De Novo Design of High-Affinity HER2-Targeting Protein Minibinders
Abstract
1. Introduction
2. Materials and Methods
2.1. Hydrophobicity Analysis of the Target Protein
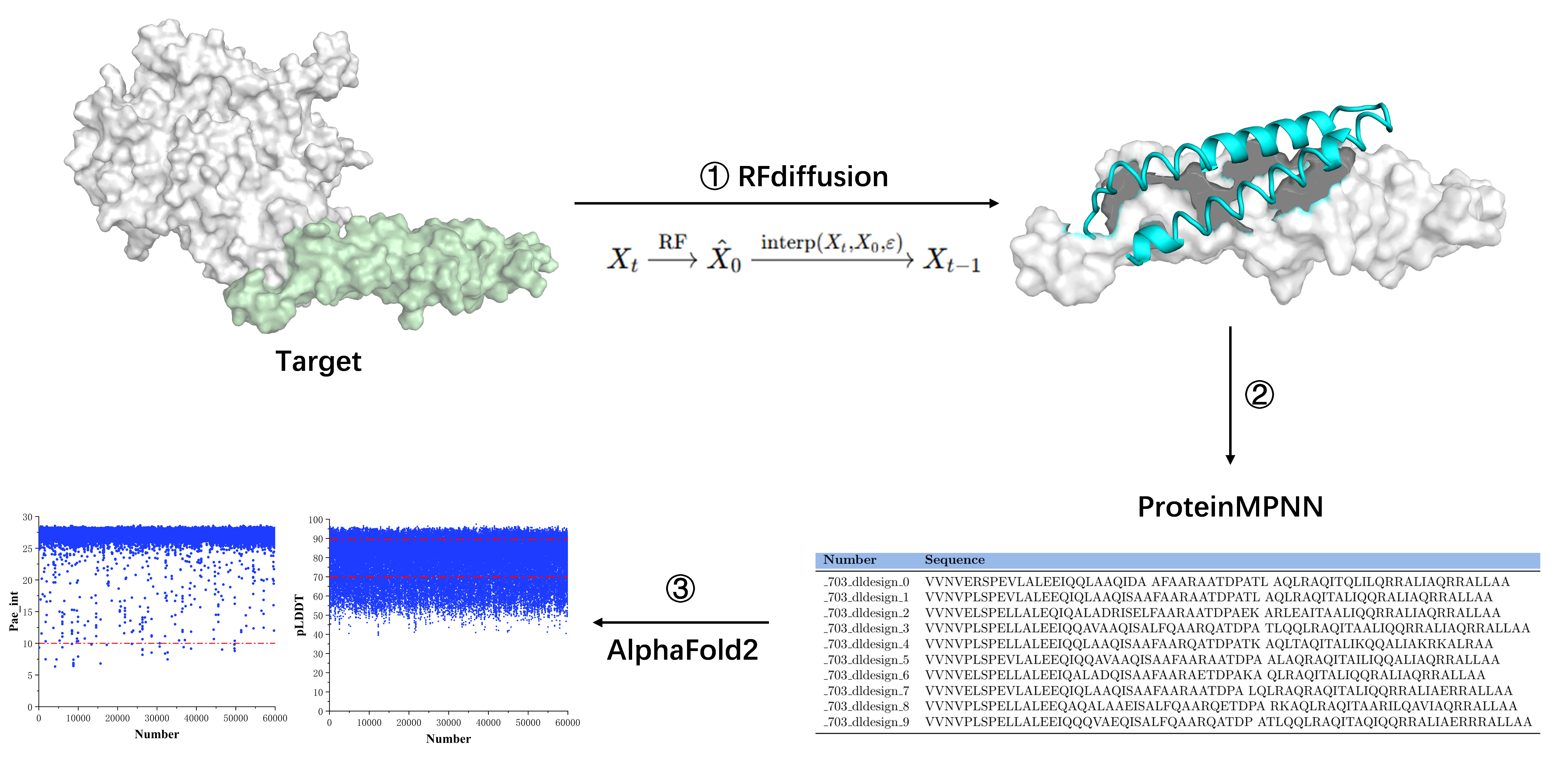
2.2. De Novo Design of Minibinder
2.3. Molecular Dynamics Simulation and Binding Free Energy Analysis
2.4. Protein’s Expression and Purification
2.5. Flow Cytometry Analysis via Vesicle Adapted System
2.6. Circular Dichroism
2.7. Isothermal Titration Calorimetry (ITC)
2.8. Cell Culture
2.9. Confocal Microscopy
3. Results
3.1. De Novo Design of Protein Minibinders Targeting on HER2 Domain IV
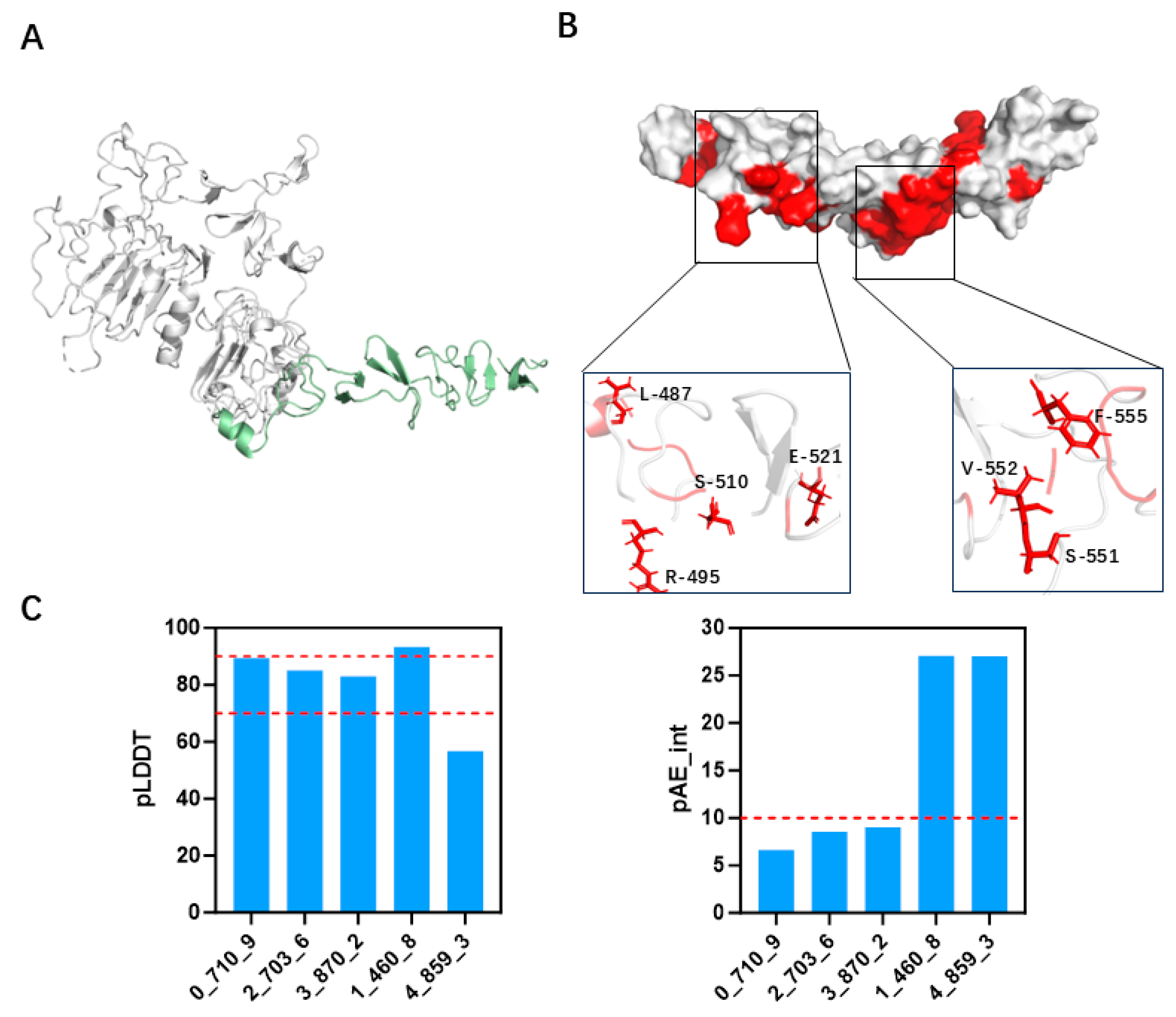
3.2. Screening and Specificity Assessment of Minibinders via Surface Display Technology
3.3. Binding Stability Differences Revealed by Molecular Dynamics Simulations
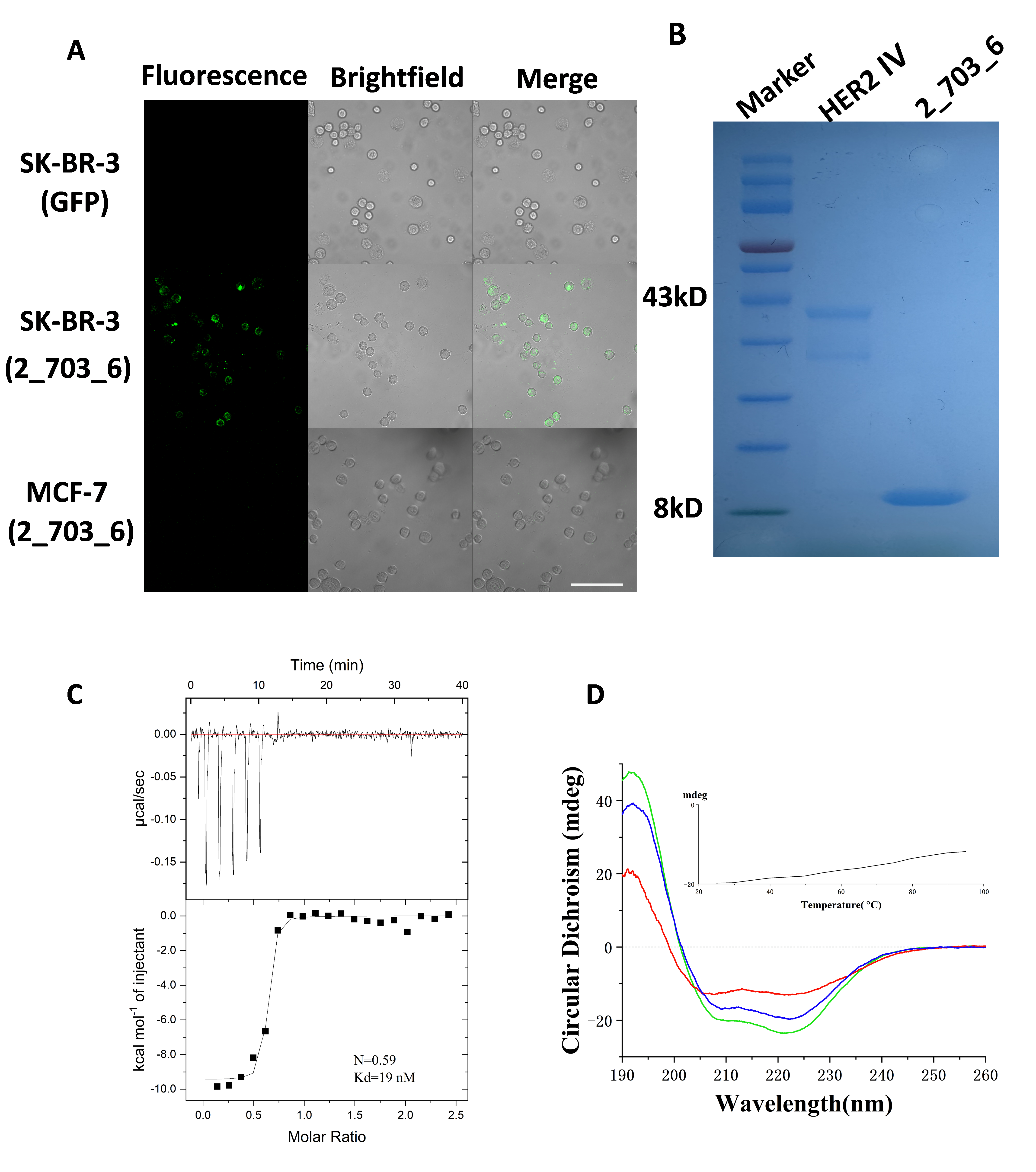
3.4. Specific Binding of 2_703_6 to HER2 Confirmed by Cellular and In Vitro Assays
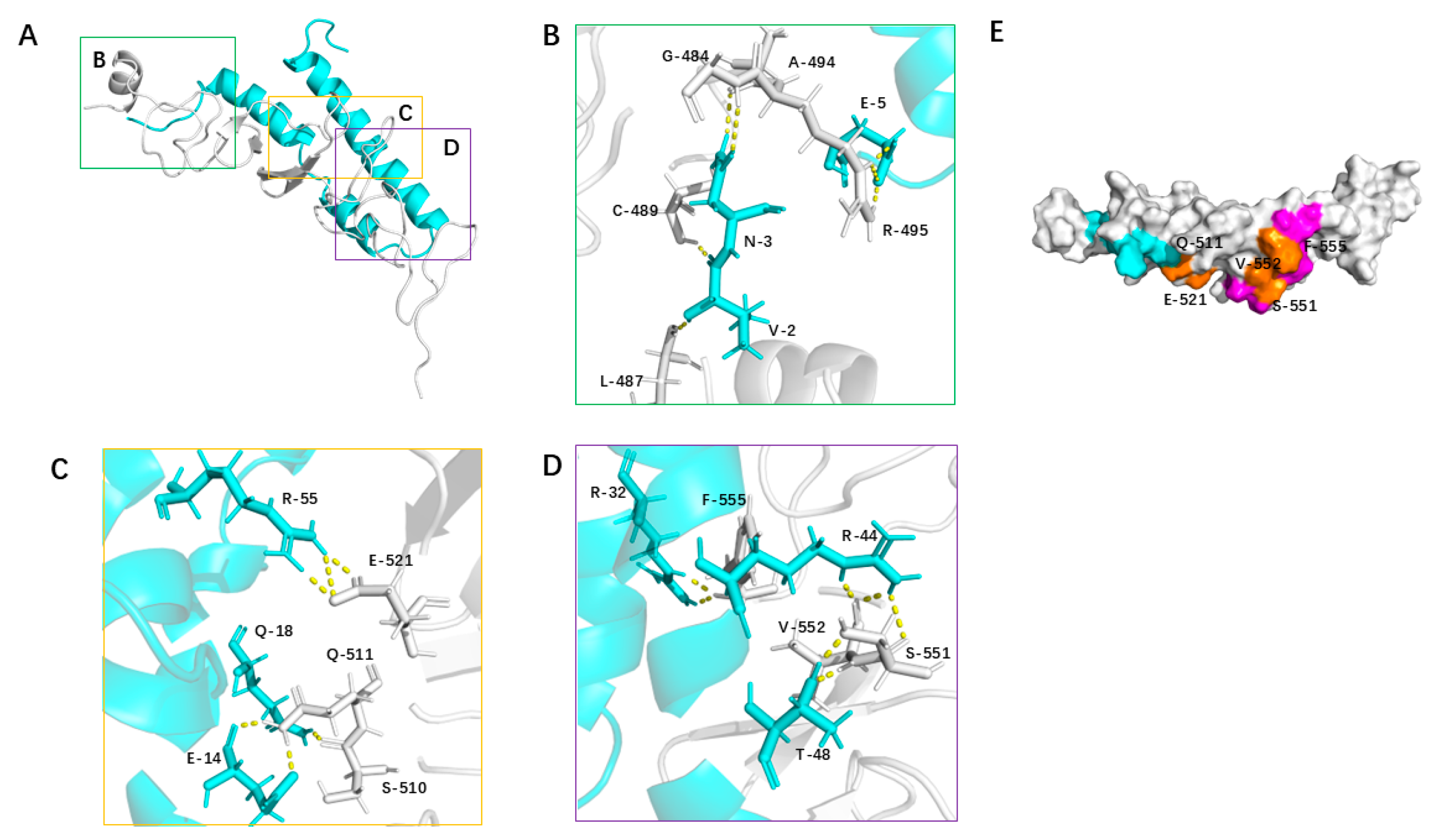
3.5. Structural Analysis of the 2_703_6–HER2 Domain IV Complex
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, X. A Comprehensive Review of HER2 in Cancer Biology and Therapeutics. Genes 2024, 15, 903. [Google Scholar] [CrossRef]
- Chao, W.R.; Lee, M.Y.; Lee, Y.J.; Sheu, G.T.; Chiu, H.H.; Shen, H.P.; Han, C.P. Profiling of HER2, KRAS, and PIK3CA mutations in uterine cervical neuroendocrine carcinoma and implications for oncogenic driver targeting therapy. Cancer Genet. 2025, 296–297, 9–14. [Google Scholar] [CrossRef]
- Di Lisa, D.; Cortese, K.; Chiappalone, M.; Arnaldi, P.; Martinoia, S.; Castagnola, P.; Pastorino, L. Electrophysiological and morphological modulation of neuronal-glial network by breast cancer and nontumorigenic mammary cell conditioned medium. Front. Bioeng. Biotechnol. 2024, 12, 1368851. [Google Scholar] [CrossRef] [PubMed]
- Wilcock, D.M.; Zhao, J.; Gulbahce, H.E. Resolving HER2 status in breast carcinoma patients with complete deletion of CEP17 in fluorescence in-situ hybridization assays. Cancer Genet. 2025, 296–297, 196–199. [Google Scholar] [CrossRef]
- Tapia, M.; Hernando, C.; Martínez, M.T.; Burgués, O.; Tebar-Sánchez, C.; Lameirinhas, A.; Ágreda-Roca, A.; Torres-Ruiz, S.; Garrido-Cano, I.; Lluch, A.; et al. Clinical Impact of New Treatment Strategies for HER2-Positive Metastatic Breast Cancer Patients with Resistance to Classical Anti-HER Therapies. Cancers 2023, 15, 4522. [Google Scholar] [CrossRef]
- Martínez Bedoya, D.; Dutoit, V.; Migliorini, D. Allogeneic CAR T Cells: An Alternative to Overcome Challenges of CAR T Cell Therapy in Glioblastoma. Front. Immunol. 2021, 12, 640082. [Google Scholar] [CrossRef]
- Han, X.; Cinay, G.E.; Zhao, Y.; Guo, Y.; Zhang, X.; Wang, P. Adnectin-Based Design of Chimeric Antigen Receptor for T Cell Engineering. Mol. Ther. 2017, 25, 2466–2476. [Google Scholar] [CrossRef]
- Krokhotin, A.; Du, H.; Hirabayashi, K.; Popov, K.; Kurokawa, T.; Wan, X.; Ferrone, S.; Dotti, G.; Dokholyan, N.V. Computationally Guided Design of Single-Chain Variable Fragment Improves Specificity of Chimeric Antigen Receptors. Mol. Ther. Oncolytics 2019, 15, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Landoni, E.; Fucá, G.; Wang, J.; Chirasani, V.R.; Yao, Z.; Dukhovlinova, E.; Ferrone, S.; Savoldo, B.; Hong, L.K.; Shou, P.; et al. Modifications to the Framework Regions Eliminate Chimeric Antigen Receptor Tonic Signaling. Cancer Immunol. Res. 2021, 9, 441–453. [Google Scholar] [CrossRef]
- Zahid, R.; Wang, J.; Cai, Z.; Ishtiaq, A.; Liu, M.; Ma, D.; Liang, Y.; Xu, Y. Single chain fragment variable, a new theranostic approach for cardiovascular diseases. Front. Immunol. 2024, 15, 1443290. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.Y.; Cha, J.S.; Kim, H.; Choi, Y.; Cho, H.-S.; Kim, H.-S. Computationally-guided design and affinity improvement of a protein binder targeting a specific site on HER2. Comput. Struct. Biotechnol. J. 2021, 19, 1325–1334. [Google Scholar] [CrossRef]
- Xia, Z.; Jin, Q.; Long, Z.; He, Y.; Liu, F.; Sun, C.; Liao, J.; Wang, C.; Wang, C.; Zheng, J.; et al. Targeting overexpressed antigens in glioblastoma via CAR T cells with computationally designed high-affinity protein binders. Nat. Biomed. Eng. 2024, 8, 1634–1650. [Google Scholar] [CrossRef]
- Wei, L.; Hu, Y.; Liu, Y.; Xing, B.; Wang, K.; Weng, J.; Liu, Z.; Fang, Y.; Ming, K.; Mei, M.; et al. De novo design mini-binder proteins targeting the glycoproteins D to inhibit PRV replication in PK15 cells. Int. J. Biol. Macromol. 2025, 315 Pt 1, 144403. [Google Scholar] [CrossRef]
- Berger, S.; Seeger, F.; Yu, T.Y.; Aydin, M.; Yang, H.; Rosenblum, D.; Guenin-Macé, L.; Glassman, C.; Arguinchona, L.; Sniezek, C.; et al. Preclinical proof of principle for orally delivered Th17 antagonist miniproteins. Cell 2024, 187, 4305–4317.e4318. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, L.; Yang, Q.Y.; Yu, K. Novel anti-HER2 nanobody-drug conjugates with enhanced penetration of solid tumor and BBB, reduced systemic exposure and superior antitumor efficacy. Acta Pharmacol. Sin. 2025, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.L.; Juergens, D.; Bennett, N.R.; Trippe, B.L.; Yim, J.; Eisenach, H.E.; Ahern, W.; Borst, A.J.; Ragotte, R.J.; Milles, L.F.; et al. De novo design of protein structure and function with RFdiffusion. Nature 2023, 620, 1089–1100. [Google Scholar] [CrossRef]
- Pacesa, M.; Nickel, L.; Schellhaas, C.; Schmidt, J.; Pyatova, E.; Kissling, L.; Barendse, P.; Choudhury, J.; Kapoor, S.; Alcaraz-Serna, A.; et al. BindCraft: One-shot design of functional protein binders. bioRxiv 2025. [Google Scholar] [CrossRef] [PubMed]
- Vázquez Torres, S.; Leung, P.J.Y.; Venkatesh, P.; Lutz, I.D.; Hink, F.; Huynh, H.H.; Becker, J.; Yeh, A.H.; Juergens, D.; Bennett, N.R.; et al. De novo design of high-affinity binders of bioactive helical peptides. Nature 2024, 626, 435–442. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, Y.; Chen, M.; Chen, X.; Ming, K.; Liu, Y.; Weng, J.; Xing, B.; Wei, L.; Wang, Z.; et al. De novo designed mini-binders targeting glyceraldehyde-3-phosphate dehydrogenase of Streptococcus equi ssp. zooepidemicus provided partial protection in mice model of infection. Int. J. Biol. Macromol. 2025, 307 Pt 3, 142293. [Google Scholar] [CrossRef] [PubMed]
- Glögl, M.; Krishnakumar, A.; Ragotte, R.J.; Goreshnik, I.; Coventry, B.; Bera, A.K.; Kang, A.; Joyce, E.; Ahn, G.; Huang, B.; et al. Target-conditioned diffusion generates potent TNFR superfamily antagonists and agonists. Science 2024, 386, 1154–1161. [Google Scholar] [CrossRef]
- Bennett, N.R.; Coventry, B.; Goreshnik, I.; Huang, B.; Allen, A.; Vafeados, D.; Peng, Y.P.; Dauparas, J.; Baek, M.; Stewart, L.; et al. Improving de novo protein binder design with deep learning. Nat. Commun. 2023, 14, 2625. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Arai, S.; Suzuki, H. Immobilization of E. coli expressing gamma-glutamyltranspeptidase on its surface for gamma-glutamyl compound production. AMB Express 2023, 13, 27. [Google Scholar] [CrossRef]
- Lei, S.; Zheng, R.; Zhang, S.; Chen, R.; Wang, S.; Sun, K.; Zeng, H.; Wei, W.; He, J. Breast cancer incidence and mortality in women in China: Temporal trends and projections to 2030. Cancer Biol. Med. 2021, 18, 900–909. [Google Scholar] [CrossRef]
- Shagufta; Ahmad, I.; Nelson, D.J.; Hussain, M.I.; Nasar, N.A. Potential of covalently linked tamoxifen hybrids for cancer treatment: Recent update. RSC Med. Chem. 2024, 15, 1877–1898. [Google Scholar] [CrossRef] [PubMed]
- Jordan, V.C. Tamoxifen: Toxicities and drug resistance during the treatment and prevention of breast cancer. Annu. Rev. Pharmacol. Toxicol. 1995, 35, 195–211. [Google Scholar] [CrossRef] [PubMed]
- Bisht, A.; Avinash, D.; Sahu, K.K.; Patel, P.; Das Gupta, G.; Kurmi, B.D. A comprehensive review on doxorubicin: Mechanisms, toxicity, clinical trials, combination therapies and nanoformulations in breast cancer. Drug Deliv. Transl. Res. 2025, 15, 102–133. [Google Scholar] [CrossRef]
- Gautam, S.; Maurya, R.; Vikal, A.; Patel, P.; Thakur, S.; Singh, A.; Gupta, G.D.; Kurmi, B.D. Understanding drug resistance in breast cancer: Mechanisms and emerging therapeutic strategies. Med. Drug Discov. 2025, 26, 100210. [Google Scholar] [CrossRef]
- Jallah, J.K.; Dweh, T.J.; Anjankar, A.; Palma, O. A Review of the Advancements in Targeted Therapies for Breast Cancer. Cureus 2023, 15, e47847. [Google Scholar] [CrossRef]
- Swain, S.M.; Shastry, M.; Hamilton, E. Targeting HER2-positive breast cancer: Advances and future directions. Nat. Rev. Drug Discov. 2023, 22, 101–126. [Google Scholar] [CrossRef]
- Kunte, S.; Abraham, J.; Montero, A.J. Novel HER2-targeted therapies for HER2-positive metastatic breast cancer. Cancer 2020, 126, 4278–4288. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Goreshnik, I.; Coventry, B.; Case, J.B.; Miller, L.; Kozodoy, L.; Chen, R.E.; Carter, L.; Walls, A.C.; Park, Y.-J.; et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 2020, 370, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Sonkin, D.; Thomas, A.; Teicher, B.A. Cancer treatments: Past, present, and future. Cancer Genet. 2024, 286–287, 18–24. [Google Scholar] [CrossRef]
- Cao, L.; Coventry, B.; Goreshnik, I.; Huang, B.; Sheffler, W.; Park, J.S.; Jude, K.M.; Marković, I.; Kadam, R.U.; Verschueren, K.H.G.; et al. Design of protein-binding proteins from the target structure alone. Nature 2022, 605, 551–560. [Google Scholar] [CrossRef]
- Zheng, M.; Li, C.; Zhou, M.; Jia, R.; She, F.; Wei, L.; Cheng, F.; Li, Q.; Cai, J.; Wang, Y. Peptidomimetic-based antibody surrogate for HER2. Acta Pharm. Sin. B 2021, 11, 2645–2654. [Google Scholar] [CrossRef]
- Rodak, M.; Dekempeneer, Y.; Wojewodzka, M.; Caveliers, V.; Covens, P.; Miller, B.W.; Sevenois, M.B.; Bruchertseifer, F.; Morgenstern, A.; Lahoutte, T.; et al. Preclinical Evaluation of 225Ac-Labeled Single-Domain Antibody for the Treatment of HER2pos Cancer. Mol. Cancer Ther. 2022, 21, 1835–1845. [Google Scholar] [CrossRef]
- Oroujeni, M.; Westerlund, K.; Papalanis, E.; van Deventer, A.; Liu, Y.; Clinton, J.; Wang, Z.; Zelepukin, I.; Orlova, A.; Tolmachev, V.; et al. Designed Ankyrin Repeat Protein-Mediated Peptide Nucleic Acid-Based Pretargeting: A Proof-of-Principle Study. J. Nucl. Med. 2025, 66, 1105–1111. [Google Scholar] [CrossRef]
- Adams, C.S.; Kim, H.; Burtner, A.E.; Lee, D.S.; Dobbins, C.; Criswell, C.; Coventry, B.; Tran-Pearson, A.; Kim, H.M.; King, N.P. De novo design of protein minibinder agonists of TLR3. Nat. Commun. 2025, 16, 1234. [Google Scholar] [CrossRef]
- Liu, H.; Hamaia, S.W.; Dobson, L.; Weng, J.; Hernández, F.L.; Beaudoin, C.A.; Salvage, S.C.; Huang, C.L.; Machesky, L.M.; Jackson, A.P. The voltage-gated sodium channel β3 subunit modulates C6 glioma cell motility independently of channel activity. Biochim. Biophys. Acta Mol. Basis Dis. 2025, 1871, 167844. [Google Scholar] [CrossRef]
- Weng, J.; Geng, M.; Hu, X.; Hu, Y.; Yang, Y.; Xing, B.; Wu, Z.; Wei, Z. Design of minibinder proteins specific to TNFR1. Int. J. Biol. Macromol. 2025, 293, 139403. [Google Scholar] [CrossRef]
- Chen, J.; Qiu, S.; Li, W.; Wang, K.; Zhang, Y.; Yang, H.; Liu, B.; Li, G.; Li, L.; Chen, M.; et al. Tuning charge density of chimeric antigen receptor optimizes tonic signaling and CAR-T cell fitness. Cell Res. 2023, 33, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Snapper, C.M. Distinct Immunologic Properties of Soluble Versus Particulate Antigens. Front. Immunol. 2018, 9, 598. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.K.; Andreatta, M.; Marcatili, P.; Buus, S.; Greenbaum, J.A.; Yan, Z.; Sette, A.; Peters, B.; Nielsen, M. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 2018, 154, 394–406. [Google Scholar] [CrossRef] [PubMed]


| Number | Sequence | Molecular Weight (kDa) |
|---|---|---|
| 0_293_7 | AREAARQAAIDEISALFARARANPDLSPAERAALAAAITAAYRRYRALL | 6.12 |
| 0_324_7 | SAAEAATRARIRARAAEVAEQIKDLSPEERKALYLQAFQEFSGESQRQLNLLHSLLNERWAELQ | 8.45 |
| 0_459_7 | STAAAIAAARAEVENLPYEEALARADELAERLGYEKRTVNGGTIYLPA | 5.95 |
| 0_679_6 | AERQQKIAQLKALLATAEGYLKLLKDNEEAQIRVAEHLDKQLAEVLGEKEVKEGITLEEMIARIQAEIAKLEA | 9.48 |
| 0_710_3 | AAQTAALIAQIATLNAAQNIALISQLFAAARSDPSLDRAELATQITAAYQQYRALL | 6.88 |
| 0_710_5 | SAETEATIERIKTMNREENIELISEAFARARSDPSLDRAALARIITAAYQQYRKLL | 7.39 |
| 0_710_6 | SAATAATIARIATLNAEENIALISQLFAQARTDPSLDRAALATQITAAYRQYRALL | 6.95 |
| 0_710_7 | SEETAATIARIETMTAEENIALISELFERARTDPSLDRAELATMITAAVQRYRRLL | 7.30 |
| 0_710_9 | DAATAATIARIATLDAAGNIALISELFAQARSDPSLDRAALARQITAAYQRYRALL | 6.89 |
| 0_818_7 | SEALLARLREVAATKSDEEFARELARVDAEGLGREAAREAFRERAASLPTNAAVNRLMALYSEALAEERARLAAEAAAA | 9.89 |
| 0_1007_6 | AAKAAQIATLNAIRDALKAGDLETAKALIAATSLSNTVANHVLNALSYAERAKADPATAAADAAEMDAEFAKALAAVNAE | 9.42 |
| 0_1156_1 | MKVVVLPASAPVEEKRRVAQELVAQYGSLVVTTVDDVPVEEAVAIAKRDIAAIKRAYPQVKAILVITSDEAAKALKA | 9.58 |
| 1_250_0 | RAALLERAITAFDLAAYAVRQGNRAAAAQALAHALALLEEAGLAVSEELRRLLELPPEEALALLARERARLRAL | 9.29 |
| 1_394_0 | PVHVPLSAVASTPEELRARVREIVAEFSIEEVTASALRIQPLLPPELFAAILATLEELREEAEKA | 8.27 |
| 1_968_7 | ARYRVTFHDTGLTYETDKAAAERDVALAKEKGLRVTVEPVVT | 5.42 |
| 1_1124_9 | PHHRQLPAAELRALAQAELARAQAAGQVAQAQRHQAILAQVDAGQPLTYIALPP | 6.68 |
| 2_128_0 | PLHVLIFKPGKVLYRVETPEITREAEIDLESALALTRVLVRAGARVVVENGEEALKEAEKTNKKDAELIREILELVK | 10.03 |
| 2_128_4 | PLIVVELRPGRVLLTVVTPTITKTAEIDLESALDLVRVLKRAGVKVEVRGGDAALALAKRTDPAAAALIEAILAEVA | 9.39 |
| 2_128_8 | ALHVVRVEPGRITLVTRTPTITRSAEIDAESALALVRVLRRAGATVELENLDAGIAAAAATDPATAALLRAIAAEAA | 9.22 |
| 2_337_7 | ARVEATVSFPDREAAYAVLREVNVRFIVDFQPDGTVVLIAPESDRAGLEAEAAKLKALVAARA | 7.87 |
| 2_520_2 | RWVYRELTVAELEALRAREASPVDQPRWDAVLAQMRASGQPALYVQRLP | 6.53 |
| 2_520_5 | RWVYRELTVEELKAIAARETLPVDAPRWAAVLAQAEASGQPVWYVRRLP | 6.50 |
| 2_645_0 | TVLSPEELIRRLYRRAAELIATLPPSVQLTVAVLATPTSVEVRVTAPAEAAAHAEALRAELEAEIA | 8.19 |
| 2_703_2 | VVNVELSPELLALQEQIQALADRISELFARARAATDPAEKARLRAEITAAILQRRALIAQRRALLAA | 8.53 |
| 2_703_6 | VVNVELSPELLALEAEIQALADQISAAFAAARAETDPAAKAALRAQITALIQQRRALIAQRRALLAA | 8.24 |
| 3_15_0 | SATRELNRLLGEASLFIEELRKRAPELSPEELKKILEEKVEEYAKINPLVAAYMKHQVEKLL | 8.31 |
| 3_870_2 | MTPEELEALRARILAMSPEEIRALFRADAQLAAAALEAANLAAAAAAGLTVQGGVVRPEP | 7.22 |
| 3_1069_8 | MSRFTEKLLALYRTNVAAYTAAGNAANIEKVTAYYKAEAASLPDAAEVNAELDAAKAAALAALAAA | 7.92 |
| 4_114_3 | PVLRHRTITEAEALAITARTAATLSPLQAAALRARTAALLAAARAAGTPLTYISLE | 6.76 |
| 4_114_4 | PRLVHREVTEAEAQAILAATAASLSPLQAALLAQRHAAILAQARAAGLPLTYTSLE | 6.81 |
| 4_1157_3 | MRRVVLDRLAPGRWRLTVTTPERTLVFELSTEVARALWRRYTADEIAARPEALVAEALAAA | 7.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Wei, W.; Cheng, Z.; Yang, M.; Yan, Y. De Novo Design of High-Affinity HER2-Targeting Protein Minibinders. Biomolecules 2025, 15, 1587. https://doi.org/10.3390/biom15111587
Zhao Y, Wei W, Cheng Z, Yang M, Yan Y. De Novo Design of High-Affinity HER2-Targeting Protein Minibinders. Biomolecules. 2025; 15(11):1587. https://doi.org/10.3390/biom15111587
Chicago/Turabian StyleZhao, Yize, Wenping Wei, Zijun Cheng, Min Yang, and Yunjun Yan. 2025. "De Novo Design of High-Affinity HER2-Targeting Protein Minibinders" Biomolecules 15, no. 11: 1587. https://doi.org/10.3390/biom15111587
APA StyleZhao, Y., Wei, W., Cheng, Z., Yang, M., & Yan, Y. (2025). De Novo Design of High-Affinity HER2-Targeting Protein Minibinders. Biomolecules, 15(11), 1587. https://doi.org/10.3390/biom15111587







