Ecological History Shapes Transcriptome Variation in Quiescent Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Strains
2.2. Fractionation Assay
2.3. RNA Extraction and Purification from Quiescent Cells
2.4. Transcriptome Analysis
3. Results
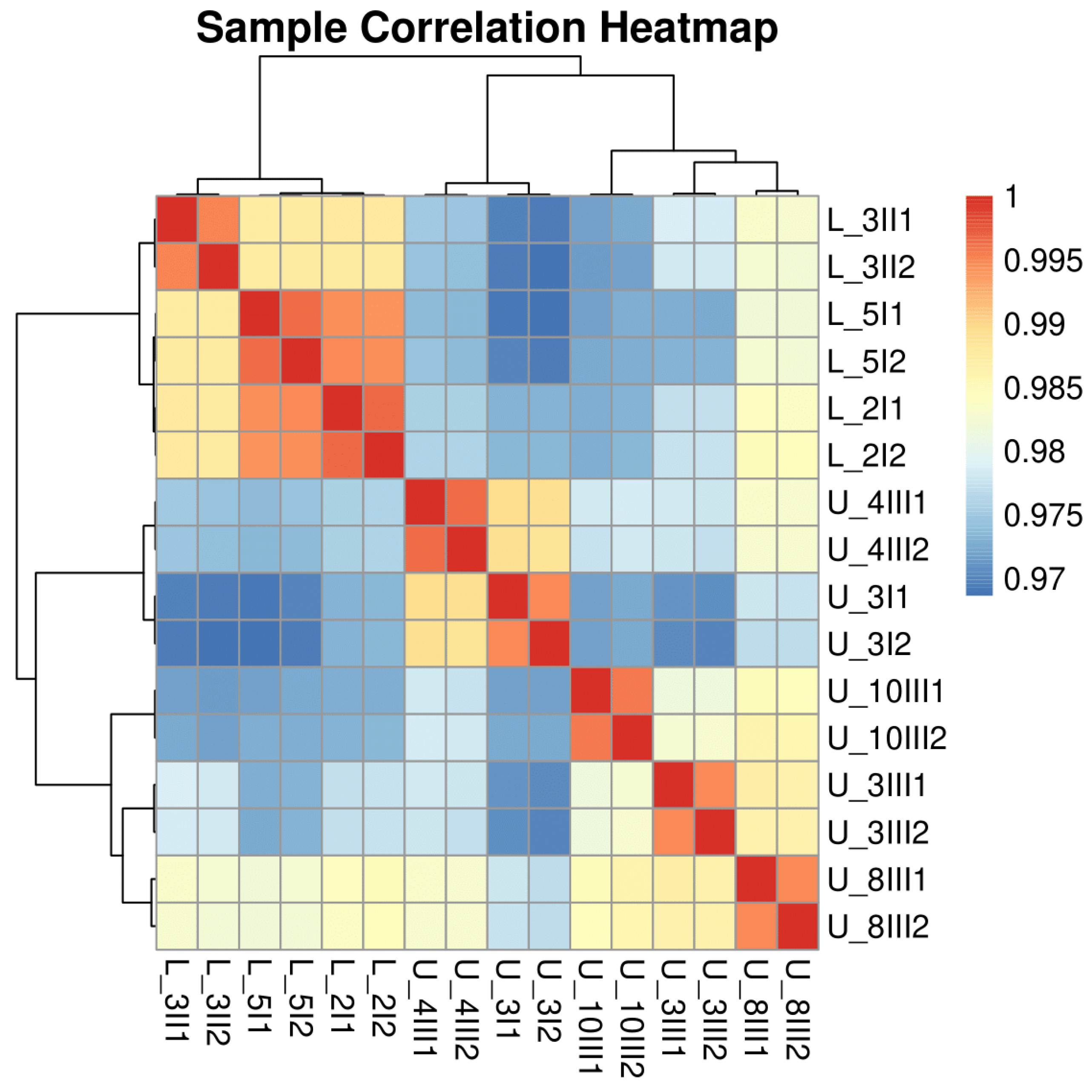
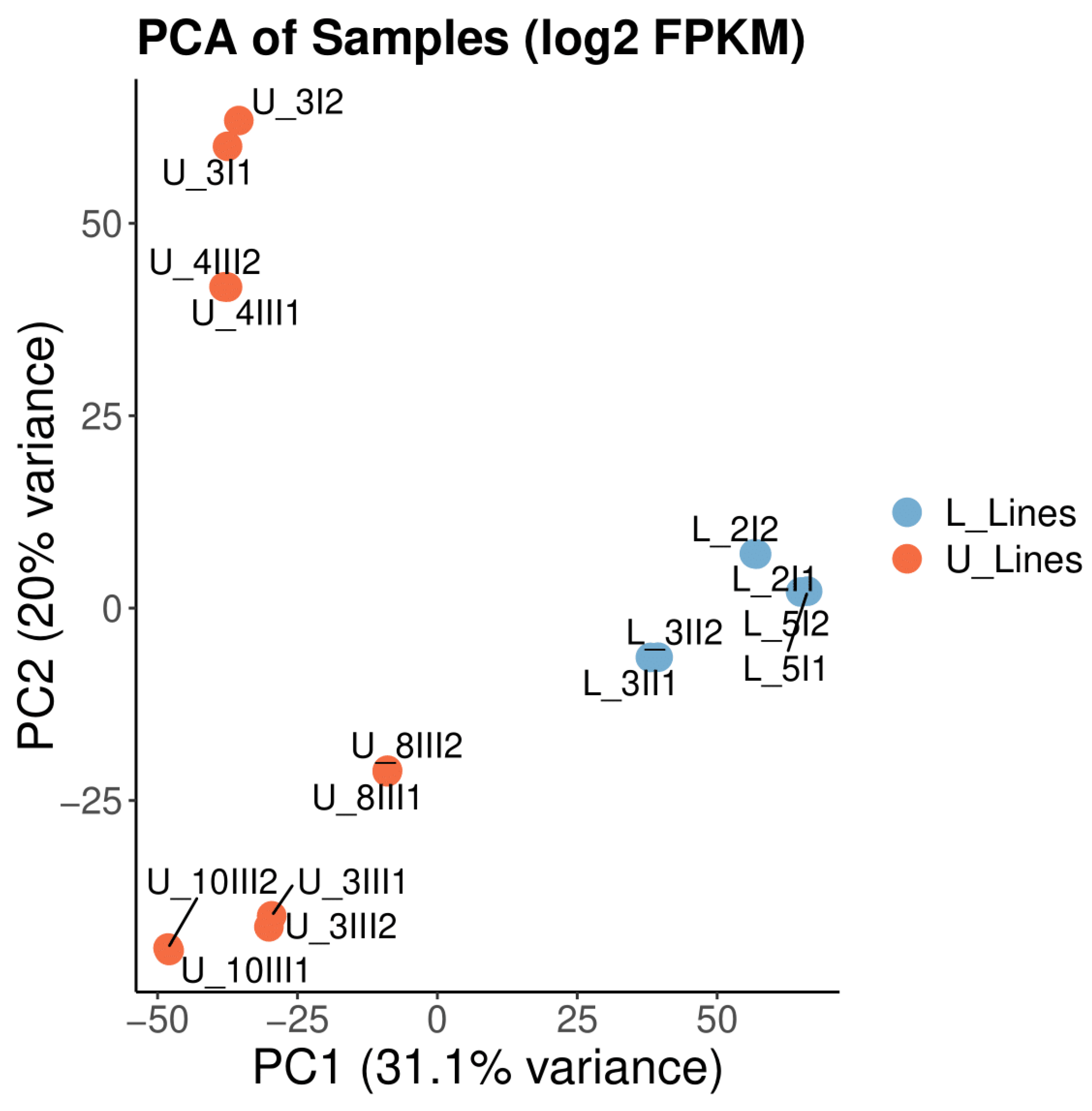
3.1. Data Quality
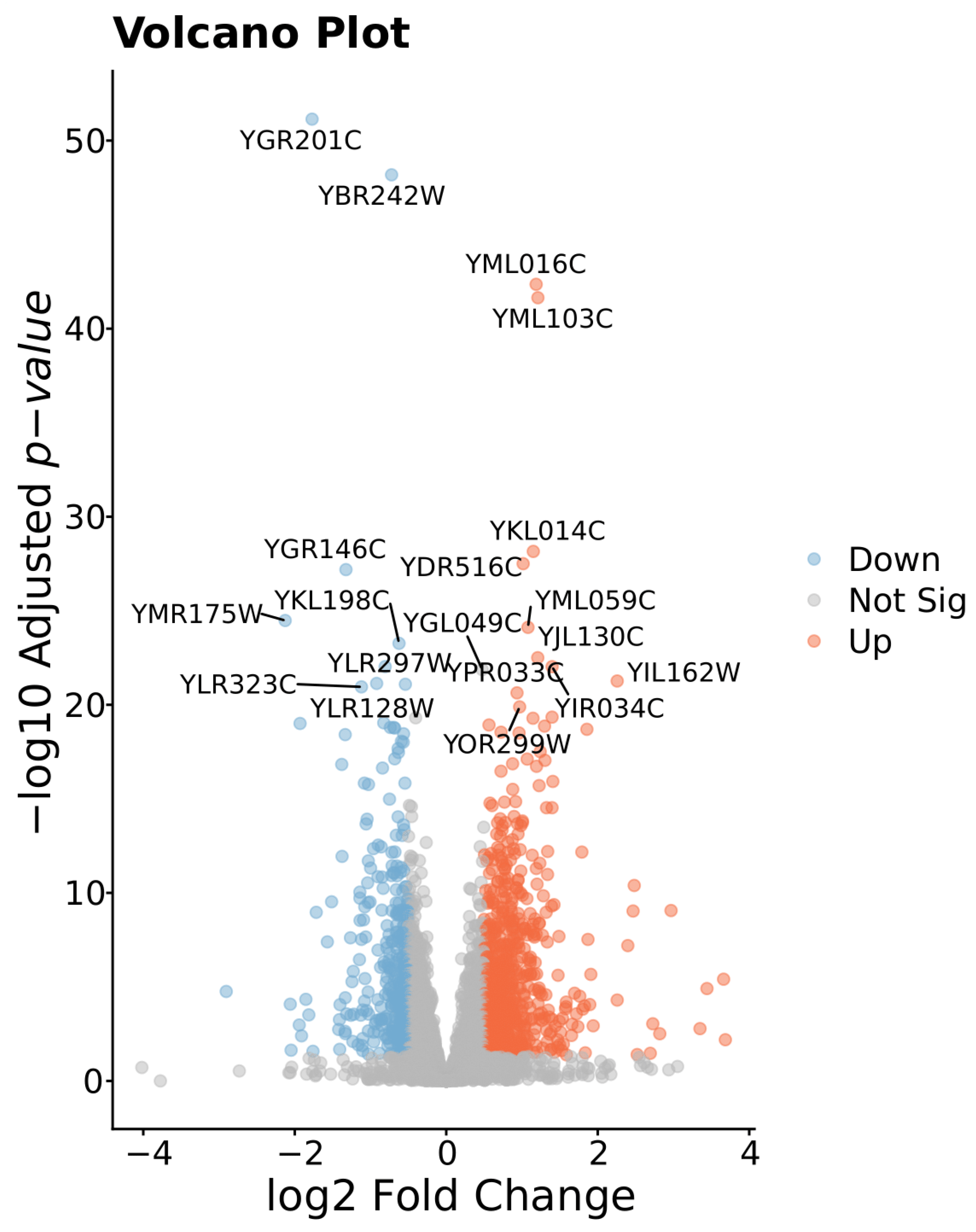
3.2. Differentially Expressed Genes
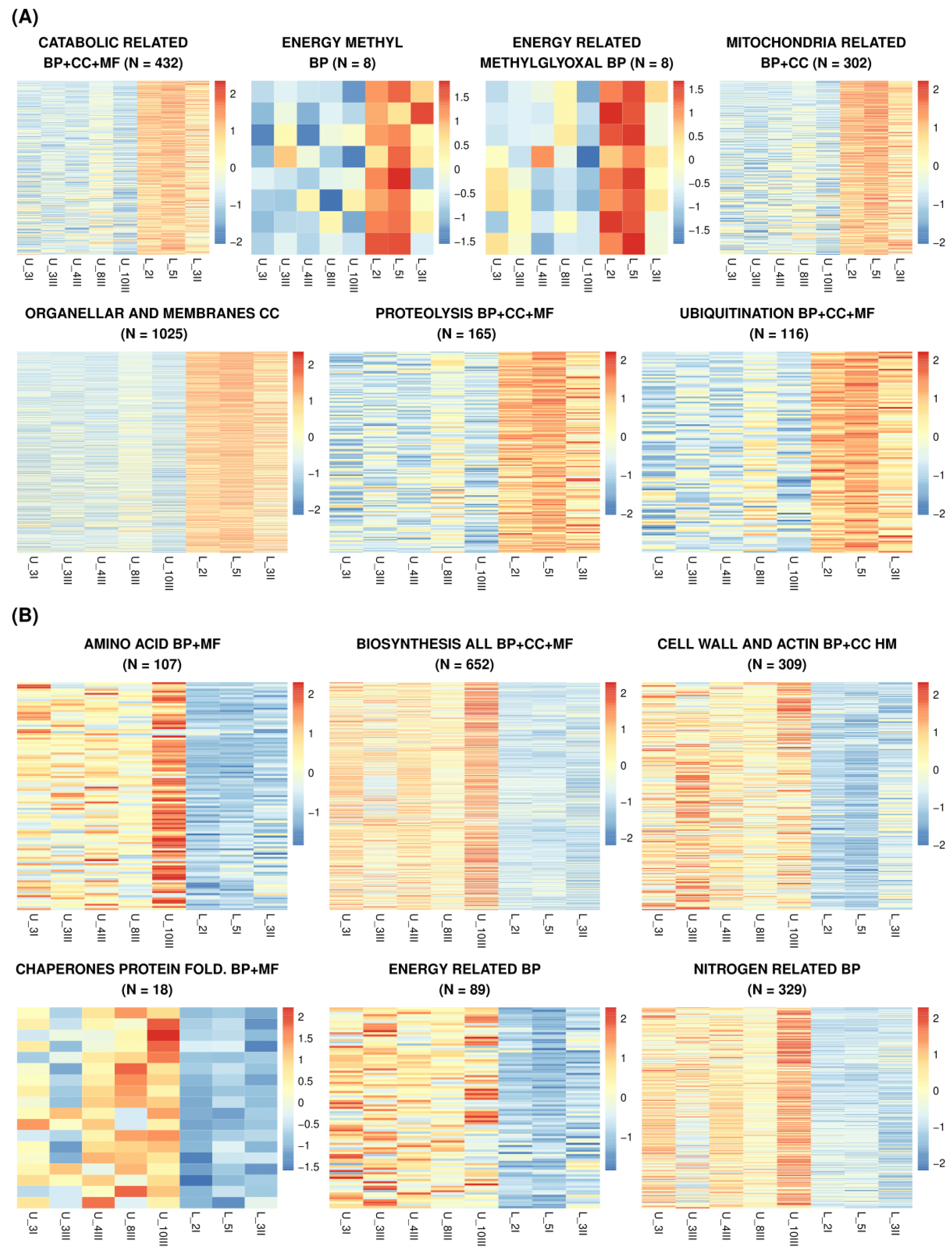
3.3. Ontology of the RNA-Seq Upregulated in L_ or U_line Genes Show Differences Between Quiescence Cells from Populations of Different Enrichment Histories
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Daignan-Fornier, B.; Laporte, D.; Sagot, I. Quiescence through the prism of evolution. Front. Cell Dev. Biol. 2021, 9, 745069. [Google Scholar] [CrossRef]
- Valcourt, J.R.; Lemons, J.M.; Haley, E.M.; Kojima, M.; Demuren, O.O.; Coller, H.A. Staying alive: Metabolic adaptations to quiescence. Cell Cycle 2012, 11, 1680–1696. [Google Scholar] [CrossRef] [PubMed]
- Bradley, J.A. Microbial dormancy as an ecological and biogeochemical regulator on Earth. Nat. Commun. 2025, 16, 3909. [Google Scholar] [CrossRef]
- Cheung, T.H.; Rando, T.A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 2013, 14, 329–340. [Google Scholar] [CrossRef]
- Coller, H.A.; Sang, L.; Roberts, J.M. A new description of cellular quiescence. PLoS Biol. 2006, 4, e83. [Google Scholar] [CrossRef] [PubMed]
- De Virgilio, C. The essence of yeast quiescence. FEMS Microbiol. Rev. 2012, 36, 306–339. [Google Scholar] [CrossRef] [PubMed]
- Mitra, M.; Batista, S.L.; Coller, H.A. Transcription factor networks in cellular quiescence. Nat. Cell Biol. 2025, 27, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Laporte, D.; Massoni-Laporte, A.; LEfranc, C.; Dompierre, J.; Mauboules, D.; Nsamba, E.T.; Royou, A.; Gal, L.; Schuldiner, M.; Gupta, M.L., Jr. A stable microtubule bundle formed through an orchestrated multistep process controls quiescence exit. eLife 2024, 12, RP89958. [Google Scholar] [CrossRef]
- Balayan, V.; Guddati, A.K. Tumor dormancy: Biologic and therapeutic implications. World J. Oncol. 2022, 13, 8. [Google Scholar] [CrossRef]
- Gray, J.V.; Petsko, G.A.; Johnston, G.C.; Ringe, D.; Singer, R.A.; Werner-Washburne, M. “Sleeping beauty”: Quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2004, 68, 187–206. [Google Scholar] [CrossRef]
- Allen, C.; Buttner, S.; Aragon, A.D.; Thomas, J.A.; Meirelles, O.; Jaetao, J.E.; Benn, D.; Ruby, S.W.; Veenhuis, M.; Madeo, F.; et al. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 2006, 174, 89–100. [Google Scholar] [CrossRef]
- Sun, S.; Gresham, D. Cellular quiescence in budding yeast. Yeast 2021, 38, 12–29. [Google Scholar] [CrossRef]
- Breeden, L.L.; Tsukiyama, T. Quiescence in Saccharomyces cerevisiae. Annu. Rev. Genet. 2022, 56, 253–278. [Google Scholar] [CrossRef]
- Opalek, M.; Smug, B.; Doebeli, M.; Wloch-Salamon, D. On the Ecological Significance of Phenotypic Heterogeneity in Microbial Populations Undergoing Starvation. Microbiol. Spectr. 2022, 10, e00450-21. [Google Scholar] [CrossRef] [PubMed]
- Sagot, I.; Laporte, D. The cell biology of quiescent yeast–a diversity of individual scenarios. J. Cell Sci. 2019, 132, jcs213025. [Google Scholar] [CrossRef] [PubMed]
- Opalek, M.; Tutaj, H.; Pirog, A.; Smug, B.J.; Rutkowska, J.; Wloch-Salamon, D. A Systematic Review on Quiescent State Research Approaches in S. cerevisiae. Cells 2023, 12, 1608. [Google Scholar] [CrossRef] [PubMed]
- Aragon, A.D.; Rodriguez, A.L.; Meirelles, O.; Roy, S.; Davidson, G.S.; Tapia, P.H.; Allen, C.; Joe, R.; Benn, D.; Werner-Washburne, M. Characterization of differentiated quiescent and nonquiescent cells in yeast stationary-phase cultures. Mol. Biol. Cell 2008, 19, 1271–1280. [Google Scholar] [CrossRef]
- Wloch-Salamon, D.M.; Tomala, K.; Aggeli, D.; Dunn, B. Adaptive roles of SSY1 and SIR3 during cycles of growth and starvation in Saccharomyces cerevisiae populations enriched for quiescent or nonquiescent cells. G3 Genes Genomes Genet. 2017, 7, 1899–1911. [Google Scholar] [CrossRef]
- Klosinska, M.M.; Crutchfield, C.A.; Bradley, P.H.; Rabinowitz, J.D.; Broach, J.R. Yeast cells can access distinct quiescent states. Genes Dev. 2011, 25, 336–349. [Google Scholar] [CrossRef]
- Cap, M.; Stepanek, L.; Harant, K.; Vachova, L.; Palkova, Z. Cell differentiation within a yeast colony: Metabolic and regulatory parallels with a tumor-affected organism. Mol. Cell 2012, 46, 436–448. [Google Scholar] [CrossRef]
- Greenlaw, A.C.; Alavattam, K.G.; Tsukiyama, T. Post-transcriptional regulation shapes the transcriptome of quiescent budding yeast. Nucleic Acids Res. 2024, 52, 1043–1063. [Google Scholar] [CrossRef]
- Marek, A.; Opalek, M.; Kałdon, A.; Mickowska, B.; Wloch-Salamon, D. Hypersensitive SSY1 mutations negatively influence transition to quiescence in yeast Saccharomyces cerevisiae. Yeast 2021, 38, 102–116. [Google Scholar] [CrossRef]
- Cubillos, F.A.; Louis, E.J.; Liti, G. Generation of a large set of genetically tractable haploid and diploid Saccharomyces strains. Fems Yeast Res. 2009, 9, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Azuma, K.; Ohtsuka, H.; Murakami, H.; Aiba, H. Extension of chronological lifespan by ScEcl1 depends on mitochondria in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2012, 76, 1938–1942. [Google Scholar] [CrossRef]
- Caligaris, M.; Nicastro, R.; Hu, Z.; Tripodi, F.; Hummel, J.E.; Pillet, B.; Deprez, M.-A.; Winderickx, J.; Rospert, S.; Coccetti, P.; et al. Snf1/AMPK fine-tunes TORC1 signaling in response to glucose starvation. eLife 2023, 12, e84319. [Google Scholar] [CrossRef]
- Smets, B.; Ghillebert, R.; De Snijder, P.; Binda, M.; Swinnen, E.; De Virgilio, C.; Winderickx, J. Life in the midst of scarcity: Adaptations to nutrient availability in Saccharomyces cerevisiae. Curr. Genet. 2010, 56, 1–32. [Google Scholar] [CrossRef]
- Kurz, T.; Ozlü, N.; Rudolf, F.; O’Rourke, S.M.; Luke, B.; Hofmann, K.; Hyman, A.A.; Bowerman, B.; Peter, M. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 2005, 435, 1257–1261. [Google Scholar] [CrossRef]
- Zhang, S.; Yu, Q.; Li, Z.; Zhao, Y.; Sun, Y. Protein neddylation and its role in health and diseases. Signal Transduct. Target. Ther. 2024, 9, 85. [Google Scholar] [CrossRef]
- Murakami, K.; Gibbons, B.J.; Davis, R.E.; Nagai, S.; Liu, X.; Robinson, P.J.; Wu, T.; Kaplan, C.D.; Kornberg, R.D. Tfb6, a previously unidentified subunit of the general transcription factor TFIIH, facilitates dissociation of Ssl2 helicase after transcription initiation. Proc. Natl. Acad. Sci. USA 2012, 109, 4816–4821. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Miles, S.; Melville, Z.; Prasad, A.; Bradley, G.; Breeden, L.L. Key events during the transition from rapid growth to quiescence in budding yeast require posttranscriptional regulators. Mol. Biol. Cell 2013, 24, 3697–3709. [Google Scholar] [CrossRef] [PubMed]
- Coltri, P.P.; Oliveira, C.C. Cwc24p is a general Saccharomyces cerevisiae splicing factor required for the stable U2 snRNP binding to primary transcripts. PLoS ONE 2012, 7, e45678. [Google Scholar] [CrossRef]
- Shen, W.; Gao, Z.; Chen, K.; Zhao, A.; Ouyang, Q.; Luo, C. The regulatory mechanism of the yeast osmoresponse under different glucose concentrations. iScience 2023, 26, 105809. [Google Scholar] [CrossRef]
- de Mendoza, D.; Pilon, M. Control of membrane lipid homeostasis by lipid-bilayer associated sensors: A mechanism conserved from bacteria to humans. Prog. Lipid. Res. 2019, 76, 100996. [Google Scholar] [CrossRef]
- Persson, S.; Welkenhuysen, N.; Shashkova, S.; Cvijovic, M. Fine-Tuning of Energy Levels Regulates SUC2 via a SNF1-Dependent Feedback Loop. Front. Physiol. 2020, 11, 954. [Google Scholar] [CrossRef]
- Yenush, L.; Mulet, J.M.; Ariño, J.; Serrano, R. The Ppz protein phosphatases are key regulators of K+ and pH homeostasis: Implications for salt tolerance, cell wall integrity and cell cycle progression. Embo J. 2002, 21, 920–929. [Google Scholar] [CrossRef]
- Yang, G.; Schmid-Siegel, M.; Heissenberger, C.; Kos-Braun, I.C.; Prechtl, M.; Meca-Laguna, G.; Rocha, M.; Wagner-Schrittwieser, A.; Pils, V.; Meixner, B.; et al. 2′-O-ribose methylation levels of ribosomal RNA distinguish different types of growth arrest in human dermal fibroblasts. J. Cell Sci. 2024, 137, jcs261930. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Barna, M. Specialized ribosomes: A new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012, 13, 355–369. [Google Scholar] [CrossRef]
- Ljungdahl, P.O.; Daignan-Fornier, B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 2012, 190, 885–929. [Google Scholar] [CrossRef] [PubMed]
- Shimoi, H.; Kitagaki, H.; Ohmori, H.; Iimura, Y.; Ito, K. Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. J. Bacteriol. 1998, 180, 3381–3387. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marek, A.; Tomala, K.; Wloch-Salamon, D. Ecological History Shapes Transcriptome Variation in Quiescent Saccharomyces cerevisiae. Biomolecules 2025, 15, 1588. https://doi.org/10.3390/biom15111588
Marek A, Tomala K, Wloch-Salamon D. Ecological History Shapes Transcriptome Variation in Quiescent Saccharomyces cerevisiae. Biomolecules. 2025; 15(11):1588. https://doi.org/10.3390/biom15111588
Chicago/Turabian StyleMarek, Agnieszka, Katarzyna Tomala, and Dominika Wloch-Salamon. 2025. "Ecological History Shapes Transcriptome Variation in Quiescent Saccharomyces cerevisiae" Biomolecules 15, no. 11: 1588. https://doi.org/10.3390/biom15111588
APA StyleMarek, A., Tomala, K., & Wloch-Salamon, D. (2025). Ecological History Shapes Transcriptome Variation in Quiescent Saccharomyces cerevisiae. Biomolecules, 15(11), 1588. https://doi.org/10.3390/biom15111588





