Mitochondrial Translation Inhibition Triggers an Rst2-Controlled Transcriptional Reprogramming of Carbon Metabolism in Stationary-Phase Cells of Fission Yeast
Abstract
1. Introduction
2. Materials and Methods
2.1. Fission Yeast Strains and Growth Media
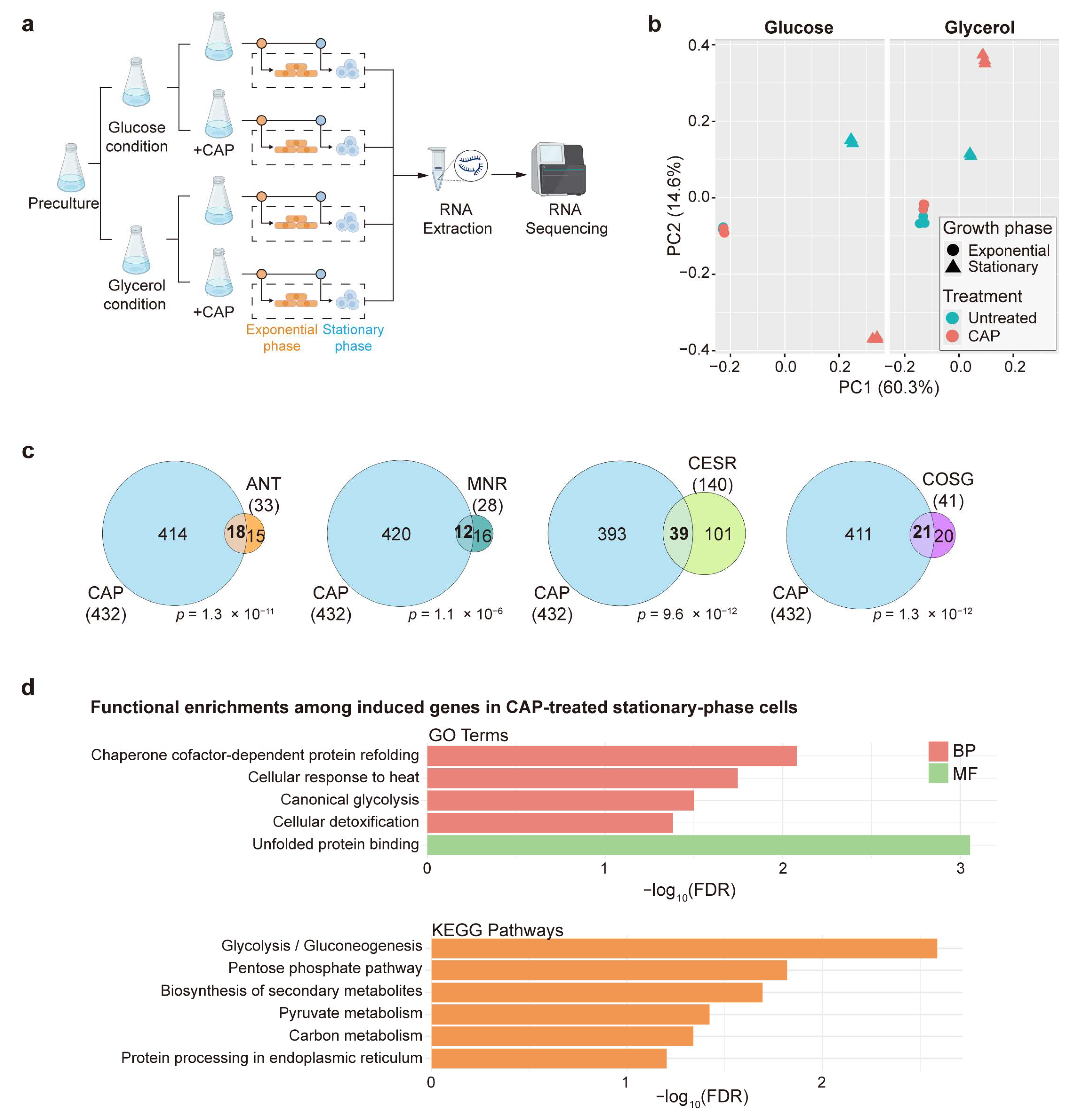
2.2. RNA-Sequencing Experiments
2.3. Quantitative Cell Growth Assays in Liquid Culture
2.4. Spot Assays
2.5. Chronological Lifespan (CLS) Assays
2.6. Synthetic Genetic Array (SGA) Analysis
2.7. Fluorescence Microscopy
3. Results
3.1. Mitochondrial Translation Inhibition Affects Genome Regulation Mainly During Stationary Phase
3.2. Mitochondrial Translation Inhibition During Stationary Phase Induces Genes Involved in the General Stress and Retrograde Responses
3.3. Mitochondrial Translation Inhibition During Stationary Phase Induces Carbon Metabolism Genes Functioning in Non-Mitochondrial Pathways
3.4. The Scr1 and Rst2 Transcription Factors Regulate Common Metabolic Genes upon Inhibition of Mitochondrial Translation in Stationary-Phase Cells
3.5. Scr1 and Rst2 Antagonistically Affect Cellular Growth and Viability upon Inhibition of Mitochondrial Translation
3.6. Rst2 Activates Carbon Metabolism Genes in Stationary-Phase Cells upon Inhibition of Mitochondrial Translation
3.7. Rst2 Genetically Interacts with Genes Involved in Stress Protection and Nutrient Response
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| mtDNA | Mitochondrial DNA |
| ETC | Electron transport chain |
| CCR | Carbon catabolite repression |
| AMPK | AMP-activated protein kinase |
| GO | Gene ontology |
| ANT | Antimycin A treatment |
| CESR | Core environmental stress response |
| COSG | Core oxidative stress gene |
| MNR | Mitonuclear retrograde |
| TCA | Tricarboxylic acid |
| CLS | Chronological lifespan |
| FYPO | Fission yeast phenotype ontology |
References
- Yan, C.; Duanmu, X.; Zeng, L.; Liu, B.; Song, Z. Mitochondrial DNA: Distribution, Mutations, and Elimination. Cells 2019, 8, 379. [Google Scholar] [CrossRef]
- Gao, M.; Yi, J.; Zhu, J.; Minikes, A.M.; Monian, P.; Thompson, C.B.; Jiang, X. Role of Mitochondria in Ferroptosis. Mol. Cell 2019, 73, 354–363.e3. [Google Scholar] [CrossRef]
- Mishra, P.; Chan, D.C. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 2016, 212, 379–387. [Google Scholar] [CrossRef]
- Herbert, C.J.; Labarre-Mariotte, S.; Cornu, D.; Sophie, C.; Panozzo, C.; Michel, T.; Dujardin, G.; Bonnefoy, N. Translational activators and mitoribosomal isoforms cooperate to mediate mRNA-specific translation in Schizosaccharomyces pombe mitochondria. Nucleic Acids Res. 2021, 49, 11145–11166. [Google Scholar] [CrossRef] [PubMed]
- Lightowlers, R.N.; Rozanska, A.; Chrzanowska-Lightowlers, Z.M. Mitochondrial protein synthesis: Figuring the fundamentals, complexities and complications, of mammalian mitochondrial translation. FEBS Lett. 2014, 588, 2496–2503. [Google Scholar] [CrossRef] [PubMed]
- Kuzmenko, A.; Atkinson, G.C.; Levitskii, S.; Zenkin, N.; Tenson, T.; Hauryliuk, V.; Kamenski, P. Mitochondrial translation initiation machinery: Conservation and diversification. Biochimie 2014, 100, 132–140. [Google Scholar] [CrossRef]
- Agirrezabala, X.; Frank, J. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q. Rev. Biophys. 2009, 42, 159–200. [Google Scholar] [CrossRef] [PubMed]
- Kummer, E.; Ban, N. Mechanisms and regulation of protein synthesis in mitochondria. Nat. Rev. Mol. Cell Biol. 2021, 22, 307–325. [Google Scholar] [CrossRef] [PubMed]
- Kummer, E.; Leibundgut, M.; Rackham, O.; Lee, R.G.; Boehringer, D.; Filipovska, A.; Ban, N. Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. Nature 2018, 560, 263–267. [Google Scholar] [CrossRef]
- Luo, Y.; Su, R.; Wang, Y.; Xie, W.; Liu, Z.; Huang, Y. Schizosaccharomyces pombe Mti2 and Mti3 act in conjunction during mitochondrial translation initiation. FEBS J. 2019, 286, 4542–4553. [Google Scholar] [CrossRef]
- Luo, Y.; Bähler, J.; Huang, Y. The Insertion Domain of Mti2 Facilitates the Association of Mitochondrial Initiation Factors with Mitoribosomes in Schizosaccharomyces pombe. Biomolecules 2025, 15, 695. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yan, J.; Zhang, Q.; Ma, X.; Zhang, J.; Su, M.; Wang, X.; Huang, Y. The Schizosaccharomyces pombe PPR protein Ppr10 associates with a novel protein Mpa1 and acts as a mitochondrial translational activator. Nucleic Acids Res. 2017, 45, 3323–3340. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wang, Y.; Huang, Y. Schizosaccharomyces pombe Ppr10 and Mpa1 together mediate mitochondrial translational initiation. J. Biol. Chem. 2021, 297, 100869. [Google Scholar] [CrossRef]
- Zuo, L.; Zhou, T.; Chuang, C.-C. The Consequences of Damaged Mitochondrial DNA. In Mitochondrial Mechanisms of Degeneration and Repair in Parkinson’s Disease; Buhlman, L.M., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 49–61. [Google Scholar]
- Kotiadis, V.N.; Duchen, M.R.; Osellame, L.D. Mitochondrial quality control and communications with the nucleus are important in maintaining mitochondrial function and cell health. Biochim. Biophys. Acta 2014, 1840, 1254–1265. [Google Scholar] [CrossRef] [PubMed]
- Malecki, M.; Bitton, D.A.; Rodríguez-López, M.; Rallis, C.; Calavia, N.G.; Smith, G.C.; Bähler, J. Functional and regulatory profiling of energy metabolism in fission yeast. Genome Biol. 2016, 17, 240. [Google Scholar] [CrossRef]
- Desai, R.; East, D.A.; Hardy, L.; Faccenda, D.; Rigon, M.; Crosby, J.; Alvarez, M.S.; Singh, A.; Mainenti, M.; Hussey, L.K.; et al. Mitochondria form contact sites with the nucleus to couple prosurvival retrograde response. Sci. Adv. 2020, 6, eabc9955. [Google Scholar] [CrossRef] [PubMed]
- Trendeleva, T.A.; Zvyagilskaya, R.A. Retrograde Signaling as a Mechanism of Yeast Adaptation to Unfavorable Factors. Biochemistry 2018, 83, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Hunt, R.J.; Bateman, J.M. Mitochondrial retrograde signaling in the nervous system. FEBS Lett. 2018, 592, 663–678. [Google Scholar] [CrossRef]
- Richter, A.S.; Nägele, T.; Grimm, B.; Kaufmann, K.; Schroda, M.; Leister, D.; Kleine, T. Retrograde signaling in plants: A critical review focusing on the GUN pathway and beyond. Plant Commun. 2023, 4, 100511. [Google Scholar] [CrossRef]
- Epstein, C.B.; Waddle, J.A.; Hale, W.T.; Davé, V.; Thornton, J.; Macatee, T.L.; Garner, H.R.; Butow, R.A. Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell 2001, 12, 297–308. [Google Scholar] [CrossRef]
- Guha, S.; López-Maury, L.; Shaw, M.; Bähler, J.; Norbury, C.J.; Agashe, V.R. Transcriptional and cellular responses to defective mitochondrial proteolysis in fission yeast. J. Mol. Biol. 2011, 408, 222–237. [Google Scholar] [CrossRef]
- Broach, J.R. Nutritional control of growth and development in yeast. Genetics 2012, 192, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Bahn, Y.S.; Xue, C.; Idnurm, A.; Rutherford, J.C.; Heitman, J.; Cardenas, M.E. Sensing the environment: Lessons from fungi. Nat. Rev. Microbiol. 2007, 5, 57–69. [Google Scholar] [CrossRef]
- Vassiliadis, D.; Wong, K.H.; Andrianopoulos, A.; Monahan, B.J. A genome-wide analysis of carbon catabolite repression in Schizosaccharomyces pombe. BMC Genom. 2019, 20, 251. [Google Scholar] [CrossRef] [PubMed]
- Matsuzawa, T.; Fujita, Y.; Tohda, H.; Takegawa, K. Snf1-like protein kinase Ssp2 regulates glucose derepression in Schizosaccharomyces pombe. Eukaryot. Cell 2012, 11, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Hoffman, C.S.; Ohta, K. Reciprocal nuclear shuttling of two antagonizing Zn finger proteins modulates Tup family corepressor function to repress chromatin remodeling. Eukaryot. Cell 2006, 5, 1980–1989. [Google Scholar] [CrossRef][Green Version]
- Mukai, Y.; Matsuo, E.; Roth, S.Y.; Harashima, S. Conservation of histone binding and transcriptional repressor functions in a Schizosaccharomyces pombe Tup1p homolog. Mol. Cell Biol. 1999, 19, 8461–8468. [Google Scholar] [CrossRef]
- Bähler, J.; Wu, J.Q.; Longtine, M.S.; Shah, N.G.; McKenzie, A., 3rd; Steever, A.B.; Wach, A.; Philippsen, P.; Pringle, J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 1998, 14, 943–951. [Google Scholar] [CrossRef]
- Kim, D.U.; Hayles, J.; Kim, D.; Wood, V.; Park, H.O.; Won, M.; Yoo, H.S.; Duhig, T.; Nam, M.; Palmer, G.; et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2010, 28, 617–623. [Google Scholar] [CrossRef]
- Anver, S.; Sumit, A.F.; Sun, X.M.; Hatimy, A.; Thalassinos, K.; Marguerat, S.; Alic, N.; Bähler, J. Ageing-associated long non-coding RNA extends lifespan and reduces translation in non-dividing cells. EMBO Rep. 2024, 25, 4921–4949. [Google Scholar] [CrossRef]
- Luo, Y.; Xu, Y.; Ahmad, F.; Feng, G.; Huang, Y. Characterization of Shy1, the Schizosaccharomyces pombe homolog of human SURF1. Sci. Rep. 2024, 14, 21678. [Google Scholar] [CrossRef] [PubMed]
- Lera-Ramírez, M.; Bähler, J.; Mata, J.; Rutherford, K.; Hoffman, C.S.; Lambert, S.; Oliferenko, S.; Martin, S.G.; Gould, K.L.; Du, L.L.; et al. Revised fission yeast gene and allele nomenclature guidelines for machine readability. Genetics 2023, 225, iyad143. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Lock, A.; Rutherford, K.; Harris, M.A.; Hayles, J.; Oliver, S.G.; Bähler, J.; Wood, V. PomBase 2018: User-driven reimplementation of the fission yeast database provides rapid and intuitive access to diverse, interconnected information. Nucleic Acids Res. 2019, 47, D821–D827. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing. 2024. Available online: https://www.r-project.org/ (accessed on 11 July 2024).
- Shen, L. GeneOverlap: Test and visualize gene overlaps. In Bioconductor; Roswell Park Comprehensive Cancer Center: Buffalo, NY, USA, 2022. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, Y.; Zhang, B. Efficient Test and Visualization of Multi-Set Intersections. Sci. Rep. 2015, 5, 16923. [Google Scholar] [CrossRef]
- Kahm, M.; Hasenbrink, G.; Lichtenberg-Fraté, H.; Ludwig, J.; Kschischo, M. grofit: Fitting Biological Growth Curves with R. J. Stat. Softw. 2010, 33, 1–21. [Google Scholar] [CrossRef]
- Romila, C.A.; Townsend, S.; Malecki, M.; Kamrad, S.; Rodríguez-López, M.; Hillson, O.; Cotobal, C.; Ralser, M.; Bähler, J. Barcode sequencing and a high-throughput assay for chronological lifespan uncover ageing-associated genes in fission yeast. Microb. Cell 2021, 8, 146–160. [Google Scholar] [CrossRef]
- Rallis, C.; Townsend, S.; Bähler, J. Genetic interactions and functional analyses of the fission yeast gsk3 and amk2 single and double mutants defective in TORC1-dependent processes. Sci. Rep. 2017, 7, 44257. [Google Scholar] [CrossRef] [PubMed]
- Wagih, O.; Parts, L. gitter: A robust and accurate method for quantification of colony sizes from plate images. G3 Genes Genomes Genet. 2014, 4, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Wagih, O.; Usaj, M.; Baryshnikova, A.; VanderSluis, B.; Kuzmin, E.; Costanzo, M.; Myers, C.L.; Andrews, B.J.; Boone, C.M.; Parts, L. SGAtools: One-stop analysis and visualization of array-based genetic interaction screens. Nucleic Acids Res. 2013, 41, W591–W596. [Google Scholar] [CrossRef] [PubMed]
- Hamburger, K.; Kramhft, B. The Effect of Chloramphenicol on Respiration, Fermentation and Growth in Schizosaccharomyces pombe. Microbiology 1980, 120, 279–282. [Google Scholar] [CrossRef][Green Version]
- Hu, Y.; Luo, Y.; Yin, D.; Zhao, L.; Wang, Y.; Rui, Y.; Zhang, P.; Wu, X.; Li, M.; Hidalgo, E.; et al. Schizosaccharomyces pombe MAP kinase Sty1 promotes survival of Δppr10 cells with defective mitochondrial protein synthesis. Int. J. Biochem. Cell Biol. 2022, 152, 106308. [Google Scholar] [CrossRef] [PubMed]
- Vustin, M.M. The Biological Role of Glycerol in Yeast Cells. Yeast as Glycerol Producers. Appl. Biochem. Microbiol. 2021, 57, 907–916. [Google Scholar] [CrossRef]
- Chen, D.; Toone, W.M.; Mata, J.; Lyne, R.; Burns, G.; Kivinen, K.; Brazma, A.; Jones, N.; Bähler, J. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 2003, 14, 214–229. [Google Scholar] [CrossRef]
- Chen, D.; Wilkinson, C.R.; Watt, S.; Penkett, C.J.; Toone, W.M.; Jones, N.; Bähler, J. Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol. Biol. Cell 2008, 19, 308–317. [Google Scholar] [CrossRef]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic Glycolysis: Meeting the Metabolic Requirements of Cell Proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef]
- Kamrad, S.; Grossbach, J.; Rodríguez-López, M.; Mülleder, M.; Townsend, S.; Cappelletti, V.; Stojanovski, G.; Correia-Melo, C.; Picotti, P.; Beyer, A.; et al. Pyruvate kinase variant of fission yeast tunes carbon metabolism, cell regulation, growth and stress resistance. Mol. Syst. Biol. 2020, 16, e9270. [Google Scholar] [CrossRef]
- Rutherford, K.M.; Lera-Ramírez, M.; Wood, V. PomBase: A Global Core Biodata Resource-growth, collaboration, and sustainability. Genetics 2024, 227, iyae007. [Google Scholar] [CrossRef]
- Woolson, R.F. Wilcoxon Signed-Rank Test. In Wiley Encyclopedia of Clinical Trials; Wiley: Hoboken, NJ, USA, 2007; pp. 1–3. [Google Scholar]
- Takenaka, K.; Tanabe, T.; Kawamukai, M.; Matsuo, Y. Overexpression of the transcription factor Rst2 in Schizosaccharomyces pombe indicates growth defect, mitotic defects, and microtubule disorder. Biosci. Biotechnol. Biochem. 2018, 82, 247–257. [Google Scholar] [CrossRef]
- Kunitomo, H.; Higuchi, T.; Iino, Y.; Yamamoto, M. A zinc-finger protein, Rst2p, regulates transcription of the fission yeast ste11(+) gene, which encodes a pivotal transcription factor for sexual development. Mol. Biol. Cell 2000, 11, 3205–3217. [Google Scholar] [CrossRef]
- Baryshnikova, A.; Costanzo, M.; Dixon, S.; Vizeacoumar, F.J.; Myers, C.L.; Andrews, B.; Boone, C. Synthetic genetic array (SGA) analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Methods Enzymol. 2010, 470, 145–179. [Google Scholar] [CrossRef]
- Costanzo, M.; Kuzmin, E.; van Leeuwen, J.; Mair, B.; Moffat, J.; Boone, C.; Andrews, B. Global Genetic Networks and the Genotype-to-Phenotype Relationship. Cell 2019, 177, 85–100. [Google Scholar] [CrossRef]
- Bitton, D.A.; Schubert, F.; Dey, S.; Okoniewski, M.; Smith, G.C.; Khadayate, S.; Pancaldi, V.; Wood, V.; Bähler, J. AnGeLi: A Tool for the Analysis of Gene Lists from Fission Yeast. Front. Genet. 2015, 6, 330. [Google Scholar] [CrossRef]
- Harris, M.A.; Lock, A.; Bähler, J.; Oliver, S.G.; Wood, V. FYPO: The fission yeast phenotype ontology. Bioinformatics 2013, 29, 1671–1678. [Google Scholar] [CrossRef]
- Sideri, T.; Rallis, C.; Bitton, D.A.; Lages, B.M.; Suo, F.; Rodríguez-López, M.; Du, L.L.; Bähler, J. Parallel profiling of fission yeast deletion mutants for proliferation and for lifespan during long-term quiescence. G3 Genes Genomes Genet. 2014, 5, 145–155. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [CrossRef] [PubMed]
- Kühl, I.; Dujeancourt, L.; Gaisne, M.; Herbert, C.J.; Bonnefoy, N. A genome wide study in fission yeast reveals nine PPR proteins that regulate mitochondrial gene expression. Nucleic Acids Res. 2011, 39, 8029–8041. [Google Scholar] [CrossRef]
- Bonfils, S.; Rozalén, A.E.; Smith, G.R.; Moreno, S.; Martín-Castellanos, C. Functional interactions of Rec24, the fission yeast ortholog of mouse Mei4, with the meiotic recombination-initiation complex. J. Cell Sci. 2011, 124, 1328–1338. [Google Scholar] [CrossRef] [PubMed]
- Steiner, S.; Kohli, J.; Ludin, K. Functional interactions among members of the meiotic initiation complex in fission yeast. Curr. Genet. 2010, 56, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Zhang, C.Y.; Bai, X.W.; Song, H.Y.; Xiao, D.G. Effects of MIG1, TUP1 and SSN6 deletion on maltose metabolism and leavening ability of baker’s yeast in lean dough. Microb. Cell Factories 2014, 13, 93. [Google Scholar] [CrossRef] [PubMed]
- Alipourfard, I.; Bakhtiyari, S.; Datukishvili, N.; Haghani, K.; Di Renzo, L.; De Miranda, R.C.; Cioccoloni, G.; Basiratyan Yazdi, P.; Mikeladze, D. Saccharomyces cerevisiae, key role of MIG1 gene in metabolic switching: Putative fermentation/oxidation. J. Biol. Regul. Homeost. Agents. 2018, 32, 649–654. [Google Scholar]
- Laera, L.; Guaragnella, N.; Ždralević, M.; Marzulli, D.; Liu, Z.; Giannattasio, S. The transcription factors ADR1 or CAT8 are required for RTG pathway activation and evasion from yeast acetic acid-induced programmed cell death in raffinose. Microb. Cell 2016, 3, 621–631. [Google Scholar] [CrossRef]
- Higuchi, T.; Watanabe, Y.; Yamamoto, M. Protein kinase A regulates sexual development and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fission yeast. Mol. Cell Biol. 2002, 22, 1–11. [Google Scholar] [CrossRef]
- Jiang, G.; Liu, Q.; Kato, T.; Miao, H.; Gao, X.; Liu, K.; Chen, S.; Sakamoto, N.; Kuno, T.; Fang, Y. Role of mitochondrial complex III/IV in the activation of transcription factor Rst2 in Schizosaccharomyces pombe. Mol. Microbiol. 2021, 115, 1323–1338. [Google Scholar] [CrossRef]
- Gupta, D.R.; Paul, S.K.; Oowatari, Y.; Matsuo, Y.; Kawamukai, M. Multistep regulation of protein kinase A in its localization, phosphorylation and binding with a regulatory subunit in fission yeast. Curr. Genet. 2011, 57, 353–365. [Google Scholar] [CrossRef]
- Skribbe, M.; Soneson, C.; Stadler, M.B.; Schwaiger, M.; Suma Sreechakram, V.N.; Iesmantavicius, V.; Hess, D.; Moreno, E.P.F.; Braun, S.; Seebacher, J.; et al. A comprehensive Schizosaccharomyces pombe atlas of physical transcription factor interactions with proteins and chromatin. Mol. Cell 2025, 85, 1426–1444.e8. [Google Scholar] [CrossRef]




| Condition | Induced | Repressed |
|---|---|---|
| Glucose_Exponential | 9 | 2 |
| Glucose_Stationary | 432 | 456 |
| Glycerol_Exponential | 15 | 8 |
| Glycerol_Stationary | 141 | 250 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Y.; Hassan, S.; Raut, S.; Bähler, J. Mitochondrial Translation Inhibition Triggers an Rst2-Controlled Transcriptional Reprogramming of Carbon Metabolism in Stationary-Phase Cells of Fission Yeast. Biomolecules 2025, 15, 1354. https://doi.org/10.3390/biom15101354
Luo Y, Hassan S, Raut S, Bähler J. Mitochondrial Translation Inhibition Triggers an Rst2-Controlled Transcriptional Reprogramming of Carbon Metabolism in Stationary-Phase Cells of Fission Yeast. Biomolecules. 2025; 15(10):1354. https://doi.org/10.3390/biom15101354
Chicago/Turabian StyleLuo, Ying, Shaimaa Hassan, Saniya Raut, and Jürg Bähler. 2025. "Mitochondrial Translation Inhibition Triggers an Rst2-Controlled Transcriptional Reprogramming of Carbon Metabolism in Stationary-Phase Cells of Fission Yeast" Biomolecules 15, no. 10: 1354. https://doi.org/10.3390/biom15101354
APA StyleLuo, Y., Hassan, S., Raut, S., & Bähler, J. (2025). Mitochondrial Translation Inhibition Triggers an Rst2-Controlled Transcriptional Reprogramming of Carbon Metabolism in Stationary-Phase Cells of Fission Yeast. Biomolecules, 15(10), 1354. https://doi.org/10.3390/biom15101354








