Screening and Validation of Functional Residues of the Antimicrobial Peptide PpRcys1
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. MD Simulations
2.3. Heterologous Expression and Purification of Recombina PpRcys1_RMRK (rPpRcys1_RMRK)
2.4. Analysis of Physical and Chemical Properties and Structure
2.5. Assay of Antimicrobial Activities of rPpRcys and rPpRcys1_RMRK
2.6. Binding Assay for Membrane Mimetic
2.7. Microorganism-Binding Assay
2.8. Membrane Permeability Assay
2.9. Assessment of Membrane Depolarization
2.10. Scanning Electron Microscopy (SEM)
2.11. Statistical Analysis
3. Results
3.1. Comparison of the Sequences and Structures of PpRcys1_RMRK and PpRcys1
3.2. Comparison of the Physical and Chemical Properties of PpRcys1_RMRK and PpRcys1
3.3. Recombinant Expression, Purification, and Identification of PpRcys_RMRK
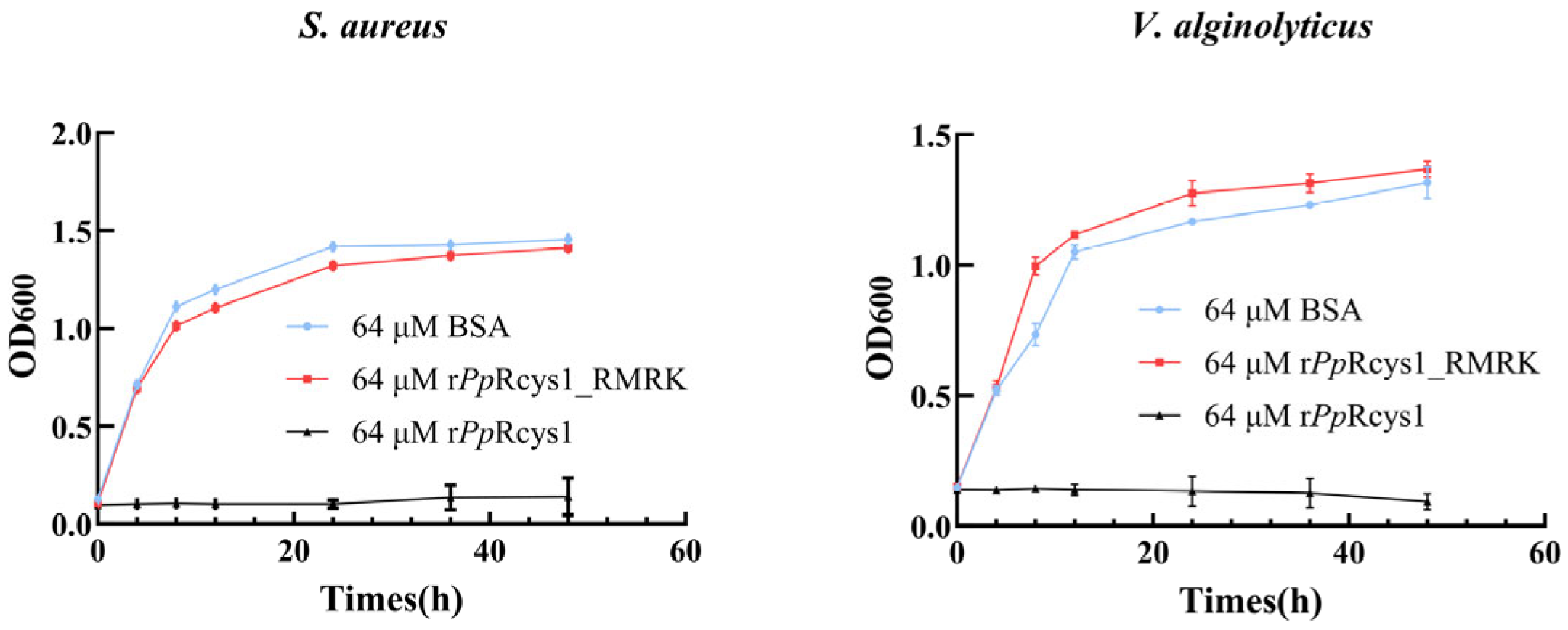
3.4. Comparison of the Antibacterial Activities of rPpRcys1_RMRK and rPpRcys1
3.5. Comparison of Membrane- and Microorganism-Binding Activities Between rPpRcys1_RMRK and rPpRcys1
3.6. Comparison of the Effects of rPpRcys1_RMRK and rPpRcys1 on Membrane Depolarization and Membrane Permeability
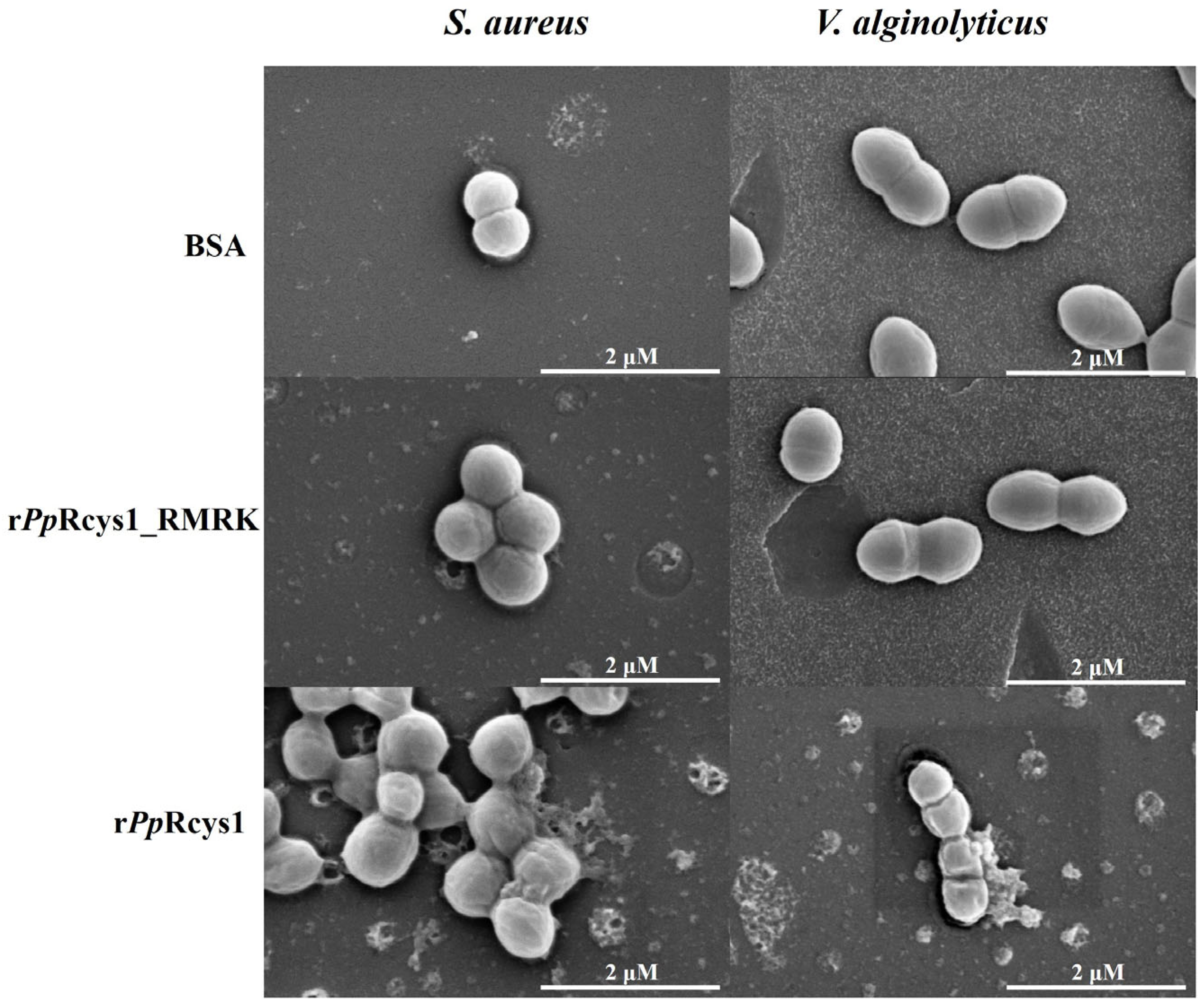
3.7. Comparison of the Effects of rPpRcys1_RMRK and rPpRcys1 on Bacterial Morphology
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manage, P.M. Heavy Use of Antibiotics in Aquaculture: Emerging Human and Animal Health Problems—A Review. Sri Lanka J. Aquat. Sci. 2018, 23, 13–27. [Google Scholar] [CrossRef]
- Rigos, G.; Kogiannou, D. Chapter 9—Antimicrobial Drugs in Aquaculture: Use and Abuse. In Present Knowledge in Food Safety; Knowles, M.E., Anelich, L.E., Boobis, A.R., Popping, B., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 142–161. ISBN 978-0-12-819470-6. [Google Scholar]
- Chen, P.; Ye, T.; Li, C.; Praveen, P.; Hu, Z.; Li, W.; Shang, C. Embracing the Era of Antimicrobial Peptides with Marine Organisms. Nat. Prod. Rep. 2024, 41, 331–346. [Google Scholar] [CrossRef]
- Li, C.; Warren, R.L.; Birol, I. Models and Data of AMPlify: A Deep Learning Tool for Antimicrobial Peptide Prediction. BMC Res. Notes 2023, 16, 11. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Oliveira Júnior, N.G.; Souza, C.M.; Buccini, D.F.; Cardoso, M.H.; Franco, O.L. Antimicrobial Peptides: Structure, Functions and Translational Applications. Nat. Rev. Microbiol. 2025, 23, 687–700. [Google Scholar] [CrossRef] [PubMed]
- Zou, F.; Tan, C.; Shinali, T.S.; Zhang, B.; Zhang, L.; Han, Z.; Shang, N. Plant Antimicrobial Peptides: A Comprehensive Review of Their Classification, Production, Mode of Action, Functions, Applications, and Challenges. Food Funct. 2023, 14, 5492–5515. [Google Scholar] [CrossRef] [PubMed]
- Barreto-Santamaría, A.; Patarroyo, M.E.; Curtidor, H. Designing and Optimizing New Antimicrobial Peptides: All Targets Are Not the Same. Crit. Rev. Clin. Lab. Sci. 2019, 56, 351–373. [Google Scholar] [CrossRef]
- Tan, P.; Lai, Z.; Zhu, Y.; Shao, C.; Akhtar, M.U.; Li, W.; Zheng, X.; Shan, A. Multiple Strategy Optimization of Specifically Targeted Antimicrobial Peptide Based on Structure–Activity Relationships to Enhance Bactericidal Efficiency. ACS Biomater. Sci. Eng. 2020, 6, 398–414. [Google Scholar] [CrossRef]
- Juba, M.L.; Porter, D.K.; Williams, E.H.; Rodriguez, C.A.; Barksdale, S.M.; Bishop, B.M. Helical Cationic Antimicrobial Peptide Length and Its Impact on Membrane Disruption. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 1081–1091. [Google Scholar] [CrossRef]
- Hall, K.; Lee, T.; Aguilar, M. The Role of Electrostatic Interactions in the Membrane Binding of Melittin. J. Mol. Recognit. 2011, 24, 108–118. [Google Scholar] [CrossRef]
- He, S.; Deber, C.M. Interaction of Designed Cationic Antimicrobial Peptides with the Outer Membrane of Gram-Negative Bacteria. Sci. Rep. 2024, 14, 1894. [Google Scholar] [CrossRef]
- Felsztyna, I.; Galassi, V.V.; Wilke, N. Selectivity of Membrane-Active Peptides: The Role of Electrostatics and Other Membrane Biophysical Properties. Biophys. Rev. 2025, 17, 591–604. [Google Scholar] [CrossRef]
- Olsen, J.V.; Ong, S.-E.; Mann, M. Trypsin Cleaves Exclusively C-Terminal to Arginine and Lysine Residues. Mol. Cell. Proteom. 2004, 3, 608–614. [Google Scholar] [CrossRef]
- Jiang, Z.; Kullberg, B.J.; Van Der Lee, H.; Vasil, A.I.; Hale, J.D.; Mant, C.T.; Hancock, R.E.W.; Vasil, M.L.; Netea, M.G.; Hodges, R.S. Effects of Hydrophobicity on the Antifungal Activity of A-helical Antimicrobial Peptides. Chem. Biol. Drug Des. 2008, 72, 483–495. [Google Scholar] [CrossRef]
- He, S.; Stone, T.A.; Deber, C.M. Uncoupling Amphipathicity and Hydrophobicity: Role of Charge Clustering in Membrane Interactions of Cationic Antimicrobial Peptides. Biochemistry 2021, 60, 2586–2592. [Google Scholar] [CrossRef]
- Yin, L.M.; Edwards, M.A.; Li, J.; Yip, C.M.; Deber, C.M. Roles of Hydrophobicity and Charge Distribution of Cationic Antimicrobial Peptides in Peptide-Membrane Interactions. J. Biol. Chem. 2012, 287, 7738–7745. [Google Scholar] [CrossRef]
- Ruiz, J.; Calderon, J.; Rondón-Villarreal, P.; Torres, R. Analysis of Structure and Hemolytic Activity Relationships of Antimicrobial Peptides (AMPs). In Advances in Computational Biology, Proceedings of the 2nd Colombian Congress on Computational Biology and Bioinformatics (CCBCOL), Manizales, Colombia, 25–27 September 2013; Springer: London, UK, 2014; pp. 253–258. [Google Scholar]
- Ciulla, M.G.; Gelain, F. Structure–Activity Relationships of Antibacterial Peptides. Microb. Biotechnol. 2023, 16, 757–777. [Google Scholar] [CrossRef]
- Ahn, H.; Cho, W.; Kang, S.-H.; Ko, S.-S.; Park, M.-S.; Cho, H.; Lee, K.-H. Design and Synthesis of Novel Antimicrobial Peptides on the Basis of α Helical Domain of Tenecin 1, an Insect Defensin Protein, and Structure–Activity Relationship Study. Peptides 2006, 27, 640–648. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, T.; Wei, D.; Strandberg, E.; Ulrich, A.S.; Ulmschneider, J.P. How Reliable Are Molecular Dynamics Simulations of Membrane Active Antimicrobial Peptides? Biochim. Biophys. Acta (BBA)-Biomembr. 2014, 1838, 2280–2288. [Google Scholar] [CrossRef]
- Ulmschneider, J.P.; Ulmschneider, M.B. Molecular Dynamics Simulations Are Redefining Our View of Peptides Interacting with Biological Membranes. Acc. Chem. Res. 2018, 51, 1106–1116. [Google Scholar] [CrossRef]
- Yuan, H.; Lyu, Y.; Cui, X.; Zhang, C.; Meng, Q. How Antimicrobial Peptide Indolicidin and Its Derivatives Interact with Phospholipid Membranes: Molecular Dynamics Simulation. J. Mol. Struct. 2024, 1312, 138625. [Google Scholar] [CrossRef]
- Nosé, S. A Molecular Dynamics Method for Simulations in the Canonical Ensemble. Mol. Phys. 1984, 52, 255–268. [Google Scholar] [CrossRef]
- Cao, Q.; Ge, C.; Wang, X.; Harvey, P.J.; Zhang, Z.; Ma, Y.; Wang, X.; Jia, X.; Mobli, M.; Craik, D.J. Designing Antimicrobial Peptides Using Deep Learning and Molecular Dynamic Simulations. Brief. Bioinform. 2023, 24, bbad058. [Google Scholar] [CrossRef]
- Sun, D.; Peyear, T.A.; Bennett, W.F.D.; Andersen, O.S.; Lightstone, F.C.; Ingólfsson, H.I. Molecular Mechanism for Gramicidin Dimerization and Dissociation in Bilayers of Different Thickness. Biophys. J. 2019, 117, 1831–1844. [Google Scholar] [CrossRef]
- Chen, R.; Mark, A.E. The Effect of Membrane Curvature on the Conformation of Antimicrobial Peptides: Implications for Binding and the Mechanism of Action. Eur. Biophys. J. 2011, 40, 545–553. [Google Scholar] [CrossRef]
- He, Z.; Fei, Z.; Shi, H.; Huang, M.; Wei, L.; Wang, J.; He, P.; Zhang, W. Heterologous Expression and Antimicrobial Mechanism of a Cysteine-Rich Peptide from Barnacle Pollicipes Pollicipes. Microorganisms 2025, 13, 1381. [Google Scholar] [CrossRef]
- Wan, H.; Li, Y.; Fan, Y.; Meng, F.; Chen, C.; Zhou, Q. A Site-Directed Mutagenesis Method Particularly Useful for Creating Otherwise Difficult-to-Make Mutants and Alanine Scanning. Anal. Biochem. 2012, 420, 163–170. [Google Scholar] [CrossRef]
- Etayash, H.; Azmi, S.; Dangeti, R.; Kaur, K. Peptide Bacteriocins-Structure Activity Relationships. Curr. Top. Med. Chem. 2016, 16, 220–241. [Google Scholar] [CrossRef]
- Avitabile, C.; Netti, F.; Orefice, G.; Palmieri, M.; Nocerino, N.; Malgieri, G.; D’Andrea, L.D.; Capparelli, R.; Fattorusso, R.; Romanelli, A. Design, Structural and Functional Characterization of a Temporin-1b Analog Active against Gram-Negative Bacteria. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2013, 1830, 3767–3775. [Google Scholar] [CrossRef]
- Zhang, W.; Wei, L.; Chen, P.; Ning, B.; Wang, J.; He, P.; Shang, C.; Yu, D. Discovery and Characterization of an Atypical Crustin Antimicrobial Peptide from Pollicipes Pollicipes. Mar. Drugs 2024, 22, 526. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell Jr, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain Χ1 and Χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- Murzyn, K.; Róg, T.; Pasenkiewicz-Gierula, M. Phosphatidylethanolamine-Phosphatidylglycerol Bilayer as a Model of the Inner Bacterial Membrane. Biophys. J. 2005, 88, 1091–1103. [Google Scholar] [CrossRef]
- Balatti, G.E.; Martini, M.F.; Pickholz, M. A Coarse-Grained Approach to Studying the Interactions of the Antimicrobial Peptides Aurein 1.2 and Maculatin 1.1 with POPG/POPE Lipid Mixtures. J. Mol. Model. 2018, 24, 208. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Bernetti, M.; Bussi, G. Pressure Control Using Stochastic Cell Rescaling. J. Chem. Phys. 2020, 153, 114107. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Homeyer, N.; Gohlke, H. Free Energy Calculations by the Molecular Mechanics Poisson− Boltzmann Surface Area Method. Mol. Inf. 2012, 31, 114–122. [Google Scholar] [CrossRef]
- Cramer, P. AlphaFold2 and the Future of Structural Biology. Nat. Struct. Mol. Biol. 2021, 28, 704–705. [Google Scholar]
- Cornett, J.B.; Shockman, G.D. Cellular Lysis of Streptococcus Faecalis Induced with Triton X-100. J. Bacteriol. 1978, 135, 153–160. [Google Scholar] [CrossRef]
- Sung, K.; Khan, S.A.; Nawaz, M.S.; Khan, A.A. A Simple and Efficient Triton X-100 Boiling and Chloroform Extraction Method of RNA Isolation from Gram-Positive and Gram-Negative Bacteria. FEMS Microbiol. Lett. 2003, 229, 97–101. [Google Scholar] [CrossRef]
- Te Winkel, J.D.; Gray, D.A.; Seistrup, K.H.; Hamoen, L.W.; Strahl, H. Analysis of Antimicrobial-Triggered Membrane Depolarization Using Voltage Sensitive Dyes. Front. Cell Dev. Biol. 2016, 4, 29. [Google Scholar] [CrossRef]
- Epand, R.F.; Pollard, J.E.; Wright, J.O.; Savage, P.B.; Epand, R.M. Depolarization, Bacterial Membrane Composition, and the Antimicrobial Action of Ceragenins. Antimicrob. Agents Chemother. 2010, 54, 3708–3713. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Chen, Y.; Yi, M.; Wang, Y.; Yao, L.; Ji, G.; Gao, Z. Identification of a Novel Antimicrobial Peptide from Amphioxus Ribosomal Protein L27. Fish. Shellfish Immunol. 2025, 157, 110063. [Google Scholar] [CrossRef]
- Li, Y.; Yu, J. Research Progress in Structure-Activity Relationship of Bioactive Peptides. J. Med. Food 2015, 18, 147–156. [Google Scholar] [CrossRef]
- Bakare, O.O.; Gokul, A.; Fadaka, A.O.; Wu, R.; Niekerk, L.-A.; Barker, A.M.; Keyster, M.; Klein, A. Plant Antimicrobial Peptides (PAMPs): Features, Applications, Production, Expression, and Challenges. Molecules 2022, 27, 3703. [Google Scholar] [CrossRef]
- Karagöl, A.; Karagöl, T.; Smorodina, E.; Zhang, S. Structural Bioinformatics Studies of Glutamate Transporters and Their AlphaFold2 Predicted Water-Soluble QTY Variants and Uncovering the Natural Mutations of L-> Q, I-> T, F-> Y and Q-> L, T-> I and Y-> F. PLoS ONE 2024, 19, e0289644. [Google Scholar] [CrossRef]
- Buel, G.R.; Walters, K.J. Can AlphaFold2 Predict the Impact of Missense Mutations on Structure? Nat. Struct. Mol. Biol. 2022, 29, 1–2. [Google Scholar] [CrossRef]
- Li, L.; Vorobyov, I.; Allen, T.W. The Different Interactions of Lysine and Arginine Side Chains with Lipid Membranes. J. Phys. Chem. B 2013, 117, 11906–11920. [Google Scholar] [CrossRef]
- Gisdon, F.J.; Bombarda, E.; Ullmann, G.M. Serine and Cysteine Peptidases: So Similar, yet Different. How the Active-Site Electrostatics Facilitates Different Reaction Mechanisms. J. Phys. Chem. B 2022, 126, 4035–4048. [Google Scholar] [CrossRef]
- Moreira, I.S.; Fernandes, P.A.; Ramos, M.J. Computational Alanine Scanning Mutagenesis—An Improved Methodological Approach. J. Comput. Chem. 2007, 28, 644–654. [Google Scholar] [CrossRef]
- Trevino, S.R.; Scholtz, J.M.; Pace, C.N. Amino Acid Contribution to Protein Solubility: Asp, Glu, and Ser Contribute More Favorably than the Other Hydrophilic Amino Acids in RNase Sa. J. Mol. Biol. 2007, 366, 449–460. [Google Scholar] [CrossRef]
- Mitaku, S.; Hirokawa, T.; Tsuji, T. Amphiphilicity Index of Polar Amino Acids as an Aid in the Characterization of Amino Acid Preference at Membrane–Water Interfaces. Bioinformatics 2002, 18, 608–616. [Google Scholar] [CrossRef]
- Conti, E.; Kuriyan, J. Crystallographic Analysis of the Specific yet Versatile Recognition of Distinct Nuclear Localization Signals by Karyopherin α. Structure 2000, 8, 329–338. [Google Scholar] [CrossRef]
- Sulea, T.; Hussack, G.; Ryan, S.; Tanha, J.; Purisima, E.O. Application of Assisted Design of Antibody and Protein Therapeutics (ADAPT) Improves Efficacy of a Clostridium Difficile Toxin A Single-Domain Antibody. Sci. Rep. 2018, 8, 2260. [Google Scholar] [CrossRef]
- Myung, Y.; Pires, D.E.V.; Ascher, D.B. MmCSM-AB: Guiding Rational Antibody Engineering through Multiple Point Mutations. Nucleic Acids Res. 2020, 48, W125–W131. [Google Scholar] [CrossRef]
- Huang, J.; Xie, X.; Zheng, W.; Xu, L.; Yan, J.; Wu, Y.; Yang, M.; Yan, Y. In Silico Design of Multipoint Mutants for Enhanced Performance of Thermomyces Lanuginosus Lipase for Efficient Biodiesel Production. Biotechnol. Biofuels Bioprod. 2024, 17, 33. [Google Scholar] [CrossRef]
- Dempsey, C.E. The Actions of Melittin on Membranes. Biochim. Biophys. Acta (BBA)-Rev. Biomembr. 1990, 1031, 143–161. [Google Scholar] [CrossRef]
- Elssner, A.; Duncan, M.; Gavrilin, M.; Wewers, M.D. A Novel P2X7 Receptor Activator, the Human Cathelicidin-Derived Peptide LL37, Induces IL-1β Processing and Release. J. Immunol. 2004, 172, 4987–4994. [Google Scholar]
- Torrent, M.; Andreu, D.; Nogués, V.M.; Boix, E. Connecting Peptide Physicochemical and Antimicrobial Properties by a Rational Prediction Model. PLoS ONE 2011, 6, e16968. [Google Scholar]
- Giangaspero, A.; Sandri, L.; Tossi, A. Amphipathic α Helical Antimicrobial Peptides. A Systematic Study of the Effects of Structural and Physical Properties on Biological Activity. Eur. J. Biochem. 2001, 268, 5589–5600. [Google Scholar]
- Travkova, O.G.; Moehwald, H.; Brezesinski, G. The Interaction of Antimicrobial Peptides with Membranes. Adv. Colloid Interface Sci. 2017, 247, 521–532. [Google Scholar] [CrossRef]
- Schmidtchen, A.; Pasupuleti, M.; Malmsten, M. Effect of Hydrophobic Modifications in Antimicrobial Peptides. Adv. Colloid Interface Sci. 2014, 205, 265–274. [Google Scholar] [CrossRef]
- Gagat, P.; Ostrówka, M.; Duda-Madej, A.; Mackiewicz, P. Enhancing Antimicrobial Peptide Activity through Modifications of Charge, Hydrophobicity, and Structure. Int. J. Mol. Sci. 2024, 25, 10821. [Google Scholar] [CrossRef]
- Schmidtchen, A.; Pasupuleti, M.; Mörgelin, M.; Davoudi, M.; Alenfall, J.; Chalupka, A.; Malmsten, M. Boosting Antimicrobial Peptides by Hydrophobic Oligopeptide End Tags. J. Biol. Chem. 2009, 284, 17584–17594. [Google Scholar] [CrossRef]
- Jindal, M.H.; Le, C.F.; Mohd Yusof, M.Y.; Sekaran, S.D. Net Charge, Hydrophobicity and Specific Amino Acids Contribute to the Activity of Antimicrobial Peptides. J. Health Transl. Med. 2014, 17, 1–7. [Google Scholar]
- Pirtskhalava, M.; Vishnepolsky, B.; Grigolava, M.; Managadze, G. Physicochemical Features and Peculiarities of Interaction of AMP with the Membrane. Pharmaceuticals 2021, 14, 471. [Google Scholar] [CrossRef]
- Espeche, J.C.; Varas, R.; Maturana, P.; Cutro, A.C.; Maffía, P.C.; Hollmann, A. Membrane Permeability and Antimicrobial Peptides: Much More than Just Making a Hole. Pept. Sci. 2024, 116, e24305. [Google Scholar] [CrossRef]
- Penyige, A.; Matkó, J.; Deák, E.; Bodnár, A.; Barabás, G. Depolarization of the Membrane Potential by β-Lactams as a Signal to Induce Autolysis. Biochem. Biophys. Res. Commun. 2002, 290, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xu, X.; Zhang, J.; Ye, T.; Zhou, Q.; Xu, Y.; Li, W.; Hu, Z.; Shang, C. Discovery and Characterization of a New Crustin Antimicrobial Peptide from Amphibalanus Amphitrite. Pharmaceutics 2022, 14, 413. [Google Scholar] [CrossRef] [PubMed]
- Lubec, G.; Afjehi-Sadat, L. Limitations and Pitfalls in Protein Identification by Mass Spectrometry. Chem. Rev. 2007, 107, 3568–3584. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, M.; Muhammad, I.; Cui, Q.; Zhang, H.; Jia, Y.; Xu, Q.; Kong, L.; Ma, H. An Antibacterial Peptide with High Resistance to Trypsin Obtained by Substituting D-Amino Acids for Trypsin Cleavage Sites. Antibiotics 2021, 10, 1465. [Google Scholar] [CrossRef]
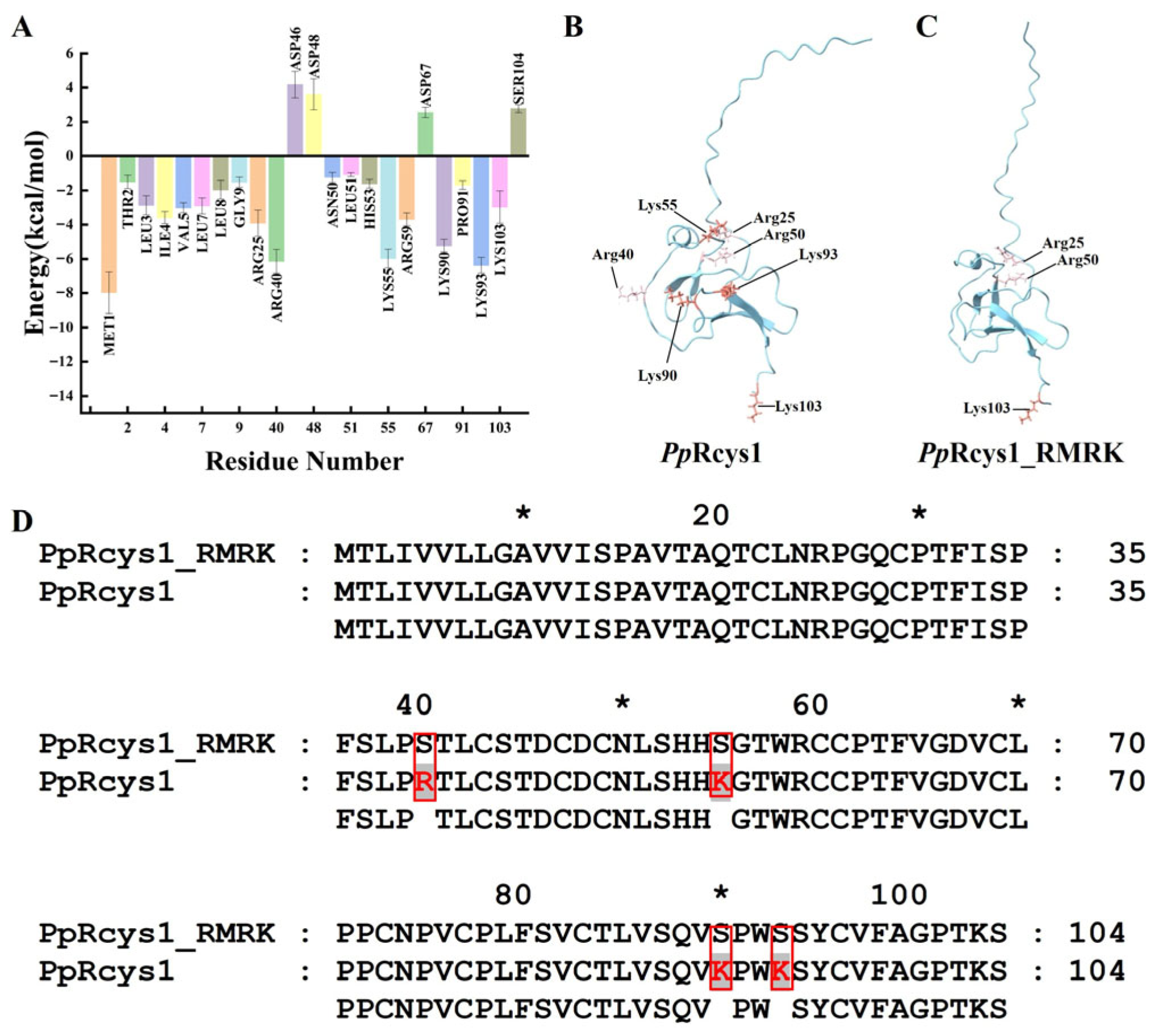




| Peptides | Molecular Weight (kDa) | Protein Isoelectric Point | Net Charge | Grand Average Hydropathy | Wimley–White Whole-Residue Hydrophobicity | Boman Index |
|---|---|---|---|---|---|---|
| PpRcys1 | 11.18 | 8.50 | +4.5 | 0.48 | 3.03 | 0.32 |
| PpRcys1_RMRK | 10.98 | 6.65 | +0.5 | 0.60 | −0.23 | 0.15 |
| Microorganism | Minimal Inhibitory Concentrations (μM) | |||
|---|---|---|---|---|
| rPpRcys1 | rPpRcys1_RMRK | Ampicillin | ||
| Gram-positive bacteria | S. aureus | 8 | - | 2 |
| Bacillus sp. T2 | 8 | - | ||
| S. agalactiae | 16 | - | 4 | |
| Gram-negative bacteria − | A. hydrophila | 32 | - | 128 |
| Acinetobacter sp. L32 | 32 | - | - | |
| E. coli | 16 | - | 64 | |
| V. alginolyticus | 16 | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, M.; Fei, Z.; Sun, A.; Yu, G.; Ye, H.; Shi, H.; Zhang, W.; Wang, J. Screening and Validation of Functional Residues of the Antimicrobial Peptide PpRcys1. Biomolecules 2025, 15, 1617. https://doi.org/10.3390/biom15111617
Tao M, Fei Z, Sun A, Yu G, Ye H, Shi H, Zhang W, Wang J. Screening and Validation of Functional Residues of the Antimicrobial Peptide PpRcys1. Biomolecules. 2025; 15(11):1617. https://doi.org/10.3390/biom15111617
Chicago/Turabian StyleTao, Ming, Zixun Fei, Aobo Sun, Guangming Yu, Huaiyuan Ye, Huishao Shi, Wei Zhang, and Junjian Wang. 2025. "Screening and Validation of Functional Residues of the Antimicrobial Peptide PpRcys1" Biomolecules 15, no. 11: 1617. https://doi.org/10.3390/biom15111617
APA StyleTao, M., Fei, Z., Sun, A., Yu, G., Ye, H., Shi, H., Zhang, W., & Wang, J. (2025). Screening and Validation of Functional Residues of the Antimicrobial Peptide PpRcys1. Biomolecules, 15(11), 1617. https://doi.org/10.3390/biom15111617





