Multi-Omics Analysis Reveals 1-Propanol-Induced Pentadecanoic Acid Biosynthesis in Yarrowia lipolytica
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strain
2.2. Cultivation Conditions
2.3. Determination of Biomass
2.4. Determination of Total Lipids and Fatty Acid Profiles
2.5. Transcriptomic Analysis
2.6. Untargeted Metabolomic Analysis
2.7. Integrated Transcriptomic and Metabolomic Analyses
2.8. RT-qPCR
2.9. Statistical Analysis
3. Results
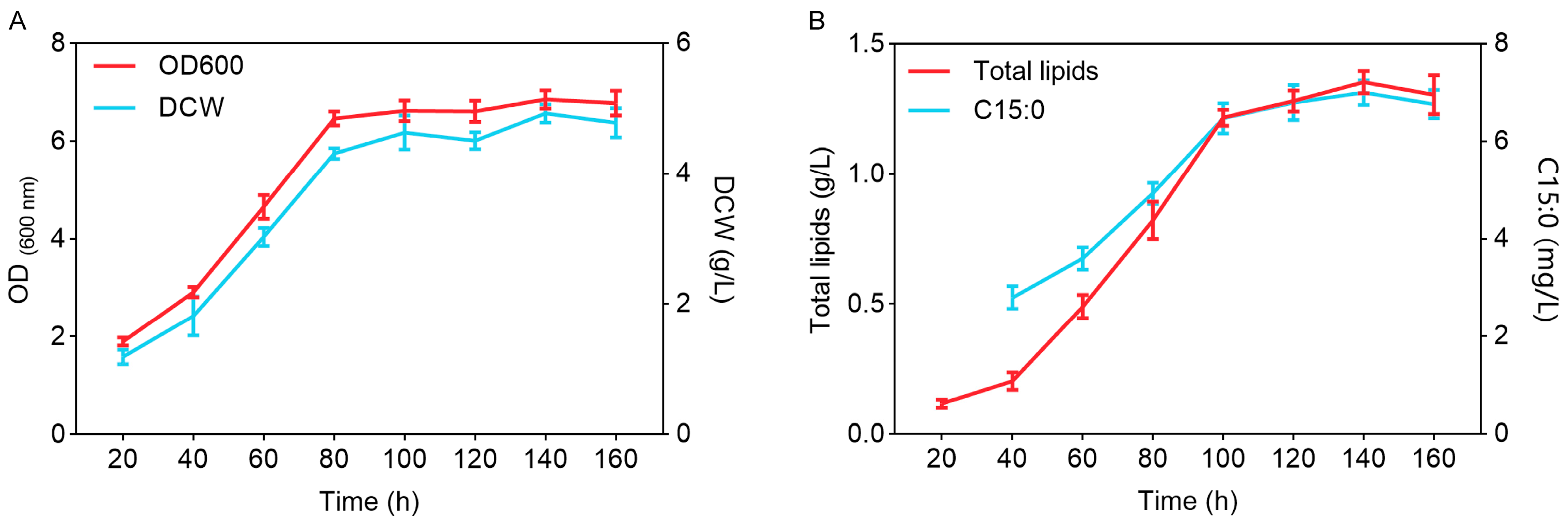
3.1. The Characteristics of Growth and Lipid Production of Y. lipolytica CICC1778
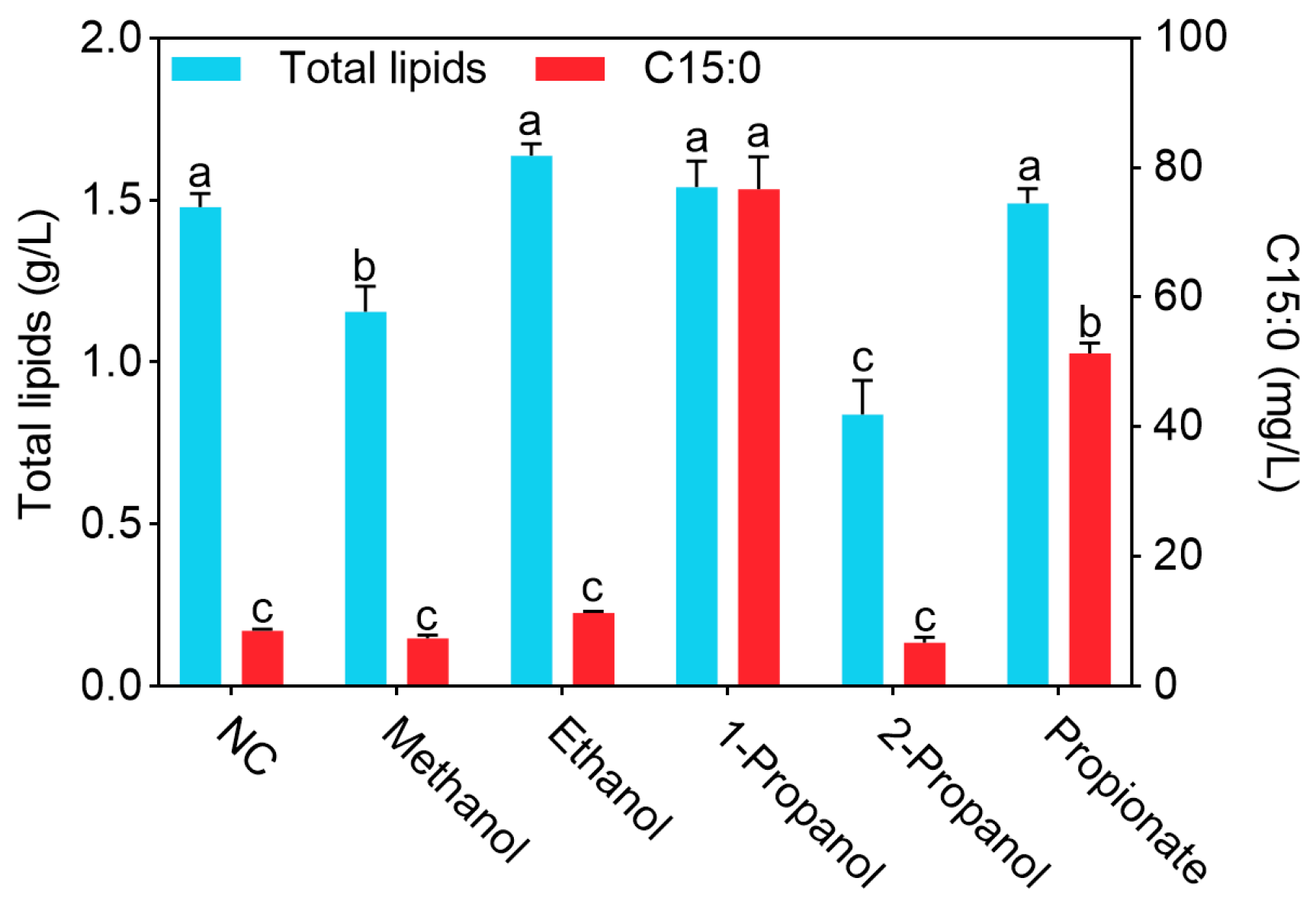
3.2. The Effects of Different Alcohols on the Growth and Lipid Production of Y. lipolytica CICC1778
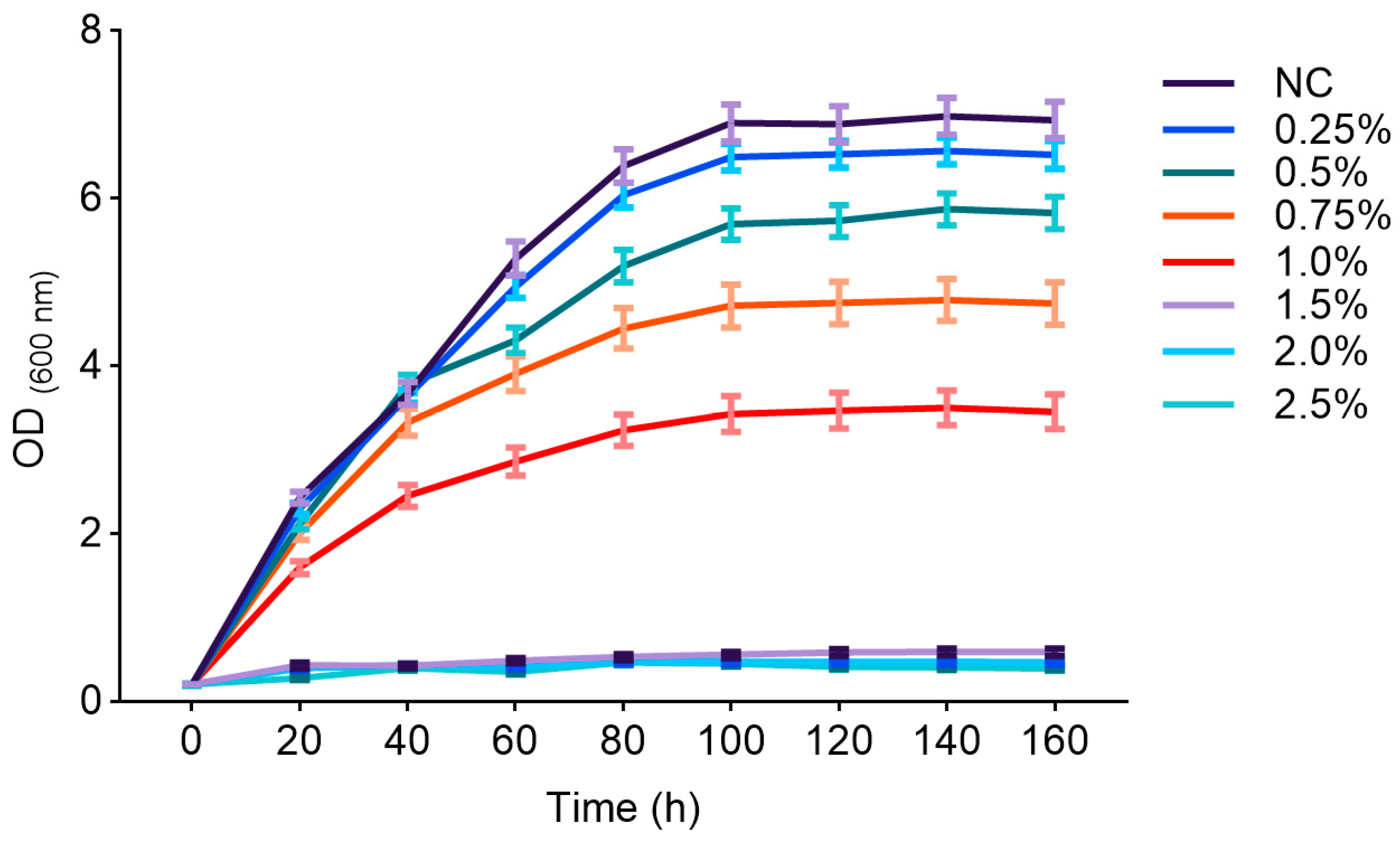
3.3. The Effects of Different Concentrations of 1-Propanol on the Growth and Lipid Production of Y. lipolytica CICC1778
3.4. Transcriptomic Analysis
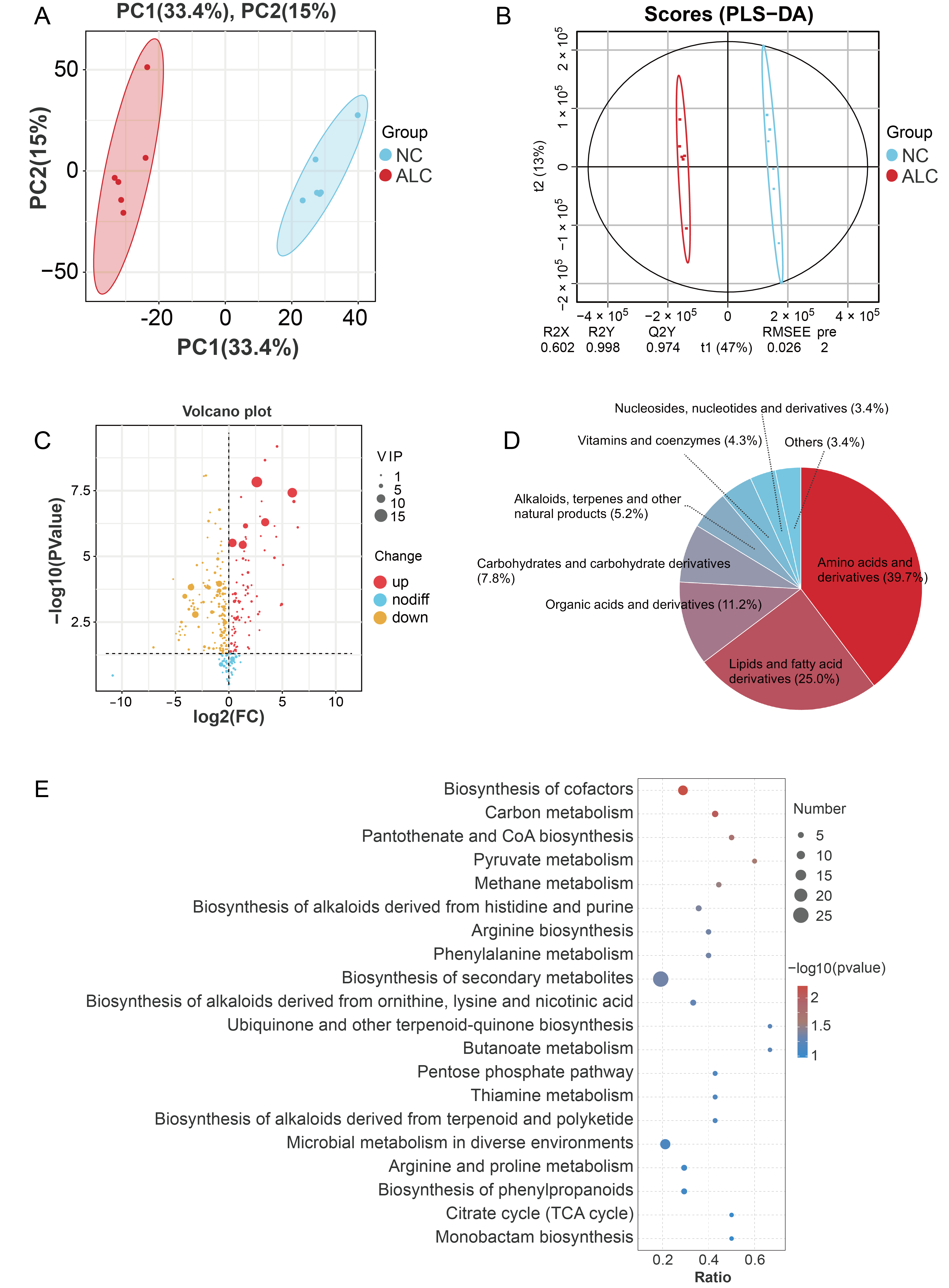
3.5. Untargeted Metabolomic Analysis
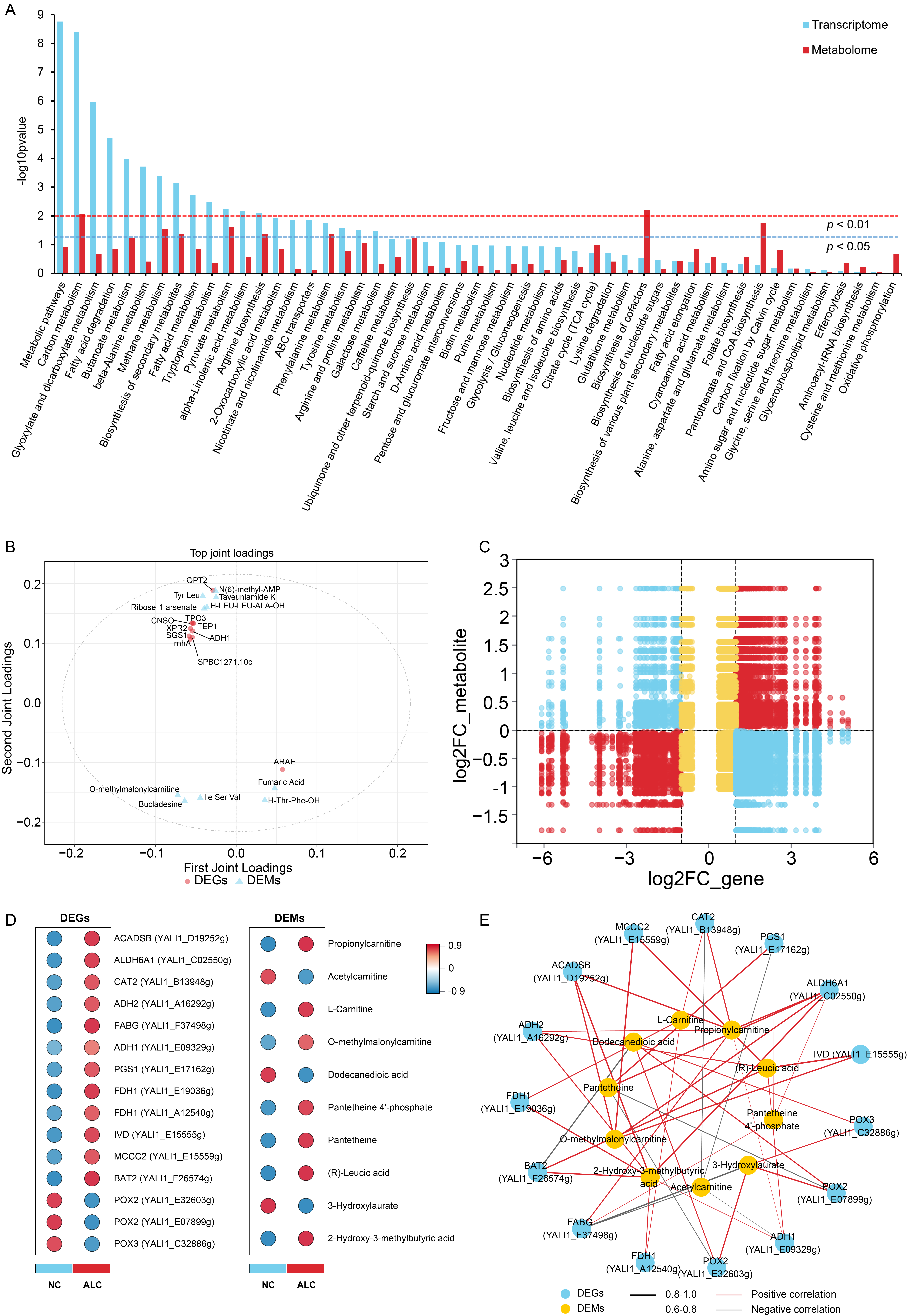
3.6. Integrated Transcriptomic and Metabolomic Analyses
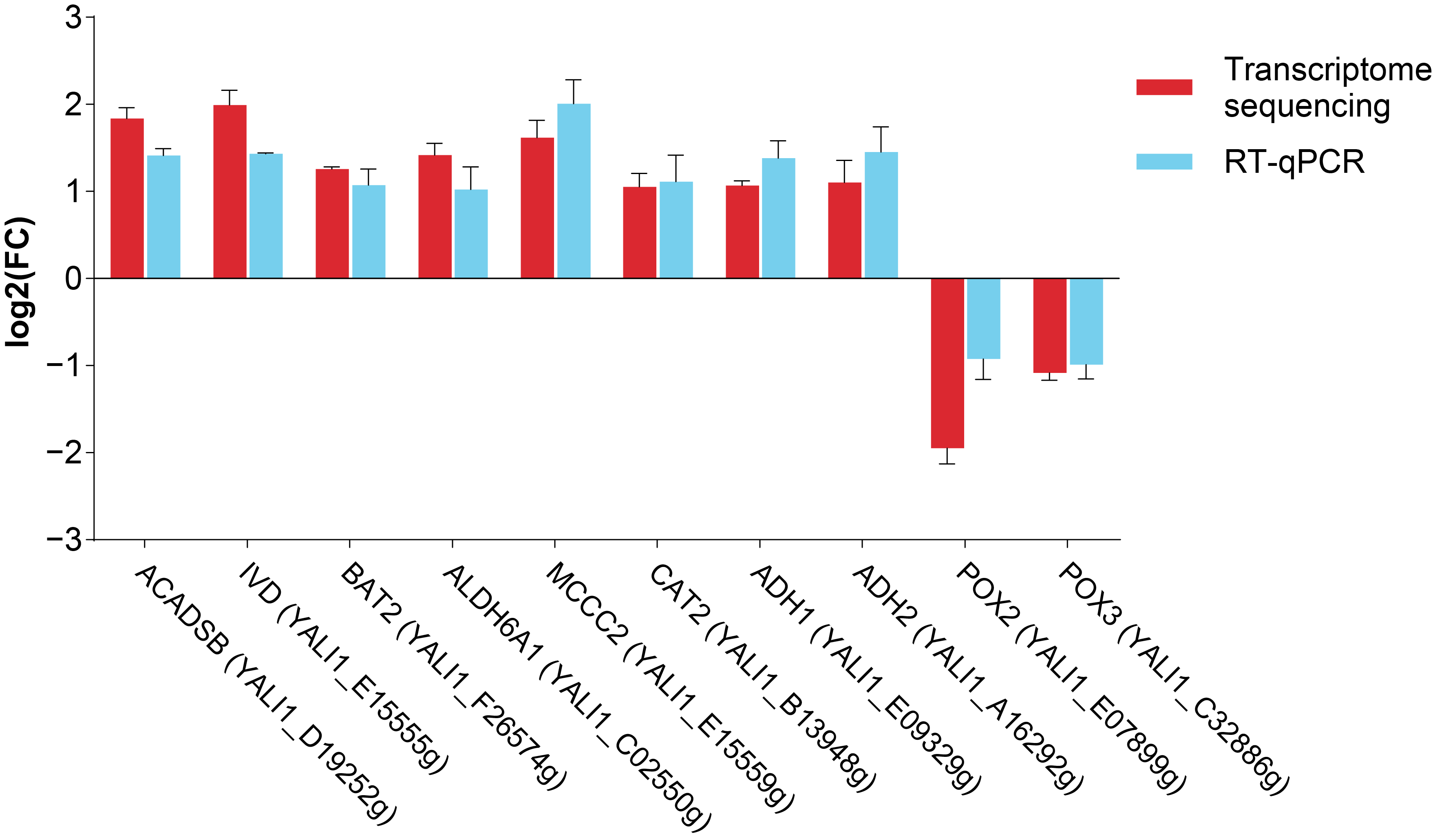
3.7. RT-qPCR Validation Analysis
4. Discussion
4.1. Effects of Different Alcohols on the Growth and Lipid Production of Y. lipolytica CICC1778
4.2. The Effects of Different Concentrations of 1-Propanol on the Growth and Lipid Production of Y. lipolytica CICC1778
4.3. Mechanism of C15:0 Synthetic Regulation by 1-Propanol in Y. lipolytica CICC1778
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Venn-Watson, S.; Lumpkin, R.; Dennis, E.A. Efficacy of dietary odd-chain saturated fatty acid pentadecanoic acid parallels broad associated health benefits in humans: Could it be essential? Sci. Rep. 2020, 10, 8161. [Google Scholar] [CrossRef]
- Carta, S.; Correddu, F.; Battacone, G.; Pulina, G.; Nudda, A. Comparison of Milk Odd- and Branched-Chain Fatty Acids among Human, Dairy Species and Artificial Substitutes. Foods 2022, 11, 4118. [Google Scholar] [CrossRef]
- Venn-Watson, S.; Schork, N.J. Pentadecanoic Acid (C15:0), an Essential Fatty Acid, Shares Clinically Relevant Cell-Based Activities with Leading Longevity-Enhancing Compounds. Nutrients 2023, 15, 4607. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, B.; West, J.A.; Koulman, A. A review of odd-chain fatty acid metabolism and the role of pentadecanoic Acid (c15:0) and heptadecanoic Acid (c17:0) in health and disease. Molecules 2015, 20, 2425–2444. [Google Scholar] [CrossRef]
- Zhang, L.S.; Liang, S.; Zong, M.H.; Yang, J.G.; Lou, W.Y. Microbial synthesis of functional odd-chain fatty acids: A review. World J. Microbiol. Biotechnol. 2020, 36, 35. [Google Scholar] [CrossRef]
- Qin, N.; Li, L.; Wang, Z.; Shi, S. Microbial production of odd-chain fatty acids. Biotechnol. Bioeng. 2023, 120, 917–931. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.K.; Dulermo, T.; Ledesma-Amaro, R.; Nicaud, J.M. Optimization of odd chain fatty acid production by Yarrowia lipolytica. Biotechnol. Biofuels 2018, 11, 158. [Google Scholar] [CrossRef]
- Zhang, L.S.; Xu, P.; Chu, M.Y.; Zong, M.H.; Yang, J.G.; Lou, W.Y. Using 1-propanol to significantly enhance the production of valuable odd-chain fatty acids by Rhodococcus opacus PD630. World J. Microbiol. Biotechnol. 2019, 35, 164. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.K.; Ledesma-Amaro, R.; Nicaud, J.M. De novo Biosynthesis of Odd-Chain Fatty Acids in Yarrowia lipolytica Enabled by Modular Pathway Engineering. Front. Bioeng. Biotechnol. 2019, 7, 484. [Google Scholar] [CrossRef]
- Zieniuk, B.; Fabiszewska, A. Yarrowia lipolytica: A beneficious yeast in biotechnology as a rare opportunistic fungal pathogen: A minireview. World J. Microbiol. Biotechnol. 2018, 35, 10. [Google Scholar] [CrossRef]
- Mamaev, D.; Zvyagilskaya, R. Yarrowia lipolytica: A multitalented yeast species of ecological significance. FEMS Yeast Res. 2021, 21, foab008. [Google Scholar] [CrossRef]
- Ullah, A.; Orij, R.; Brul, S.; Smits, G.J. Quantitative analysis of the modes of growth inhibition by weak organic acids in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2012, 78, 8377–8387. [Google Scholar] [CrossRef]
- Iwama, R.; Kobayashi, S.; Ohta, A.; Horiuchi, H.; Fukuda, R. Alcohol dehydrogenases and an alcohol oxidase involved in the assimilation of exogenous fatty alcohols in Yarrowia lipolytica. FEMS Yeast Res. 2015, 15, fov014. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Pir, P.; Kırdar, B.; Hayes, A.; Önsan, Z.İ.; Ülgen, K.Ö.; Oliver, S.G. Integrative investigation of metabolic and transcriptomic data. BMC Bioinform. 2006, 7, 203. [Google Scholar] [CrossRef][Green Version]
- Cavill, R.; Jennen, D.; Kleinjans, J.; Briedé, J.J. Transcriptomic and metabolomic data integration. Brief. Bioinform. 2016, 17, 891–901. [Google Scholar] [CrossRef] [PubMed]
- Pomraning, K.R.; Wei, S.; Karagiosis, S.A.; Kim, Y.M.; Dohnalkova, A.C.; Arey, B.W.; Bredeweg, E.L.; Orr, G.; Metz, T.O.; Baker, S.E. Comprehensive Metabolomic, Lipidomic and Microscopic Profiling of Yarrowia lipolytica during Lipid Accumulation Identifies Targets for Increased Lipogenesis. PLoS ONE 2015, 10, e0123188. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, Z.; Tian, J.; Miao, Z.; Liang, W.; Wang, G. Metabolome and Transcriptome Profiling Reveal Carbon Metabolic Flux Changes in Yarrowia lipolytica Cells to Rapamycin. J. Fungi 2022, 8, 939. [Google Scholar] [CrossRef]
- Back, A.; Rossignol, T.; Krier, F.; Nicaud, J.M.; Dhulster, P. High-throughput fermentation screening for the yeast Yarrowia lipolytica with real-time monitoring of biomass and lipid production. Microb. Cell Factories 2016, 15, 147. [Google Scholar] [CrossRef]
- Gajdoš, P.; Ledesma-Amaro, R.; Nicaud, J.M.; Čertík, M.; Rossignol, T. Overexpression of diacylglycerol acyltransferase in Yarrowia lipolytica affects lipid body size, number and distribution. FEMS Yeast Res. 2016, 16, fow062. [Google Scholar] [CrossRef]
- Liu, L.; Pan, A.; Spofford, C.; Zhou, N.; Alper, H.S. An evolutionary metabolic engineering approach for enhancing lipogenesis in Yarrowia lipolytica. Metab. Eng. 2015, 29, 36–45. [Google Scholar] [CrossRef]
- Kolouchová, I.; Schreiberová, O.; Sigler, K.; Masák, J.; Řezanka, T. Biotransformation of volatile fatty acids by oleaginous and non-oleaginous yeast species. FEMS Yeast Res. 2015, 15, fov076. [Google Scholar] [CrossRef]
- Yuan, M.; Breitkopf, S.B.; Yang, X.; Asara, J.M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012, 7, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shen, Z.; Yang, Z.; Jiang, H.; Chu, S.; Mao, Y.; Aboragah, A.; Loor, J.; Yang, Z. Abundance of solute carrier family 27 member 6 (SLC27A6) in the bovine mammary gland alters fatty acid metabolism. Food Funct. 2021, 12, 4909–4920. [Google Scholar] [CrossRef]
- Wilbanks, B.; Trinh, C.T. Comprehensive characterization of toxicity of fermentative metabolites on microbial growth. Biotechnol. Biofuels 2017, 10, 262. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mawgoud, A.M.; Markham, K.A.; Palmer, C.M.; Liu, N.; Stephanopoulos, G.; Alper, H.S. Metabolic engineering in the host Yarrowia lipolytica. Metab. Eng. 2018, 50, 192–208. [Google Scholar] [CrossRef]
- Beopoulos, A.; Chardot, T.; Nicaud, J.M. Yarrowia lipolytica: A model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie 2009, 91, 692–696. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Aggelis, G. Lipids of oleaginous yeasts. Part I: Biochemistry of single cell oil production. Eur. J. Lipid Sci. Technol. 2011, 113, 1031–1051. [Google Scholar] [CrossRef]
- Ratledge, C.; Wynn, J.P. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv. Appl. Microbiol. 2002, 51, 1–51. [Google Scholar] [CrossRef]
- Bonzanini, V.; Haddad Momeni, M.; Olofsson, K.; Olsson, L.; Geijer, C. Impact of glucose and propionic acid on even and odd chain fatty acid profiles of oleaginous yeasts. BMC Microbiol. 2025, 25, 79. [Google Scholar] [CrossRef] [PubMed]
- Huffer, S.; Clark, M.E.; Ning, J.C.; Blanch, H.W.; Clark, D.S. Role of alcohols in growth, lipid composition, and membrane fluidity of yeasts, bacteria, and archaea. Appl. Environ. Microbiol. 2011, 77, 6400–6408. [Google Scholar] [CrossRef]
- Xiao, Z.; Xiong, X.; Sun, Y.; Tourang, M.; Chen, S.; Tang, Y.J. Metabolic analyses of Yarrowia lipolytica for biopolymer production reveals roadblocks and strategies for microbial utilizing volatile fatty acids as sustainable feedstocks. Bioresour. Technol. 2025, 417, 131855. [Google Scholar] [CrossRef]
- Stanley, D.; Bandara, A.; Fraser, S.; Chambers, P.J.; Stanley, G.A. The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J. Appl. Microbiol. 2010, 109, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Dong, G.; Xu, S.; Li, L.; Shi, S. Engineering propionyl-CoA pools for de novo biosynthesis of odd-chain fatty acids in microbial cell factories. Crit. Rev. Biotechnol. 2023, 43, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Horswill, A.R.; Escalante-Semerena, J.C. Characterization of the propionyl-CoA synthetase (PrpE) enzyme of Salmonella enterica: Residue Lys592 is required for propionyl-AMP synthesis. Biochemistry 2002, 41, 2379–2387. [Google Scholar] [CrossRef]
- Il’chenko, A.; Chernyavskaya, O.; Shishkanova, N.; Finogenova, T. Metabolism of Yarrowia lipolytica Grown on Ethanol under Conditions Promoting the Production of α-Ketoglutaric and Citric Acids: A Comparative Study of the Central Metabolism Enzymes. Microbiology 2002, 71, 269–274. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, F.; Jiang, Y.; Yang, Q.; Yang, S.; Ma, J.; Xin, F.; Hasunuma, T.; Kondo, A.; Zhang, W.; et al. Improving methanol assimilation in Yarrowia lipolytica via systematic metabolic engineering combined with compartmentalization. Green Chem. 2022, 25, 183–195. [Google Scholar] [CrossRef]
- Mira, N.P.; Teixeira, M.C.; Sá-Correia, I. Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: A genome-wide view. OMICS A J. Integr. Biol. 2010, 14, 525–540. [Google Scholar] [CrossRef]
- van Rossum, H.M.; Kozak, B.U.; Niemeijer, M.S.; Dykstra, J.C.; Luttik, M.A.; Daran, J.M.; van Maris, A.J.; Pronk, J.T. Requirements for Carnitine Shuttle-Mediated Translocation of Mitochondrial Acetyl Moieties to the Yeast Cytosol. mBio 2016, 7, e00520-16. [Google Scholar] [CrossRef]
- Messina, E.; de Souza, C.P.; Cappella, C.; Barile, S.N.; Scarcia, P.; Pisano, I.; Palmieri, L.; Nicaud, J.M.; Agrimi, G. Genetic inactivation of the Carnitine/Acetyl-Carnitine mitochondrial carrier of Yarrowia lipolytica leads to enhanced odd-chain fatty acid production. Microb. Cell Factories 2023, 22, 128. [Google Scholar] [CrossRef]
- Longo, N.; Frigeni, M.; Pasquali, M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta 2016, 1863, 2422–2435. [Google Scholar] [CrossRef]
- van Roermund, C.W.; Hettema, E.H.; van den Berg, M.; Tabak, H.F.; Wanders, R.J. Molecular characterization of carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria in Saccharomyces cerevisiae and identification of a plasma membrane carnitine transporter, Agp2p. EMBO J. 1999, 18, 5843–5852. [Google Scholar] [CrossRef]
- Mutka, S.C.; Bondi, S.M.; Carney, J.R.; Da Silva, N.A.; Kealey, J.T. Metabolic pathway engineering for complex polyketide biosynthesis in Saccharomyces cerevisiae. FEMS Yeast Res. 2006, 6, 40–47. [Google Scholar] [CrossRef]
- Datta, S.; Annapure, U.S.; Timson, D.J. Different specificities of two aldehyde dehydrogenases from Saccharomyces cerevisiae var. boulardii. Biosci. Rep. 2017, 37, BSR20160529. [Google Scholar] [CrossRef]
- Dickinson, F.M.; Monger, G.P. A study of the kinetics and mechanism of yeast alcohol dehydrogenase with a variety of substrates. Biochem. J. 1973, 131, 261–270. [Google Scholar] [CrossRef]
- Xie, N.; Zhang, L.; Gao, W.; Huang, C.; Huber, P.E.; Zhou, X.; Li, C.; Shen, G.; Zou, B. NAD+ metabolism: Pathophysiologic mechanisms and therapeutic potential. Signal Transduct. Target. Ther. 2020, 5, 227. [Google Scholar] [CrossRef] [PubMed]
- Sophos, N.A.; Pappa, A.; Ziegler, T.L.; Vasiliou, V. Aldehyde dehydrogenase gene superfamily: The 2000 update. Chem.-Biol. Interact. 2001, 130–132, 323–337. [Google Scholar] [CrossRef]
- Plapp, B.V.; Leidal, K.G.; Murch, B.P.; Green, D.W. Contribution of liver alcohol dehydrogenase to metabolism of alcohols in rats. Chem.-Biol. Interact. 2015, 234, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Park, K.C.; Crump, N.T.; Louwman, N.; Krywawych, S.; Cheong, Y.J.; Vendrell, I.; Gill, E.K.; Gunadasa-Rohling, M.; Ford, K.L.; Hauton, D.; et al. Disrupted propionate metabolism evokes transcriptional changes in the heart by increasing histone acetylation and propionylation. Nat. Cardiovasc. Res. 2023, 2, 1221–1245. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Christopher, B.A.; Wilson, K.A.; Muoio, D.; McGarrah, R.W.; Brunengraber, H.; Zhang, G.F. Propionate-induced changes in cardiac metabolism, notably CoA trapping, are not altered by l-carnitine. Am. J. Physiol. Endocrinol. Metab. 2018, 315, E622–E633. [Google Scholar] [CrossRef]
- Marchuk, H.; Wang, Y.; Ladd, Z.A.; Chen, X.; Zhang, G.F. Pathophysiological mechanisms of complications associated with propionic acidemia. Pharmacol. Ther. 2023, 249, 108501. [Google Scholar] [CrossRef]
- Crown, S.B.; Marze, N.; Antoniewicz, M.R. Catabolism of Branched Chain Amino Acids Contributes Significantly to Synthesis of Odd-Chain and Even-Chain Fatty Acids in 3T3-L1 Adipocytes. PLoS ONE 2015, 10, e0145850. [Google Scholar] [CrossRef]
- Santos, L.P.A.; Assunção, L.D.P.; Lima, P.S.; Tristão, G.B.; Brock, M.; Borges, C.L.; Silva-Bailão, M.G.; Soares, C.M.A.; Bailão, A.M. Propionate metabolism in a human pathogenic fungus: Proteomic and biochemical analyses. IMA Fungus 2020, 11, 9. [Google Scholar] [CrossRef]
- Reger, A.S.; Carney, J.M.; Gulick, A.M. Biochemical and crystallographic analysis of substrate binding and conformational changes in acetyl-CoA synthetase. Biochemistry 2007, 46, 6536–6546. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Wang, R.S.; Handy, D.E.; Loscalzo, J. NAD(H) and NADP(H) Redox Couples and Cellular Energy Metabolism. Antioxid. Redox Signal. 2018, 28, 251–272. [Google Scholar] [CrossRef]
- Yang, R.; Guo, Z.; Li, B. NADH reductive stress drives metabolic reprogramming. Trends Cell Biol. 2025. [Google Scholar] [CrossRef]
- Neinast, M.; Murashige, D.; Arany, Z. Branched Chain Amino Acids. Annu. Rev. Physiol. 2019, 81, 139–164. [Google Scholar] [CrossRef]
- Henriques, B.J.; Katrine Jentoft Olsen, R.; Gomes, C.M.; Bross, P. Electron transfer flavoprotein and its role in mitochondrial energy metabolism in health and disease. Gene 2021, 776, 145407. [Google Scholar] [CrossRef]
- Grünert, S.C.; Stucki, M.; Morscher, R.J.; Suormala, T.; Bürer, C.; Burda, P.; Christensen, E.; Ficicioglu, C.; Herwig, J.; Kölker, S.; et al. 3-methylcrotonyl-CoA carboxylase deficiency: Clinical, biochemical, enzymatic and molecular studies in 88 individuals. Orphanet J. Rare Dis. 2012, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Alfardan, J.; Mohsen, A.W.; Copeland, S.; Ellison, J.; Keppen-Davis, L.; Rohrbach, M.; Powell, B.R.; Gillis, J.; Matern, D.; Kant, J.; et al. Characterization of new ACADSB gene sequence mutations and clinical implications in patients with 2-methylbutyrylglycinuria identified by newborn screening. Mol. Genet. Metab. 2010, 100, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Colón, M.; Hernández, F.; López, K.; Quezada, H.; González, J.; López, G.; Aranda, C.; González, A. Saccharomyces cerevisiae Bat1 and Bat2 aminotransferases have functionally diverged from the ancestral-like Kluyveromyces lactis orthologous enzyme. PLoS ONE 2011, 6, e16099. [Google Scholar] [CrossRef] [PubMed]
- Srirangan, K.; Bruder, M.; Akawi, L.; Miscevic, D.; Kilpatrick, S.; Moo-Young, M.; Chou, C.P. Recent advances in engineering propionyl-CoA metabolism for microbial production of value-added chemicals and biofuels. Crit. Rev. Biotechnol. 2017, 37, 701–722. [Google Scholar] [CrossRef] [PubMed]
- Mlícková, K.; Roux, E.; Athenstaedt, K.; d’Andrea, S.; Daum, G.; Chardot, T.; Nicaud, J.M. Lipid accumulation, lipid body formation, and acyl coenzyme A oxidases of the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 2004, 70, 3918–3924. [Google Scholar] [CrossRef] [PubMed]



| Fatty Acids (%) | Groups | |||||
|---|---|---|---|---|---|---|
| NC | Methanol | Ethanol | 1-Propanol | 2-Propanol | Propionate | |
| C15:0 | 0.58 ± 0.01 d | 0.64 ± 0.06 d | 0.71 ± 0.03 d | 4.89 ± 0.08 a | 0.90 ± 0.07 c | 3.53 ± 0.05 b |
| C16:0 | 15.39 ± 0.12 d | 18.50 ± 0.75 b | 16.57 ± 0.36 c | 16.18 ± 0.11 d | 21.85 ± 0.94 a | 12.35 ± 0.12 e |
| C16:1 | 22.40 ± 0.23 a | 19.99 ± 0.61 b | 21.68 ± 0.35 ab | 7.51 ± 0.08 d | 20.43 ± 0.80 b | 10.74 ± 0.09 c |
| C17:0 | 2.12 ± 0.06 d | 2.27 ± 0.11 d | 2.31 ± 0.03 d | 4.42 ± 0.04 a | 2.82 ± 0.15 c | 4.00 ± 0.06 b |
| C17:1 | 0.99 ± 0.01 d | 1.10 ± 0.08 d | 1.13 ± 0.03 c | 28.00 ± 0.17 b | 1.46 ± 0.11 c | 42.81 ± 0.16 a |
| C18:1 | 29.89 ± 0.34 a | 28.32 ± 0.75 b | 29.38 ± 0.43 ab | 21.68 ± 0.12 | 28.85 ± 0.65 ab | 13.81 ± 0.09 c |
| C18:2 | 28.60 ± 0.17 a | 29.07 ± 0.56 a | 28.77 ± 0.21 a | 14.69 ± 0.12 c | 23.45 ± 0.98 b | 11.54 ± 0.09 c |
| C19:1 | 0.03 ± 0.00 e | 0.11 ± 0.01 d | 0.12 ± 0.01 d | 2.64 ± 0.01 b | 0.24 ± 0.03 c | 1.22 ± 0.01 a |
| OCFAs | 3.72 ± 0.18 d | 4.13 ± 0.24 d | 4.27 ± 0.08 d | 39.95 ± 0.05 b | 5.42 ± 0.36 c | 51.56 ± 0.07 a |
| ECFAs | 96.28 ± 0.31 a | 95.88 ± 0.24 a | 95.73 ± 0.08 a | 60.06 ± 0.05 c | 94.58 ± 0.36 b | 48.44 ± 0.07 d |
| Fatty Acids (%) | Groups | ||||
|---|---|---|---|---|---|
| NC | 0.25% | 0.50% | 0.75% | 1.00% | |
| C15:0 | 0.58 ± 0.01 d | 2.61 ± 0.06 b | 4.89 ± 0.08 a | 5.12 ± 0.17 a | 5.49 ± 0.43 a |
| C16:0 | 15.39 ± 0.12 d | 15.74 ± 0.05 cd | 16.18 ± 0.11 c | 16.93 ± 0.26 b | 18.65 ± 0.32 a |
| C16:1 | 22.40 ± 0.23 a | 14.98 ± 0.07 b | 7.51 ± 0.08 c | 6.99 ± 0.12 d | 6.53 ± 0.23 e |
| C17:0 | 2.12 ± 0.06 d | 3.25 ± 0.04 d | 4.42 ± 0.04 c | 4.60 ± 0.07 b | 5.21 ± 0.06 a |
| C17:1 | 0.99 ± 0.01 d | 14.92 ± 0.14 c | 28.00 ± 0.17 b | 29.82 ± 0.56 a | 29.94 ± 0.55 a |
| C18:1 | 29.89 ± 0.34 a | 25.91 ± 0.11 b | 21.68 ± 0.12 c | 20.06 ± 0.38 d | 19.14 ± 0.69 d |
| C18:2 | 28.60 ± 0.17 a | 21.16 ± 0.11 b | 14.69 ± 0.12 c | 13.73 ± 0.39 c | 12.29 ± 0.57 d |
| C19:1 | 0.03 ± 0.00 e | 1.43 ± 0.01 b | 2.64 ± 0.01 a | 2.82 ± 0.04 a | 2.76 ± 0.08 a |
| OCFAs | 3.72 ± 0.18 d | 22.21 ± 0.23 c | 39.95 ± 0.05 b | 42.36 ± 0.83 a | 43.39 ± 0.94 a |
| ECFAs | 96.28 ± 0.31 a | 77.79 ± 0.23 b | 60.03 ± 0.05 c | 57.64 ± 0.83 d | 56.61 ± 0.94 d |
| Pathways | Count | p-Value | Genes |
|---|---|---|---|
| Valine, leucine and isoleucine degradation | 10 | <0.01 | ECHS1, MCCA, HMGCL, ALDH6A1, ACADSB, PAT1, IVD, MCCC2, BAT2, OXCT1 |
| Carbon metabolism | 11 | <0.01 | FDH1, ECHS1, ALDH6A1, POX3, ILV1, ACO1, MCSA, POX2, PAT1 |
| Propanoate metabolism | 5 | <0.01 | ECHS1, ALDH6A1, POX3, POX2 |
| Glyoxylate and dicarboxylate metabolism | 5 | <0.01 | FDH1, ACO1, MCSA, PAT1 |
| Fatty acid degradation | 7 | <0.01 | ADH2, ECHS1, POX3, ACADSB, POX2, PAT1, POX2 |
| Butanoate metabolism | 4 | <0.01 | ECHS1, HMGCL, PAT1, OXCT1 |
| Fatty acid metabolism | 7 | <0.01 | ECHS1, POX3, ACADSB, POX2, PAT1, FABG |
| Pyruvate metabolism | 3 | 0.01 | ADH2, PAT1, ACH1 |
| Peroxisome | 6 | 0.01 | CAT2, HMGCL, POX3, POX2, YAT1 |
| 2-Oxocarboxylic acid metabolism | 3 | 0.01 | ACO1, MCSA, BAT2 |
| Biosynthesis of unsaturated fatty acids | 3 | 0.04 | POX3, POX2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cong, J.; Hu, X.; Lu, D.; Kollie, S.C.; Elolimy, A.A.; Loor, J.J.; Yang, Z.; Li, M.; Mao, Y.; Yang, Z.; et al. Multi-Omics Analysis Reveals 1-Propanol-Induced Pentadecanoic Acid Biosynthesis in Yarrowia lipolytica. Biomolecules 2025, 15, 1618. https://doi.org/10.3390/biom15111618
Cong J, Hu X, Lu D, Kollie SC, Elolimy AA, Loor JJ, Yang Z, Li M, Mao Y, Yang Z, et al. Multi-Omics Analysis Reveals 1-Propanol-Induced Pentadecanoic Acid Biosynthesis in Yarrowia lipolytica. Biomolecules. 2025; 15(11):1618. https://doi.org/10.3390/biom15111618
Chicago/Turabian StyleCong, Jiahe, Xin Hu, Dongsheng Lu, Sam C. Kollie, Ahmed A. Elolimy, Juan J. Loor, Zhendong Yang, Mingxun Li, Yongjiang Mao, Zhangping Yang, and et al. 2025. "Multi-Omics Analysis Reveals 1-Propanol-Induced Pentadecanoic Acid Biosynthesis in Yarrowia lipolytica" Biomolecules 15, no. 11: 1618. https://doi.org/10.3390/biom15111618
APA StyleCong, J., Hu, X., Lu, D., Kollie, S. C., Elolimy, A. A., Loor, J. J., Yang, Z., Li, M., Mao, Y., Yang, Z., & Zhang, H. (2025). Multi-Omics Analysis Reveals 1-Propanol-Induced Pentadecanoic Acid Biosynthesis in Yarrowia lipolytica. Biomolecules, 15(11), 1618. https://doi.org/10.3390/biom15111618







