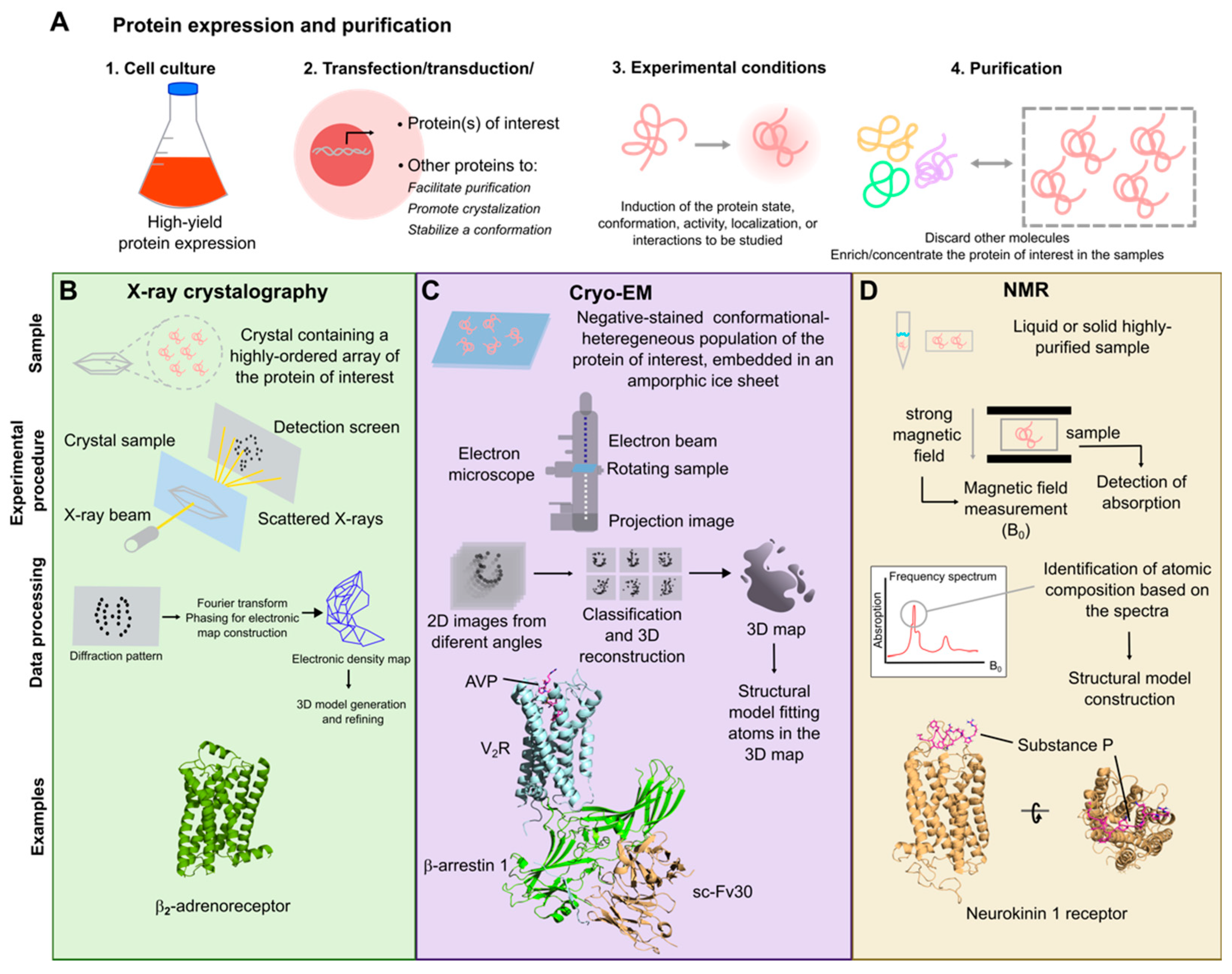
3. Experimental Structure Determination
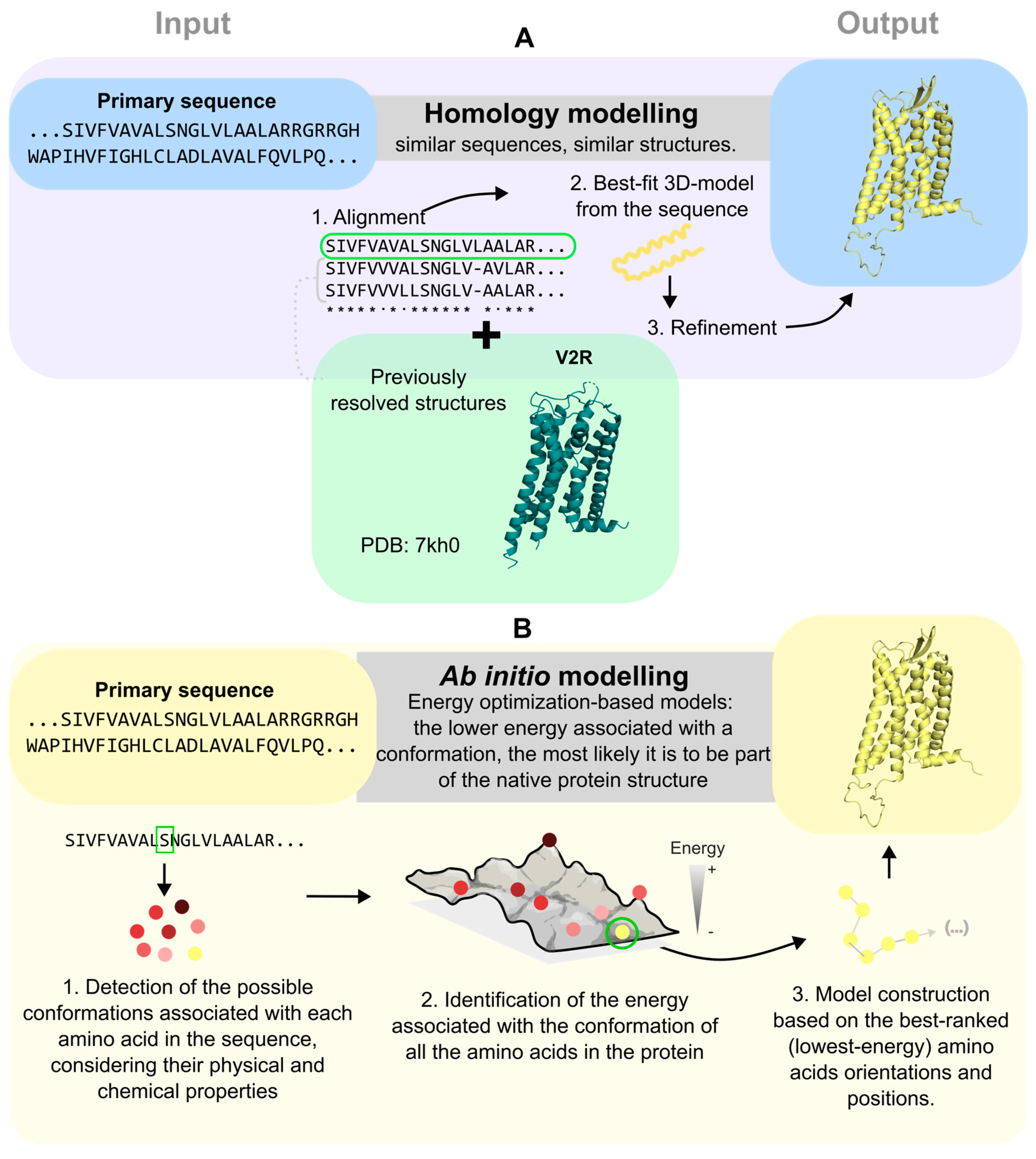
4. Computational Approaches to Predict Structural Models
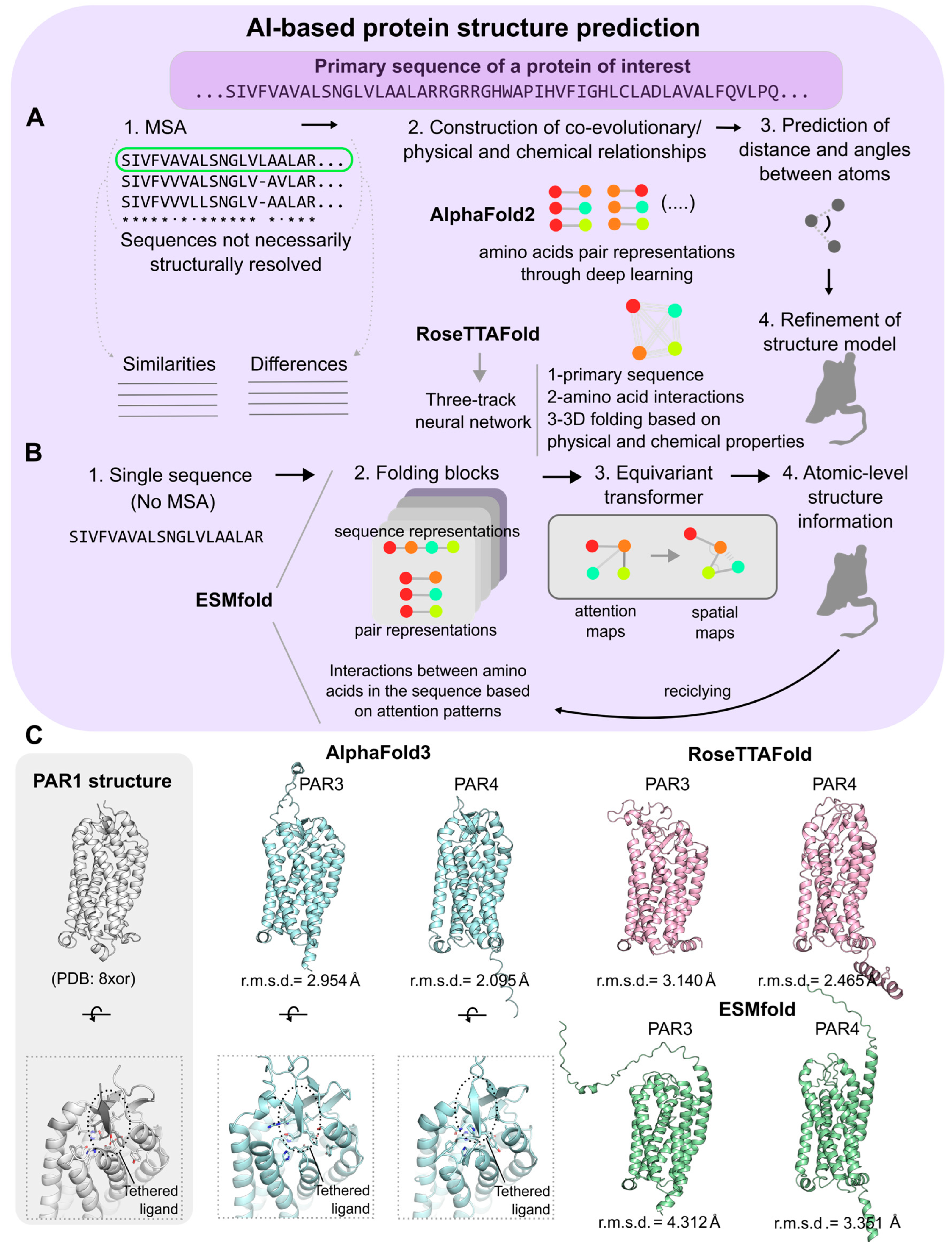
5. Structure Biology and AI/ML
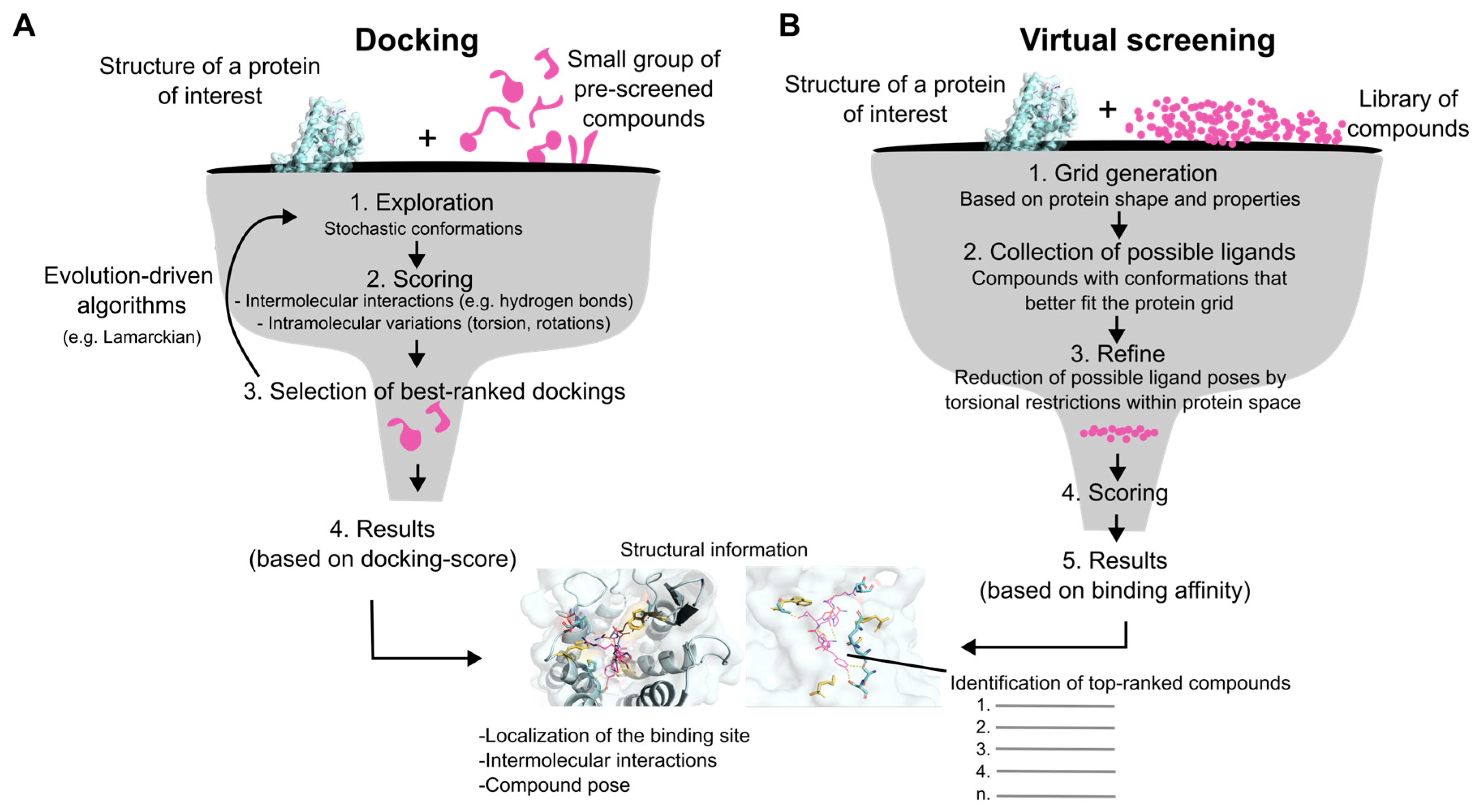
6. Computation-Based Docking/Virtual Screening
7. AI-Enabled Structure-Based GPCR Drug Discovery: Are We There Yet?
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stollar, E.J.; Smith, D.P. Uncovering protein structure. Essays Biochem. 2020, 64, 649–680. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Coventry, B.; Goreshnik, I.; Huang, B.; Sheffler, W.; Park, J.S.; Jude, K.M.; Marković, I.; Kadam, R.U.; Verschueren, K.H.G.; et al. Design of protein-binding proteins from the target structure alone. Nature 2022, 605, 551–560. [Google Scholar] [CrossRef]
- Chevalier, A.; Silva, D.-A.; Rocklin, G.J.; Hicks, D.R.; Vergara, R.; Murapa, P.; Bernard, S.M.; Zhang, L.; Lam, K.-H.; Yao, G.; et al. Massively parallel de novo protein design for targeted therapeutics. Nature 2017, 550, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Rosenbaum, D.M.; Rasmussen, S.G.F.; Kobilka, B.K. The structure and function of G-protein-coupled receptors. Nature 2009, 459, 356–363. [Google Scholar] [CrossRef]
- Katritch, V.; Cherezov, V.; Stevens, R.C. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol. Sci. 2021, 33, 17–27. [Google Scholar] [CrossRef]
- Fredriksson, R.; Lagerström, M.C.; Lundin, L.G.; Schiöth, H.B. The G-Protein-Coupled Receptors in the Human Genome Form Five Main Families. Phylogenetic Analysis, Paralogon Groups, and Fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272. [Google Scholar] [CrossRef]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schiöth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef]
- De Lean, A.; Stadel, J.M.; Lefkowitz, R.J. A ternary complex model explains the agonist-specific binding properties of the adenylate cyclase-coupled beta-adrenergic receptor. J. Biol. Chem. 1980, 255, 7108–7117. [Google Scholar] [CrossRef]
- Gilman, A.G. G proteins: Transducers of receptor-generated signals. Annu. Rev. Biochem. 1987, 56, 615–649. [Google Scholar] [CrossRef]
- Moore, C.A.; Milano, S.K.; Benovic, J.L. Regulation of receptor trafficking by GRKs and arrestins. Annu. Rev. Physiol. 2007, 69, 451–482. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.J.; Benovic, J.L.; Codina, J.; Caron, M.G.; Lefkowitz, R.J. β-Arrestin: A protein that regulates beta-adrenergic receptor function. Science 1990, 248, 1547–1550. [Google Scholar] [CrossRef]
- Goodman, O.B., Jr.; Krupnick, J.G.; Santini, F.; Gurevich, V.V.; Penn, R.B.; Gagnon, A.W.; Keen, J.H.; Benovic, J.L. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature 1996, 383, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Laporte, S.A.; Oakley, R.H.; Zhang, J.; Holt, J.A.; Ferguson, S.S.G.; Caron, M.G.; Barak, L.S. The beta2-adrenergic receptor/betaarrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proc. Natl. Acad. Sci. USA 1999, 96, 3712–3717. [Google Scholar] [CrossRef] [PubMed]
- Pierce, K.L.; Premont, R.T.; Lefkowitz, R.J. Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 2002, 3, 639–650. [Google Scholar] [CrossRef]
- Mannes, M.; Martin, C.; Menet, C.; Ballet, S. Wandering beyond small molecules: Peptides as allosteric protein modulators. Trends Pharmacol. Sci. 2022, 43, 406–423. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, T.; Lu, X.; Lan, X.; Chen, Z.; Lu, S. G protein-coupled receptors (GPCRs): Advances in structures, mechanisms and drug discovery. Signal Transduct. Target. Ther. 2024, 9, 88. [Google Scholar] [CrossRef]
- Markosian, C.; Di Costanzo, L.; Sekharan, M.; Shao, C.; Burley, S.K.; Zardecki, C. Analysis of impact metrics for the Protein Data Bank. Sci. Data 2018, 5, 180212. [Google Scholar] [CrossRef]
- Dokholyan, N.V. Experimentally-driven protein structure modeling. J. Proteom. 2020, 220, 103777. [Google Scholar] [CrossRef]
- Palczewski, K.; Kumasaka, T.; Hori, T.; Behnke, C.A.; Motoshima, H.; Fox, B.A.; Le Trong, I.; Teller, D.C.; Okada, T.; Stenkamp, R.E.; et al. Crystal Structure of Rhodopsin: A G Protein-Coupled Receptor. Science 2000, 289, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Cherezov, V.; Rosenbaum, D.M.; Hanson, M.A.; Rasmussen, S.G.F.; Thian, F.S.; Kobilka, T.S.; Choi, H.-J.; Kuhn, P.; Weis, W.I.; Kobilka, B.K.; et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 2007, 318, 1258–1265. [Google Scholar] [CrossRef]
- Rasmussen, S.G.F.; Choi, H.-J.; Rosenbaum, D.M.; Kobilka, T.S.; Thian, F.S.; Edwards, P.C.; Burghammer, M.; Ratnala, V.R.P.; Sanishvili, R.; Fischetti, R.F.; et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature 2007, 450, 383–387. [Google Scholar] [CrossRef]
- Dessau, M.A.; Modis, Y. Protein crystallization for X-ray crystallography. J. Vis. Exp. 2011, 16, e2285. [Google Scholar] [CrossRef]
- Cahill, T.J., 3rd; Thomsen, A.R.B.; Tarrasch, J.T.; Plouffe, B.; Nguyen, A.H.; Yang, F.; Huang, L.-Y.; Kahsai, A.W.; Bassoni, D.L.; Gavino, B.J.; et al. Distinct conformations of GPCR-beta-arrestin complexes mediate desensitization, signaling, and endocytosis. Proc. Natl. Acad. Sci. USA 2017, 114, 2562–2567. [Google Scholar] [CrossRef]
- Shukla, A.K.; Manglik, A.; Kruse, A.C.; Xiao, K.; Reis, R.I.; Tseng, W.-C.; Staus, D.P.; Hilger, D.; Uysal, S.; Huang, L.-Y.; et al. Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature 2013, 497, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.K.; Westfield, G.H.; Xiao, K.; Reis, R.I.; Huang, L.-Y.; Tripathi-Shukla, P.; Qian, J.; Li, S.; Blanc, A.; Oleskie, A.N.; et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature 2014, 512, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S.G.; DeVree, B.T.; Zou, Y.; Kruse, A.C.; Chung, K.Y.; Kobilka, T.S.; Thian, F.S.; Chae, P.S.; Pardon, E.; Calinski, D.; et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 2011, 477, 549–555. [Google Scholar] [CrossRef]
- Rasmussen, S.G.; Choi, H.-J.; Fung, J.J.; Pardon, E.; Casarosa, P.; Chae, P.S.; DeVree, B.T.; Rosenbaum, D.M.; Thian, F.S.; Kobilka, T.S.; et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 2011, 469, 175–180. [Google Scholar] [CrossRef]
- Staus, D.P.; Strachan, R.T.; Manglik, A.; Pani, B.; Kahsai, A.W.; Kim, T.H.; Wingler, L.M.; Ahn, S.; Chatterjee, A.; Masoudi, A.; et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature 2016, 535, 448–452. [Google Scholar] [CrossRef]
- Smyth, M.S.; Martin, J.H. X ray crystallography. Mol. Pathol. 2000, 53, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Zhou, X.E.; Gao, X.; He, Y.; Liu, W.; Ishchenko, A.; Barty, A.; White, T.A.; Yefanov, O.; Han, G.W.; et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 2015, 523, 561–567. [Google Scholar] [CrossRef]
- Callaway, E. Revolutionary cryo-EM is taking over structural biology. Nature 2020, 578, 201. [Google Scholar] [CrossRef] [PubMed]
- Sigworth, F.J. Principles of cryo-EM single-particle image processing. Microscopy 2016, 65, 57–67. [Google Scholar] [CrossRef]
- Wang, H.W.; Wang, J.W. How cryo-electron microscopy and X-ray crystallography complement each other. Protein Sci. 2017, 26, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.-L.; Khoshouei, M.; Radjainia, M.; Zhang, Y.; Glukhova, A.; Tarrasch, J.; Thal, D.M.; Furness, S.G.B.; Christopoulos, G.; Coudrat, T.; et al. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature 2017, 546, 118–123. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, B.; Feng, D.; Hu, H.; Chu, M.; Qu, Q.; Tarrasch, J.T.; Li, S.; Kobilka, T.S.; Kobilka, B.K.; et al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 2017, 546, 248–253. [Google Scholar] [CrossRef]
- Krishna Kumar, K.; Shalev-Benami, M.; Robertson, M.J.; Hu, H.; Banister, S.D.; Hollingsworth, S.A.; Latorraca, N.R.; Kato, H.E.; Hilger, D.; Maeda, S.; et al. Structure of a Signaling Cannabinoid Receptor 1-G Protein Complex. Cell 2019, 176, 448–458.e12. [Google Scholar] [CrossRef]
- Yang, F.; Guo, L.; Li, Y.; Wang, G.; Wang, J.; Zhang, C.; Fang, G.-X.; Chen, X.; Liu, L.; Yan, X.; et al. Structure, function and pharmacology of human itch receptor complexes. Nature 2021, 600, 164–169. [Google Scholar] [CrossRef]
- Xiao, P. Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes. Cell 2021, 184, 943–956.e18. [Google Scholar] [CrossRef]
- Zhao, L.-H.; Ma, S.; Sutkeviciute, I.; Shen, D.-D.; Zhou, X.E.; de Waal, P.W.; Li, C.-Y.; Kang, Y.; Clark, L.J.; Jean-Alphonse, F.G.; et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science 2019, 364, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Liu, Q.; Yuan, Q.; Ji, Y.; Zhu, S.; Tan, Y.; He, X.; Xu, Y.; Shi, J.; Cheng, X.; et al. Insights into divalent cation regulation and G13-coupling of orphan receptor GPR35. Cell Discov. 2022, 8, 135. [Google Scholar] [CrossRef]
- Seven, A.B.; Barros-Álvarez, X.; de Lapeyrière, M.; Papasergi-Scott, M.M.; Robertson, M.J.; Zhang, C.; Nwokonko, R.M.; Gao, Y.; Meyerowitz, J.G.; Rocher, J.-P.; et al. G-protein activation by a metabotropic glutamate receptor. Nature 2021, 595, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Barros-Álvarez, X.; Zhang, S.; Kim, K.; Dämgen, M.A.; Panova, O.; Suomivuori, C.-M.; Fay, J.F.; Zhong, X.; Krumm, B.E.; et al. Signaling snapshots of 5-HT2B serotonin receptor activated by the prototypical psychedelic LSD. Neuron 2022, 110, 3154–3167.e7. [Google Scholar] [CrossRef]
- Huang, W.; Masureel, M.; Qu, Q.; Janetzko, J.; Inoue, A.; Kato, H.E.; Robertson, M.J.; Nguyen, K.C.; Glenn, J.S.; Skiniotis, G.; et al. Structure of the neurotensin receptor 1 in complex with β-arrestin 1. Nature 2020, 579, 303–308. [Google Scholar] [CrossRef]
- Bous, J.; Fouillen, A.; Orcel, H.; Trapani, S.; Cong, X.; Fontanel, S.; Saint-Paul, J.; Lai-Kee-Him, J.; Urbach, S.; Sibille, N.; et al. Structure of the vasopressin hormone-V2 receptor-β-arrestin1 ternary complex. Sci. Adv. 2022, 8, eabo7761. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhang, C.; Lin, S.; Yan, X.; Cai, H.; Yi, C.; Ma, L.; Chu, X.; Liu, Y.; Zhu, Y.; et al. Tail engagement of arrestin at the glucagon receptor. Nature 2023, 620, 904–910. [Google Scholar] [CrossRef]
- Nguyen, A.; Thomsen, A.R.B.; Cahill, T.J.; Huang, R.; Huang, L.-Y.; Marcink, T.; Clarke, O.B.; Heissel, S.; Masoudi, A.; Ben-Hail, D.; et al. Structure of an Endosomal Signaling GPCR–G Protein–β-Arrestin Mega-Complex. Nat. Struct. Mol. Biol. 2019, 26, 1123–1131. [Google Scholar] [CrossRef]
- Chen, Q.; Plasencia, M.; Li, Z.; Mukherjee, S.; Patra, D.; Chen, C.-L.; Klose, T.; Yao, X.-Q.; Kossiakoff, A.A.; Chang, L.; et al. Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1. Nature 2021, 595, 600–605. [Google Scholar] [CrossRef]
- Duan, J.; Liu, H.; Zhao, F.; Yuan, Q.; Ji, Y.; Cai, X.; He, X.; Li, X.; Li, J.; Wu, K.; et al. GPCR activation and GRK2 assembly by a biased intracellular agonist. Nature 2023, 620, 676–681. [Google Scholar] [CrossRef]
- Hu, Y.; Cheng, K.; He, L.; Zhang, X.; Jiang, B.; Jiang, L.; Li, C.; Wang, G.; Yang, Y.; Liu, M. NMR-Based Methods for Protein Analysis. Anal. Chem. 2021, 93, 1866–1879. [Google Scholar] [CrossRef] [PubMed]
- Nygaard, R.; Zou, Y.; Dror, R.O.; Mildorf, T.J.; Arlow, D.H.; Manglik, A.; Pan, A.C.; Liu, C.W.; Fung, J.J.; Bokoch, M.P.; et al. The dynamic process of β2-adrenergic receptor activation. Cell 2013, 152, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Yu, X.; Liu, C.; Qu, C.-X.; Gong, Z.; Liu, H.-D.; Li, F.-H.; Wang, H.-M.; He, D.-F.; Yi, F.; et al. Phospho-selective mechanisms of arrestin conformations and functions revealed by unnatural amino acid incorporation and 19F-NMR. Nat. Commun. 2015, 6, 8202. [Google Scholar] [CrossRef]
- Gayen, A.; Goswami, S.K.; Mukhopadhyay, C. NMR evidence of GM1-induced conformational change of Substance P using isotropic bicelles. Biochim. Biophys. Act. 2011, 1808, 127–139. [Google Scholar] [CrossRef]
- Yang, Z.; Zeng, X.; Zhao, Y.; Chen, R. AlphaFold2 and its applications in the fields of biology and medicine. Signal Transduct. Target. Ther. 2023, 8, 115. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed]
- Siebenmorgen, T.Z.M. Computational prediction of protein–protein binding affinities. WIREs Comput. Mol. Sci. 2020, 10, e1448. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Huang, X.P.; Karpiak, J.; Kroeze, W.K.; Zhu, H.; Chen, X.; Moy, S.S.; Saddoris, K.A.; Nikolova, V.D.; Farrell, M.S.; Wang, S.; et al. Allosteric ligands for the pharmacologically dark receptors GPR68 and GPR65. Nature 2015, 527, 477–483. [Google Scholar] [CrossRef]
- Lansu, K.; Karpiak, J.; Liu, J.; Huang, X.-P.; McCorvy, J.D.; Kroeze, W.K.; Che, T.; Nagase, H.; I Carroll, F.; Jin, J.; et al. In silico design of novel probes for the atypical opioid receptor MRGPRX2. Nat. Chem. Biol. 2017, 13, 529–536. [Google Scholar] [CrossRef]
- Levit Kaplan, A.; Strachan, R.T.; Braz, J.M.; Craik, V.; Slocum, S.; Mangano, T.; Amabo, V.; O’donnell, H.; Lak, P.; Basbaum, A.I.; et al. Structure-Based Design of a Chemical Probe Set for the 5-HT5A Serotonin Receptor. J. Med. Chem. 2022, 65, 4201–4217. [Google Scholar] [CrossRef]
- Kaplan, A.L.; Confair, D.N.; Kim, K.; Barros-Álvarez, X.; Rodriguiz, R.M.; Yang, Y.; Kweon, O.S.; Che, T.; McCorvy, J.D.; Kamber, D.N.; et al. Bespoke library docking for 5-HT2A receptor agonists with antidepressant activity. Nature 2022, 610, 582–591. [Google Scholar] [CrossRef]
- Deng, H.; Jia, Y.; Zhang, Y. Protein structure prediction. Int. J. Mod. Phys. B 2018, 32, 1840009. [Google Scholar] [CrossRef]
- Bertoline LM, F.; Lima, A.N.; Krieger, J.E.; Teixeira, S.K. Before and after AlphaFold2: An overview of protein structure prediction. Front. Bioinform. 2023, 3, 1120370. [Google Scholar] [CrossRef]
- Dorn, M.; e Silva, M.B.; Buriol, L.S.; Lamb, L.C. Three-dimensional protein structure prediction: Methods and computational strategies. Comput. Biol. Chem. 2014, 53, 251–276. [Google Scholar] [CrossRef]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Protein structure prediction using multiple deep neural networks in the 13th Critical Assessment of Protein Structure Prediction (CASP13). Proteins 2019, 87, 1141–1148. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Chandra, A.; Tunnermann, L.; Lofstedt, T.; Gratz, R. Transformer-based deep learning for predicting protein properties in the life sciences. eLife 2023, 12, e82819. [Google Scholar] [CrossRef]
- McPartlon, M.; Xu, J. An end-to-end deep learning method for protein side-chain packing and inverse folding. Proc. Natl. Acad. Sci. USA 2023, 120, e2216438120. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Mahajan, S.P.; Sulam, J.; Gray, J.J. Deep Learning in Protein Structural Modeling and Design. Patterns 2020, 1, 100142. [Google Scholar] [CrossRef]
- Pandy-Szekeres, G.; Caroli, J.; Mamyrbekov, A.; A Kermani, A.; Keserű, G.M.; Kooistra, A.J.; E Gloriam, D. GPCRdb in 2023: State-specific structure models using AlphaFold2 and new ligand resources. Nucleic Acids Res. 2023, 51, D395–D402. [Google Scholar] [CrossRef]
- Lee, C.; Su, B.H.; Tseng, Y.J. Comparative studies of AlphaFold, RoseTTAFold and Modeller: A case study involving the use of G-protein-coupled receptors. Brief. Bioinform. 2022, 23, bbac308. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; Anishchenko, I.; Humphreys, I.R.; Cong, Q.; Baker, D.; DiMaio, F. Efficient and accurate prediction of protein structure using RoseTTAFold2. bioRxiv 2023. [Google Scholar] [CrossRef]
- Baek, M.; McHugh, R.; Anishchenko, I.; Jiang, H.; Baker, D.; DiMaio, F. Accurate prediction of protein–nucleic acid complexes using RoseTTAFoldNA. Nat. Methods 2024, 21, 117–121. [Google Scholar] [CrossRef]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; Smetanin, N.; Verkuil, R.; Kabeli, O.; Shmueli, Y.; et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 2023, 379, 1123–1130. [Google Scholar] [CrossRef]
- Ballante, F.; Kooistra, A.J.; Kampen, S.; de Graaf, C.; Carlsson, J. Structure-Based Virtual Screening for Ligands of G. Protein-Coupled Receptors: What Can Molecular Docking Do for You? Pharmacol. Rev. 2021, 73, 527–565. [Google Scholar] [CrossRef]
- Wang, C.; Chen, Y.; Zhang, Y.; Li, K.; Lin, M.; Pan, F.; Wu, W.; Zhang, J. A reinforcement learning approach for protein-ligand binding pose prediction. BMC Bioinform. 2022, 23, 368. [Google Scholar] [CrossRef]
- Pagadala, N.S.; Syed, K.; Tuszynski, J. Software for molecular docking: A review. Biophys. Rev. 2017, 9, 91–102. [Google Scholar] [CrossRef]
- Reddy, K.K.; Rathore, R.S.; Srujana, P.; Burri, R.R.; Reddy, C.R.; Sumakanth, M.; Reddanna, P.; Reddy, M.R. Performance Evaluation of Docking Programs- Glide, GOLD, AutoDock & SurflexDock, Using Free Energy Perturbation Reference Data: A Case Study of Fructose-1, 6-bisphosphatase-AMP Analogs. Mini Rev. Med. Chem. 2020, 20, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, S.M.; Shakil, S.; Haneef, M. A simple click by click protocol to perform docking: AutoDock 4.2 made easy for non-bioinformaticians. EXCLI J. 2013, 12, 831–857. [Google Scholar] [PubMed]
- Gimeno, A.; Ojeda-Montes, M.J.; Tomás-Hernández, S.; Cereto-Massagué, A.; Beltrán-Debón, R.; Mulero, M.; Pujadas, G.; Garcia-Vallvé, S. The Light and Dark Sides of Virtual Screening: What Is There to Know? Int. J. Mol. Sci. 2019, 20, 1375. [Google Scholar] [CrossRef]
- Alfonso-Prieto, M.; Navarini, L.; Carloni, P. Understanding Ligand Binding to G-Protein Coupled Receptors Using Multiscale Simulations. Front. Mol. Biosci. 2019, 6, 29. [Google Scholar] [CrossRef]
- Manglik, A.; Lin, H.; Aryal, D.K.; McCorvy, J.D.; Dengler, D.; Corder, G.; Levit, A.; Kling, R.C.; Bernat, V.; Hübner, H.; et al. Structure-based discovery of opioid analgesics with reduced side effects. Nature 2016, 537, 185–190. [Google Scholar] [CrossRef]
- Wang, S.; Wacker, D.; Levit, A.; Che, T.; Betz, R.M.; McCorvy, J.D.; Venkatakrishnan, A.J.; Huang, X.-P.; Dror, R.O.; Shoichet, B.K.; et al. D4 dopamine receptor high-resolution structures enable the discovery of selective agonists. Science 2017, 358, 381–386. [Google Scholar] [CrossRef]
- Fish, I.; Stößel, A.; Eitel, K.; Valant, C.; Albold, S.; Huebner, H.; Möller, D.; Clark, M.J.; Sunahara, R.K.; Christopoulos, A.; et al. Structure-Based Design and Discovery of New M2 Receptor Agonists. J. Med. Chem. 2017, 60, 9239–9250. [Google Scholar] [CrossRef]
- Korczynska, M.; Clark, M.J.; Valant, C.; Xu, J.; Von Moo, E.; Albold, S.; Weiss, D.R.; Torosyan, H.; Huang, W.; Kruse, A.C.; et al. Structure-based discovery of selective positive allosteric modulators of antagonists for the M2 muscarinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA 2018, 115, E2419–E2428. [Google Scholar] [CrossRef]
- Stein, R.M.; Kang, H.J.; McCorvy, J.D.; Glatfelter, G.C.; Jones, A.J.; Che, T.; Slocum, S.; Huang, X.-P.; Savych, O.; Moroz, Y.S.; et al. Virtual discovery of melatonin receptor ligands to modulate circadian rhythms. Nature 2020, 579, 609–614. [Google Scholar] [CrossRef]
- Fink, E.A.; Xu, J.; Hübner, H.; Braz, J.M.; Seemann, P.; Avet, C.; Craik, V.; Weikert, D.; Schmidt, M.F.; Webb, C.M.; et al. Structure-based discovery of nonopioid analgesics acting through the α2A-adrenergic receptor. Science 2022, 377, eabn7065. [Google Scholar] [CrossRef]
- Ujiantari, N.S.O.; Ham, S.; Nagiri, C.; Shihoya, W.; Nureki, O.; Hutchinson, D.S.; Schuster, D. Pharmacophore-guided Virtual Screening to Identify New β3-adrenergic Receptor Agonists. Mol. Inform. 2021, 41, e2100223. [Google Scholar] [CrossRef]
- Proj, M.; De Jonghe, S.; Van Loy, T.; Jukič, M.; Meden, A.; Ciber, L.; Podlipnik, Č.; Grošelj, U.; Konc, J.; Schols, D.; et al. A Set of Experimentally Validated Decoys for the Human CC Chemokine Receptor 7 (CCR7) Obtained by Virtual Screening. Front. Pharmacol. 2022, 13, 855653. [Google Scholar] [CrossRef]
- Jhinjharia, D.; Kaushik, A.C.; Sahi, S. A high-throughput structural dynamics approach for identification of potential agonists of FFAR4 for type 2 diabetes mellitus therapy. J. Biomol. Struct. Dyn. 2023, 43, 176–196. [Google Scholar] [CrossRef]
- Kampen, S.; Rodríguez, D.; Jørgensen, M.; Kruszyk-Kujawa, M.; Huang, X.; Collins, M.; Boyle, N.; Maurel, D.; Rudling, A.; Lebon, G.; et al. Structure-Based Discovery of Negative Allosteric Modulators of the Metabotropic Glutamate Receptor 5. ACS Chem. Biol. 2022, 17, 2744–2752. [Google Scholar] [CrossRef]
- Smith, S.T.; Cassada, J.B.; Von Bredow, L.; Erreger, K.; Webb, E.M.; Trombley, T.A.; Kalbfleisch, J.J.; Bender, B.J.; Zagol-Ikapitte, I.; Kramlinger, V.M.; et al. Discovery of Protease-Activated Receptor 4 (PAR4)-Tethered Ligand Antagonists Using Ultralarge Virtual Screening. ACS Pharmacol. Transl. Sci. 2024, 7, 1086–1100. [Google Scholar] [CrossRef] [PubMed]
- Garisetti, V.; Dhanabalan, A.K.; Dasararaju, G. Discovery of potential TAAR1 agonist targeting neurological and psychiatric disorders: An in silico approach. Int. J. Biol. Macromol. 2024, 264 Pt 2, 130528. [Google Scholar] [CrossRef]
- Xia, A.; Yong, X.; Zhang, C.; Lin, G.; Jia, G.; Zhao, C.; Wang, X.; Hao, Y.; Wang, Y.; Zhou, P.; et al. Cryo-EM structures of human GPR34 enable the identification of selective antagonists. Proc. Natl. Acad. Sci. USA 2023, 120, e2308435120. [Google Scholar] [CrossRef]
- Gupta, H.; Sahi, S. High-throughput virtual screening of potential inhibitors of GPR52 using docking and biased sampling method for Huntington’s disease therapy. Mol. Divers. 2024, 28, 3331–3347. [Google Scholar] [CrossRef]
- Garisetti, V.; Varughese, R.E.; Anandamurthy, A.; Haribabu, J.; Dasararaju, G. Exploring potential GPR55 agonists using virtual screening, molecular docking and dynamics simulation studies. J. Biomol. Struct. Dyn. 2024, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Garisetti, V.; Varughese, R.E.; Anandamurthy, A.; Haribabu, J.; Garrote, C.A.; Dasararaju, G. Virtual screening, molecular docking and dynamics simulation studies to identify potential agonists of orphan receptor GPR78 targeting CNS disorders. J. Recept. Signal Transduct. Res. 2024, 44, 82–96. [Google Scholar] [CrossRef] [PubMed]
- Garisetti, V.; Dhanabalan, A.K.; Dasararaju, G. Orphan receptor GPR88 as a potential therapeutic target for CNS disorders—An in silico approach. J. Biomol. Struct. Dyn. 2023, 42, 4745–4758. [Google Scholar] [CrossRef]
- Pallareti, L.; Rath, T.F.; Trapkov, B.; Tsonkov, T.; Nielsen, A.T.; Harpsøe, K.; Gentry, P.R.; Bräuner-Osborne, H.; Gloriam, D.E.; Foster, S.R. Pharmacological characterization of novel small molecule agonists and antagonists for the orphan receptor GPR139. Eur. J. Pharmacol. 2023, 943, 175553. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15--Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; de Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef]
- Grygorenko, O.O. Enamine Ltd.: The science and business of organic chemistry and beyond. Eur. J. Org. Chem. 2021, 2021, 6474–6477. [Google Scholar] [CrossRef]
- Torres PH, M.; Sodero AC, R.; Jofily, P.; Silva-Jr, F.P. Key Topics in Molecular Docking for Drug Design. Int. J. Mol. Sci. 2019, 20, 4574. [Google Scholar] [CrossRef]
- Karelina, M.; Noh, J.J.; Dror, R.O. How accurately can one predict drug binding modes using AlphaFold models? eLife 2023, 12, RP89386. [Google Scholar] [CrossRef]
- Terwilliger, T.C.; Liebschner, D.; Croll, T.I.; Williams, C.J.; McCoy, A.J.; Poon, B.K.; Afonine, P.V.; Oeffner, R.D.; Richardson, J.S.; Read, R.J.; et al. AlphaFold predictions are valuable hypotheses and accelerate but do not replace experimental structure determination. Nat. Methods 2024, 21, 110–116. [Google Scholar] [CrossRef]
- Lyu, J.; Kapolka, N.; Gumpper, R.; Alon, A.; Wang, L.; Jain, M.K.; Barros-Álvarez, X.; Sakamoto, K.; Kim, Y.; DiBerto, J.; et al. AlphaFold2 structures guide prospective ligand discovery. Science 2024, 384, eadn6354. [Google Scholar] [CrossRef]
- Krishna, R.; Wang, J.; Ahern, W.; Sturmfels, P.; Venkatesh, P.; Kalvet, I.; Lee, G.R.; Morey-Burrows, F.S.; Anishchenko, I.; Humphreys, I.R.; et al. Generalized biomolecular modeling and design with RoseTTAFold All-Atom. Science 2024, 384, eadl2528. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Qiao, Z.; Nie, W.; Vahdat, A.; Miller, T.F., III; Anandkumar, A. State-specific protein–ligand complex structure prediction with a multiscale deep generative model. Nat. Mach. Intell. 2024, 6, 195–208. [Google Scholar] [CrossRef]
- Callaway, E. Who will make AlphaFold3 open source? Scientists race to crack AI model. Nature 2024, 630, 14–15. [Google Scholar] [CrossRef]
- AlphaFold3—Why did Nature publish it without its code? Nature 2024, 629, 728. [CrossRef]
- Wankowicz, S.; Beltrao, P.; Cravatt, B.; Dunbrack, R.; Gitter, A.; Lindorff-Larsen, K.; Ovchinnikov, S.; Polizzi, N.; Shoichet, B.; Fraser, J. AlphaFold3 Transparency and Reproducibility. 2024. Available online: https://zenodo.org/records/11391920 (accessed on 1 February 2025).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chung, J.; Hahn, H.; Flores-Espinoza, E.; Thomsen, A.R.B. Artificial Intelligence: A New Tool for Structure-Based G Protein-Coupled Receptor Drug Discovery. Biomolecules 2025, 15, 423. https://doi.org/10.3390/biom15030423
Chung J, Hahn H, Flores-Espinoza E, Thomsen ARB. Artificial Intelligence: A New Tool for Structure-Based G Protein-Coupled Receptor Drug Discovery. Biomolecules. 2025; 15(3):423. https://doi.org/10.3390/biom15030423
Chicago/Turabian StyleChung, Jason, Hyunggu Hahn, Emmanuel Flores-Espinoza, and Alex R. B. Thomsen. 2025. "Artificial Intelligence: A New Tool for Structure-Based G Protein-Coupled Receptor Drug Discovery" Biomolecules 15, no. 3: 423. https://doi.org/10.3390/biom15030423
APA StyleChung, J., Hahn, H., Flores-Espinoza, E., & Thomsen, A. R. B. (2025). Artificial Intelligence: A New Tool for Structure-Based G Protein-Coupled Receptor Drug Discovery. Biomolecules, 15(3), 423. https://doi.org/10.3390/biom15030423





