Differential Metabolic Dysregulations in Hepatocellular Carcinoma and Cirrhosis: Insights into Lipidomic Signatures
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Patients’ Follow-Up
2.3. Lipidomic Analysis
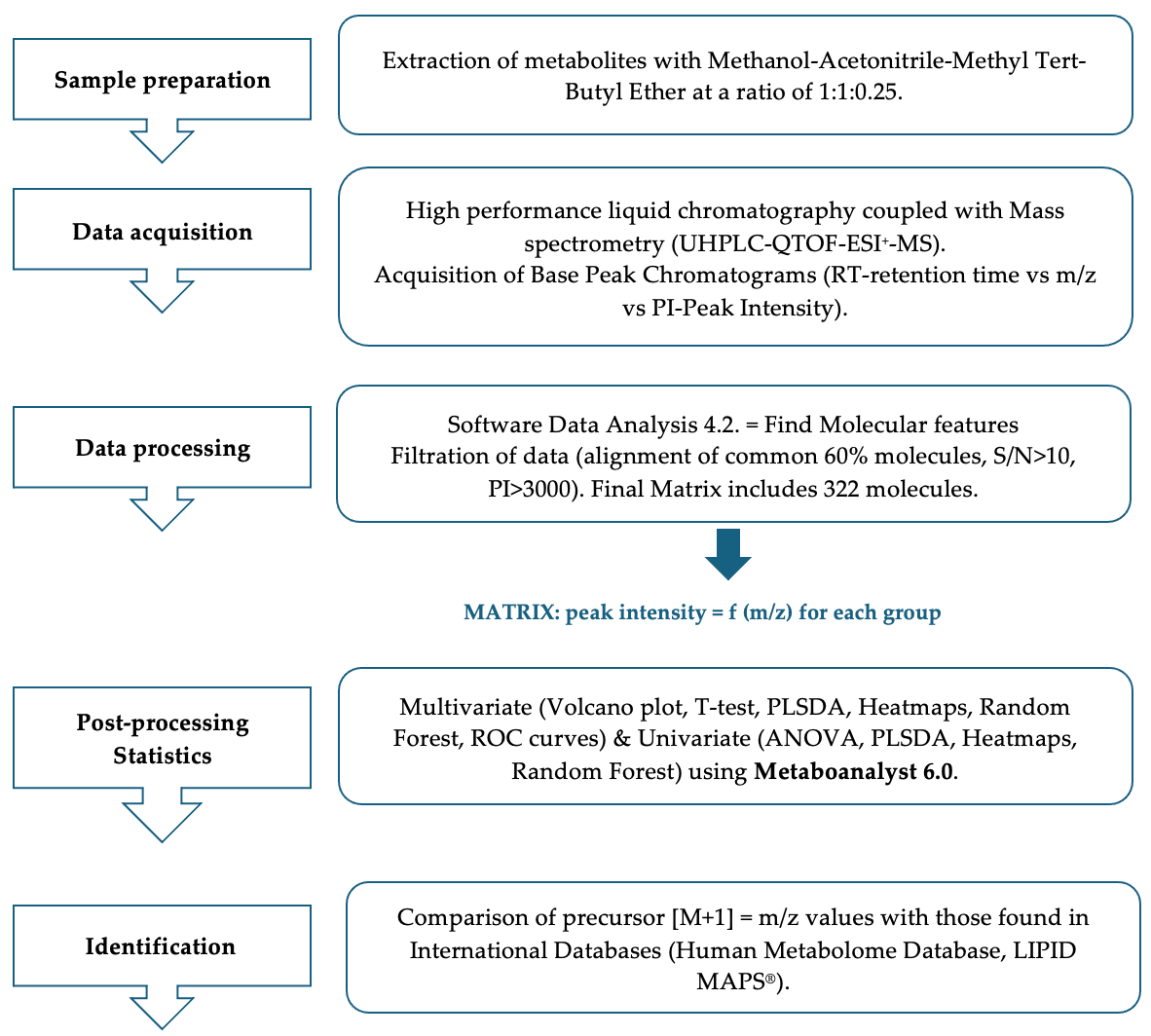
2.3.1. Sample Preparation and Processing
2.3.2. UHPLC-QTOF-ESI+-MS Analysis
2.3.3. Data Processing and Statistical Analysis
2.4. Ethical Approval
3. Results
3.1. Descriptive Characteristics of Enrolled Patients
3.2. Untargeted Multivariate Analysis of CIR vs. HCC Group
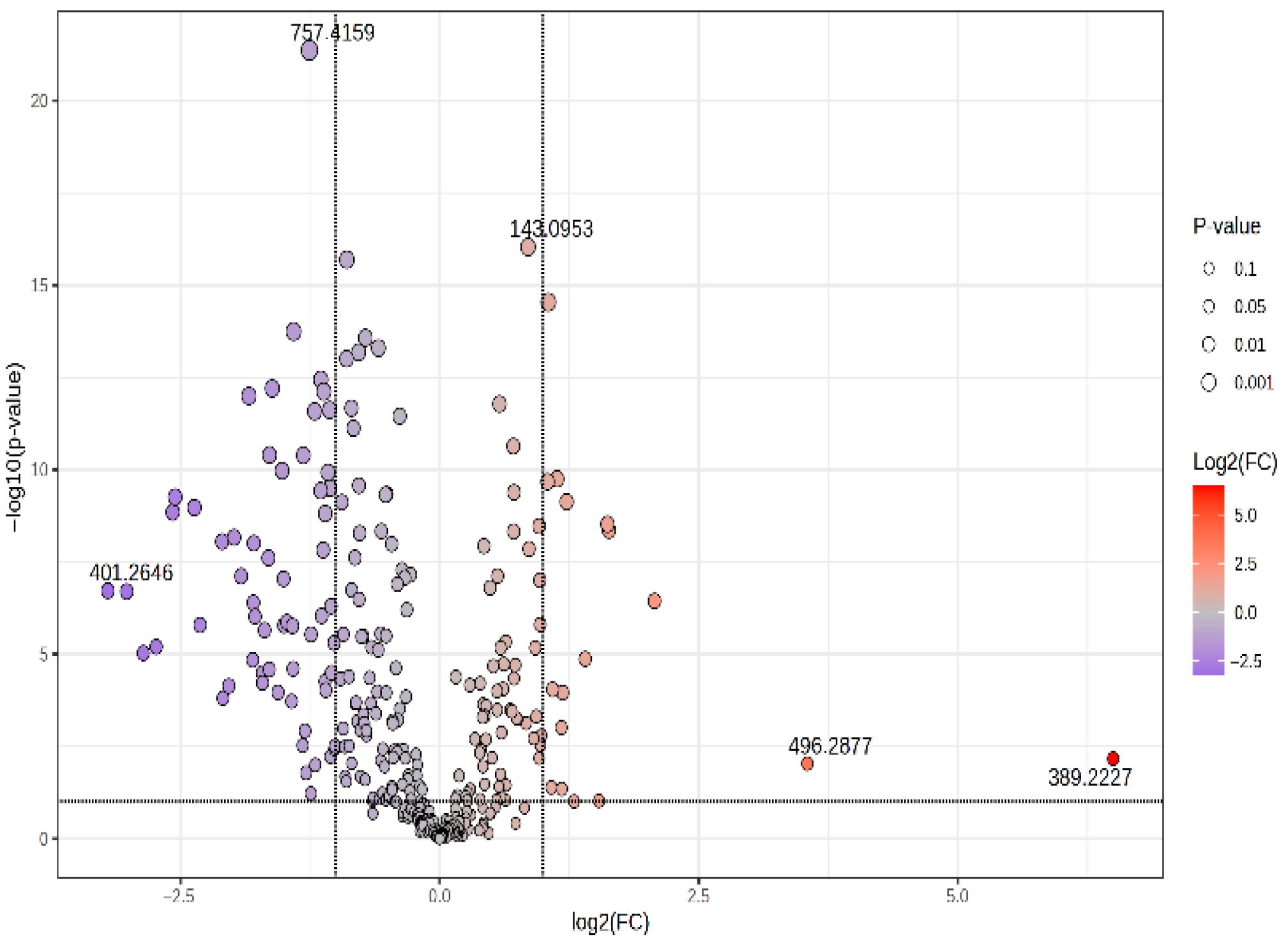
3.2.1. Volcano Plot and Correlation Plot (CIR vs. HCC)
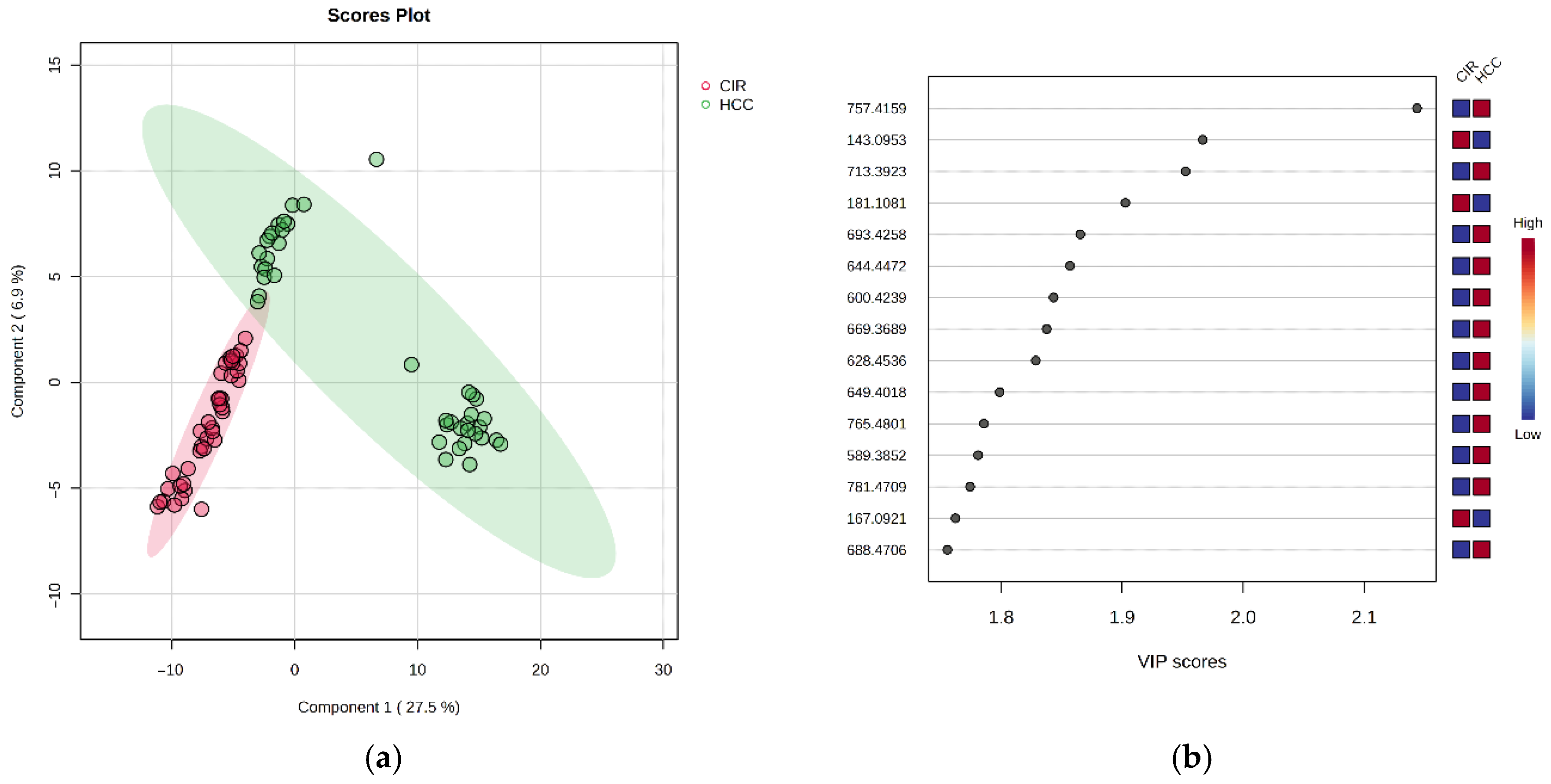
3.2.2. Discriminatory Analysis by PLSDA
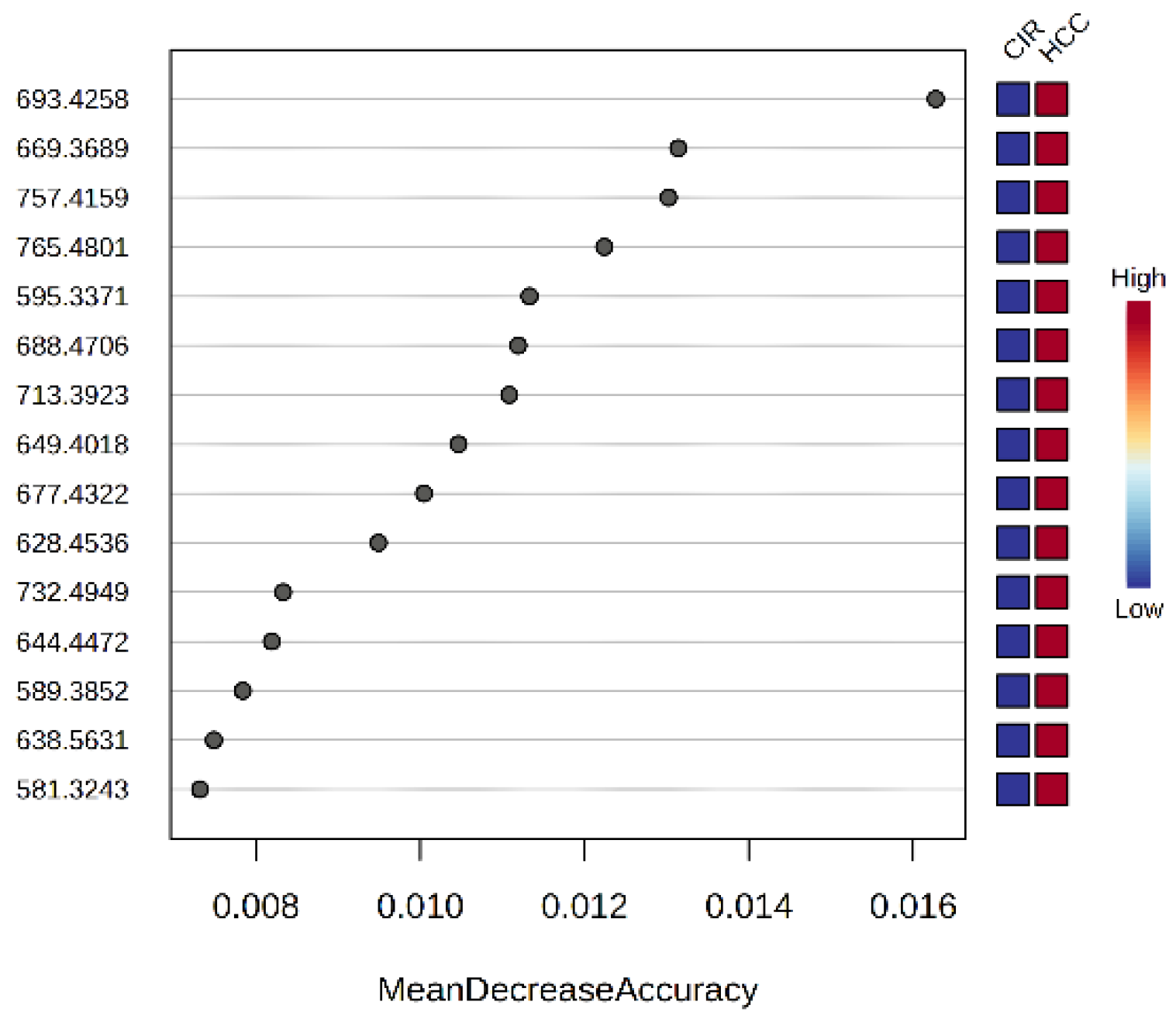
3.2.3. The Random Forest Analysis and Heatmap
3.2.4. Biomarker Analysis and Correlation Network
3.3. Semi-Targeted Metabolomic Analysis
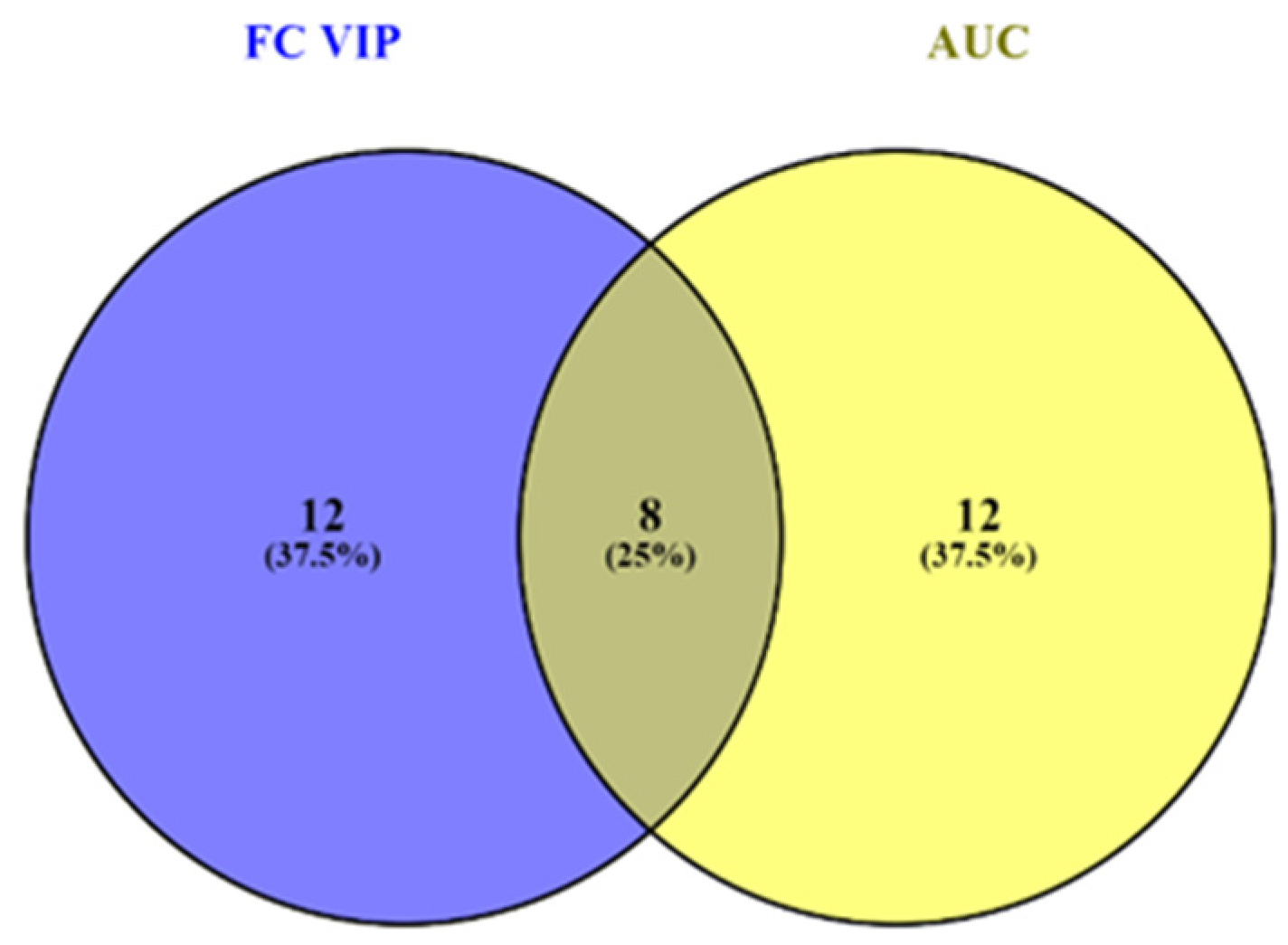
3.3.1. VIP Scores, FC Values, p-Values and Random Forest Ranking
3.3.2. Biomarker Analysis for Each Class of Metabolites
3.4. Integration of Data from Multivariate and Biomarker Analysis
4. Discussion
Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, A.; Hallouch, O.; Chernyak, V.; Kamaya, A.; Sirlin, C.B. Epidemiology of hepatocellular carcinoma: Target population for surveillance and diagnosis. Abdom. Radiol. 2018, 43, 13–25. [Google Scholar] [CrossRef]
- McGlynn, K.A.; Petrick, J.L.; El-Serag, H.B. Epidemiology of Hepatocellular Carcinoma. Hepatology 2020, 73, 4–13. [Google Scholar] [CrossRef]
- Le, D.C.; Nguyen, T.M.; Nguyen, D.H.; Nguyen, D.T.; Nguyen, L.T.M. Survival outcome and prognostic factors among patients with hepatocellular carcinoma: A hospital-based study. Clin. Med. Insights Oncol. 2023, 17, 11795549231178171. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Boyer, T.D.; Manns, M.P.; Sanyal, A.J.; Zakim, D. Zakim and Boyer’s Hepatology: A Textbook of Liver Disease, 6th ed.; Saunders/Elsevier: Philadelphia, PA, USA, 2012; pp. 1005–1031. [Google Scholar]
- Gan, C.; Yuan, Y.; Shen, H.; Gao, J.; Kong, X.; Che, Z.; Guo, Y.; Wang, H.; Dong, E.; Xiao, J. Liver diseases: Epidemiology, causes, trends and predictions. Signal Transduct. Target. Ther. 2025, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Estes, C.; Razavi, H.; Loomba, R.; Younossi, Z.; Sanyal, A.J. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 2018, 67, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Song, B.G.; Choi, S.C.; Goh, M.J.; Kang, W.; Sinn, D.H.; Gwak, G.Y.; Paik, Y.-H.; Choi, M.S.; Lee, J.H.; Paik, S.W. Metabolic dysfunction-associated fatty liver disease and the risk of hepatocellular carcinoma. JHEP Rep. 2023, 5, 100810. [Google Scholar] [CrossRef]
- Tellapuri, S.; Sutphin, P.D.; Beg, M.S.; Singal, A.G.; Kalva, S.P. Staging systems of hepatocellular carcinoma: A review. Indian J. Gastroenterol. 2018, 37, 481–491. [Google Scholar] [CrossRef]
- Reig, M.; Forner, A.; Rimola, J.; Ferrer-Fàbrega, J.; Burrel, M.; Garcia-Criado, Á.; Kelley, R.K.; Galle, P.R.; Mazzaferro, V.; Salem, R.; et al. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J. Hepatol. 2022, 76, 681–693. [Google Scholar] [CrossRef]
- Galle, P.R.; Forner, A.; Llovet, J.M.; Mazzaferro, V.; Piscaglia, F.; Raoul, J.L.; Schirmacher, P.; Vilgrain, V. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2019, 69, 182–236. [Google Scholar] [CrossRef]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Brown, Z.J.; Tsilimigras, D.I.; Ruff, S.M.; Mohseni, A.; Kamel, I.R.; Cloyd, J.M.; Pawlik, T.M. Management of hepatocellular carcinoma: A review. JAMA Surg. 2023, 158, 410–420. [Google Scholar] [CrossRef]
- Ghantus, R.; Ciocan, R.A.; Schlanger, D.; Popa, C.; Gherman, C.D.; Vaida, C.; Gherman, B.; Pîslă, D.; Al Hajjar, N.; Ciocan, A. Novel Biomarkers in Hepatocellular Carcinoma from Embryogenic Antigens to cfDNA. Biomedicines 2025, 13, 1020. [Google Scholar] [CrossRef] [PubMed]
- Hanif, H.; Ali, M.J.; Khan, I.W.; Luna-Cuadros, M.A.; Khan, M.M.; Lau, D.T.-Y. Update on the applications and limitations of alpha-fetoprotein for hepatocellular carcinoma. World J. Gastroenterol. 2022, 28, 216. [Google Scholar] [CrossRef]
- Semmler, G.; Meyer, E.L.; Kozbial, K.; Schwabl, P.; Hametner-Schreil, S.; Zanetto, A.; Bauer, D.; Chromy, D.; Simbrunner, B.; Scheiner, B.; et al. HCC risk stratification after cure of hepatitis C in patients with compensated advanced chronic liver disease. J. Hepatol. 2022, 76, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Nenu, I.; Stefanescu, H.; Procopet, B.; Sparchez, Z.; Minciuna, I.; Mocan, T.; Leucuta, D.; Morar, C.; Grigorescu, M.; Filip, G.A.; et al. Navigating through the lipid metabolism maze: Diagnosis and prognosis metabolites of hepatocellular carcinoma versus compensated cirrhosis. J. Clin. Med. 2022, 11, 1292. [Google Scholar] [CrossRef]
- Tan, S.L.W.; Israeli, E.; Ericksen, R.E.; Chow, P.K.H.; Han, W. The altered lipidome of hepatocellular carcinoma. Semin. Cancer Biol. 2022, 86, 445–456. [Google Scholar] [CrossRef]
- Bocse, H.; Ciocan, A.; Zaharie, F.V.; Ciocan, R.A.; Al Hajjar, N. Hepatocellular Carcinoma: Systematic Review of Relevant Biomarkers and the Impact on Liver Resections. Int. J. Innov. Res. Med. Sci. 2022, 7, 10. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The Warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 2016, 41, 211–218, Erratum in Trends Biochem. Sci. 2016, 41, 287. [Google Scholar] [CrossRef]
- Fukushi, A.; Kim, H.D.; Chang, Y.C.; Kim, C.H. Revisited metabolic control and reprogramming cancers by means of the Warburg effect in tumor cells. Int. J. Mol. Sci. 2022, 23, 10037. [Google Scholar] [CrossRef]
- Sangineto, M.; Villani, R.; Cavallone, F.; Romano, A.; Loizzi, D.; Serviddio, G. Lipid metabolism in development and progression of hepatocellular carcinoma. Cancers 2020, 12, 1419. [Google Scholar] [CrossRef]
- Nakagawa, H.; Hayata, Y.; Kawamura, S.; Yamada, T.; Fujiwara, N.; Koike, K. Lipid metabolic reprogramming in hepatocellular carcinoma. Cancers 2018, 10, 447. [Google Scholar] [CrossRef]
- Wang, M.; Han, J.; Xing, H.; Zhang, H.; Li, Z.; Liang, L.; Li, C.; Dai, S.; Wu, M.; Shen, F.; et al. Dysregulated fatty acid metabolism in hepatocellular carcinoma. Hepatic Oncol. 2016, 3, 241–251. [Google Scholar] [CrossRef]
- Ten Hove, M.; Pater, L.; Storm, G.; Weiskirchen, S.; Weiskirchen, R.; Lammers, T.; Bansal, R. The hepatic lipidome: From basic science to clinical translation. Adv. Drug Deliv. Rev. 2020, 159, 180–197. [Google Scholar] [CrossRef]
- Ismail, I.T.; Elfert, A.; Helal, M.; Salama, I.; El-Said, H.; Fiehn, O. Remodeling lipids in the transition from chronic liver disease to hepatocellular carcinoma. Cancers 2021, 13, 88. [Google Scholar] [CrossRef]
- Anh, N.H.; Long, N.P.; Min, Y.J.; Ki, Y.; Kim, S.J.; Jung, C.W.; Park, S.; Kwon, S.W.; Lee, S.J. Molecular and metabolic phenotyping of hepatocellular carcinoma for biomarker discovery: A meta-analysis. Metabolites 2023, 13, 1112. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chen, J.; Huang, C.; Li, N.; Zou, L.; Chia, S.E.; Chen, S.; Yu, K.; Ling, Q.; Cheng, Q.; et al. Comparison of hepatic and serum lipid signatures in hepatocellular carcinoma patients leads to the discovery of diagnostic and prognostic biomarkers. Oncotarget 2018, 9, 5032. [Google Scholar] [CrossRef]
- Powell, H.; Coarfa, C.; Ruiz-Echartea, E.; Grimm, S.L.; Najjar, O.; Yu, B.; Olivares, L.; Scheurer, M.; Ballantyne, C.; Alsarraj, A.; et al. Differences in prediagnostic serum metabolomic and lipidomic profiles between cirrhosis patients with and without incident hepatocellular carcinoma. J. Hepatocell. Carcinoma 2024, 11, 1699–1712. [Google Scholar] [CrossRef] [PubMed]
- Freiburghaus, K.; Largiadèr, C.R.; Stettler, C.; Fiedler, G.M.; Bally, L.; Bovet, C. Metabolomics by UHPLC-MS: Benefits provided by complementary use of Q-TOF and QQQ for pathway profiling. Metabolomics 2019, 15, 120. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Ali, J. UHPLC/Q-TOF-MS technique: Introduction and applications. Lett. Org. Chem. 2015, 12, 371–378. [Google Scholar] [CrossRef]
- Nishikawa, H.; Kita, R.; Kimura, T.; Ohara, Y.; Sakamoto, A.; Saito, S.; Nishijima, N.; Nasu, A.; Komekado, H.; Osaki, Y. Clinical implication of performance status in patients with hepatocellular carcinoma complicating with cirrhosis. J. Cancer 2015, 6, 394–402. [Google Scholar] [CrossRef]
- Zubrod, C.G.; Schneiderman, M.; Frei, E.; Brindley, C.; Lennard Gold, G.; Shnider, B.; Oviedo, R.; Gorman, J.; Jones, R.; Jonsson, U.; et al. Appraisal of methods for the study of chemotherapy of cancer in man: Comparative therapeutic trial of nitrogen mustard and triethylene thiophosphoramide. J. Chronic Dis. 1960, 11, 7–33. [Google Scholar] [CrossRef]
- Tsilimigras, D.I.; Hyer, J.M.; Diaz, A.; Bagante, F.; Ratti, F.; Marques, H.P.; Soubrane, O.; Lam, V.; Poultsides, G.A.; Popescu, I.; et al. Synergistic impact of alpha-fetoprotein and tumor burden on long-term outcomes following curative-intent resection of hepatocellular carcinoma. Cancers 2021, 13, 747. [Google Scholar] [CrossRef] [PubMed]
- Lima, H.A.; Endo, Y.; Moazzam, Z.; Alaimo, L.; Shaikh, C.; Munir, M.M.; Resende, V.; Guglielmi, A.; Marques, H.P.; Cauchy, F.; et al. TAC score better predicts survival than the BCLC following resection of hepatocellular carcinoma. J. Surg. Oncol. 2023, 127, 374–384. [Google Scholar] [CrossRef]
- Zeng, J.; Yin, P.; Tan, Y.; Dong, L.; Hu, C.; Huang, Q.; Lu, X.; Wang, H.; Xu, G. Metabolomics study of hepatocellular carcinoma: Discovery and validation of serum potential biomarkers by using capillary electrophoresis–mass spectrometry. J. Proteome Res. 2014, 13, 3420–3431. [Google Scholar] [CrossRef]
- Miura, K.; Nagahashi, M.; Prasoon, P.; Hirose, Y.; Kobayashi, T.; Sakata, J.; Abe, M.; Sakimura, K.; Matsuda, Y.; Butash, A.L.; et al. Dysregulation of sphingolipid metabolic enzymes leads to high levels of sphingosine-1-phosphate and ceramide in human hepatocellular carcinoma. Hepatol. Res. 2021, 51, 614–626. [Google Scholar] [CrossRef]
- Paul, B.; Lewinska, M.; Andersen, J.B. Lipid alterations in chronic liver disease and liver cancer. JHEP Rep. 2022, 4, 100479. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.; Zeng, Y.; Guo, Y.; Zhou, T.; Liu, C.; Ji, R.; Gao, Y. From cirrhosis to hepatogenous diabetes: Risk factors and glycemic management. Arch. Med. Sci. 2024, 21, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef]
- Uchida, K.; Shibata, T.; Toyokuni, S.; Daniel, B.; Zarkovic, K.; Zarkovic, N.; Sasson, S. Development of a novel monoclonal antibody against 4-hydroxy-2E,6Z-dodecadienal (4-HDDE)-protein adducts: Immunochemical application in quantitative and qualitative analyses of lipid peroxidation in vitro and ex vivo. Free Radic. Biol. Med. 2018, 124, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef]
- Kim, J.W.; Lee, S.H.; Park, Y.S.; Jeong, S.H.; Kim, N.; Lee, D.H.; Lee, H.S. Transcriptome analysis of hepatitis B virus-associated small hepatocellular carcinoma by serial analysis of gene expression. Int. J. Oncol. 2009, 35, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ding, C.; Chen, Y.; Hu, W.; Lu, Y.; Wu, W.; Zhang, Y.; Yang, B.; Wu, H.; Peng, C.; et al. ACSL4 promotes hepatocellular carcinoma progression via c-Myc stability mediated by ERK/FBW7/c-Myc axis. Oncogenesis 2020, 9, 42. [Google Scholar] [CrossRef]
- Abel, S.; De Kock, M.; van Schalkwyk, D.J.; Swanevelder, S.; Kew, M.C.; Gelderblom, W.C.A. Altered lipid profile, oxidative status and hepatitis B virus interactions in human hepatocellular carcinoma. Prostaglandins Leukot. Essent. Fat. Acids 2009, 81, 391–399. [Google Scholar] [CrossRef]
- Sun, X.; Seidman, J.S.; Zhao, P.; Troutman, T.D.; Spann, N.J.; Que, X.; Zhou, F.; Liao, Z.; Pasillas, M.; Yang, X.; et al. Neutralization of oxidized phospholipids ameliorates non-alcoholic steatohepatitis. Cell Metab. 2020, 31, 189–206. [Google Scholar] [CrossRef] [PubMed]
- Ranjpour, M.; Wajid, S.; Jain, S.K. Elevated Expression of Cytosolic Phospholipase A2 Delta Is Associated with Lipid Metabolism Dysregulation during Hepatocellular Carcinoma Progression. Cell J. 2020, 22, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Yahagi, N.; Shimano, H.; Hasegawa, K.; Ohashi, K.; Matsuzaka, T.; Najima, Y.; Sekiya, M.; Tomita, S.; Okazaki, H.; Tamura, Y.; et al. Co-ordinate activation of lipogenic enzymes in hepatocellular carcinoma. Eur. J. Cancer 2005, 41, 1316–1322. [Google Scholar] [CrossRef]
- Yang, W.S.; Stockwell, B.R. Ferroptosis: Death by lipid peroxidation. Trends Cell Biol. 2016, 26, 165–176. [Google Scholar] [CrossRef]
- Chen, J.; Li, X.; Ge, C.; Min, J.; Wang, F. The multifaceted role of ferroptosis in liver disease. Cell Death Differ. 2022, 29, 467–480. [Google Scholar] [CrossRef]
- Arana, L.; Gangoiti, P.; Ouro, A.; Trueba, M.; Gómez-Muñoz, A. Ceramide and ceramide 1-phosphate in health and disease. Lipids Health Dis. 2010, 9, 15. [Google Scholar] [CrossRef]
- Kurz, J.; Parnham, M.J.; Geisslinger, G.; Schiffmann, S. Ceramides as novel disease biomarkers. Trends Mol. Med. 2019, 25, 20–32. [Google Scholar] [CrossRef]
- Li, Z.; Guan, M.; Lin, Y.; Cui, X.; Zhang, Y.; Zhao, Z.; Zhu, J. Aberrant lipid metabolism in hepatocellular carcinoma revealed by liver lipidomics. Int. J. Mol. Sci. 2017, 18, 2550. [Google Scholar] [CrossRef]
- Krautbauer, S.; Meier, E.M.; Rein-Fischboeck, L.; Pohl, R.; Weiss, T.S.; Sigruener, A.; Aslanidis, C.; Liebisch, G.; Buechler, C. Ceramide and polyunsaturated phospholipids are strongly reduced in human hepatocellular carcinoma. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2016, 1861, 1767–1774. [Google Scholar] [CrossRef]
- Uranbileg, B.; Ikeda, H.; Kurano, M.; Enooku, K.; Sato, M.; Saigusa, D.; Aoki, J.; Ishizawa, T.; Hasegawa, K.; Kokudo, N.; et al. Increased mRNA levels of sphingosine kinases and S1P lyase and reduced levels of S1P were observed in hepatocellular carcinoma in association with poorer differentiation and earlier recurrence. PLoS ONE 2016, 11, e0149462. [Google Scholar] [CrossRef]
- Grammatikos, G.; Schoell, N.; Ferreirós, N.; Bon, D.; Herrmann, E.; Farnik, H.; Köberle, V.; Piiper, A.; Zeuzem, S.; Kronenberger, B.; et al. Serum sphingolipidomic analyses reveal an upregulation of C16-ceramide and sphingosine-1-phosphate in hepatocellular carcinoma. Oncotarget 2016, 7, 18095–18105. [Google Scholar] [CrossRef]
- Cai, H.; Xie, X.; Ji, L.; Ruan, X.; Zheng, Z. Sphingosine kinase 1: A novel independent prognosis biomarker in hepatocellular carcinoma. Oncol. Lett. 2017, 13, 2316–2322. [Google Scholar] [CrossRef]
- Maceyka, M.; Rohrbach, T.; Milstien, S.; Spiegel, S. Role of sphingosine kinase 1 and sphingosine-1-phosphate axis in hepatocellular carcinoma. Handb. Exp. Pharmacol. 2020, 259, 3–17. [Google Scholar] [CrossRef]
- Rashid, M.M.; Varghese, R.S.; Ding, Y.; Ressom, H.W. Biomarker discovery for hepatocellular carcinoma in patients with liver cirrhosis using untargeted metabolomics and lipidomics studies. Metabolites 2023, 13, 1047. [Google Scholar] [CrossRef] [PubMed]
- Belhaj, M.R.; Lawler, N.G.; Hoffman, N.J. Metabolomics and lipidomics: Expanding the molecular landscape of exercise biology. Metabolites 2021, 11, 151. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Yin, D.; Yang, X.; Liu, D.; Shan, H.; Luo, J.; Li, X.; Yin, Y. Plasma lipidomic analysis reveals disruption of ether phosphatidylcholine biosynthesis and facilitates early detection of hepatitis B-related hepatocellular carcinoma. Lipids Health Dis. 2025, 24, 69. [Google Scholar] [CrossRef]
- Wei, F.; Lamichhane, S.; Orešič, M.; Hyötyläinen, T. Lipidomes in health and disease: Analytical strategies and considerations. TrAC Trends Anal. Chem. 2019, 120, 115664. [Google Scholar] [CrossRef]
- von Gerichten, J.; Saunders, K.; Bailey, M.J.; Gethings, L.A.; Onoja, A.; Geifman, N.; Spick, M. Challenges in lipidomics biomarker identification: Avoiding the pitfalls and improving reproducibility. Metabolites 2024, 14, 461. [Google Scholar] [CrossRef] [PubMed]
- Cruz, A.L.S.; Barreto, E.d.A.; Fazolini, N.P.B.; Bozza, P.T. Lipid droplets: Platforms with multiple functions in cancer hallmarks. Cell Death Dis 2020, 11, 105. [Google Scholar] [CrossRef]
- Petan, T.; Jarc, E.; Jusović, M. Lipid Droplets in Cancer: Guardians of Fat in a Stressful World. Molecules 2018, 23, 1941. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Tang, O.; Vernon, S.T.; Kott, K.A.; Koay, Y.C.; Park, J.; James, D.E.; Grieve, S.M.; Speed, T.P.; Yang, P.; et al. A hierarchical approach to removal of unwanted variation for large-scale metabolomics data. Nat. Commun. 2021, 12, 4992. [Google Scholar] [CrossRef] [PubMed]
| Untargeted multivariate analysis: comparison of metabolic fingerprints between the CIR and HCC groups (322 metabolites with MW 120–760 Da) |
|
| Semi-targeted analysis: comparison of the profiles of 11 metabolite classes. Free fatty acids (FFAs) (n = 53) Fatty acid derivatives (n = 17) Glycerophospholipids (n = 36) Lysophospholipids (n = 32) Acylcarnitines (n = 25) Mono- and diglycerides (n = 22) Sphingolipids (n = 37) Sterol lipids (n = 33) Oxilipins (n = 11) Antioxidant lipids (n = 5) Polar molecules (n = 51) |
| Parameters | HCC Group (n = 41) | Cirrhosis Group (n = 40) | p-Value |
|---|---|---|---|
| Age | 66.95 (±9.02) | 60.03 (±8.87) | 0.001 |
| Sex, no (%) ** | 0.94 | ||
| Female | 11 (28.6%) | 11 (27.5%) | |
| Male | 30 (73.2%) | 29 (72.5%) | |
| Environment (%) ** | 0.58 | ||
| Rural | 12 (29.3%) | 14 (35%) | |
| Urban | 29 (70.7%) | 26 (65%) | |
| Etiology (%) ** | 0.001 | ||
| HCV | 11 (26.8%) | 5 (12.5%) | |
| HBV | 7 (17.1%) | 5 (12.5%) | |
| Alcohol intake | 8 (19.5%) | 30 (75%) | |
| Deceased (%) | 0.58 | ||
| Yes | 9 (21.9%) | 7 (17.5%) | |
| No | 32 (78.1%) | 33 (82.5%) | |
| ALAT * | 26.3 (19.7–46.5) | 28.8 (21–51.9) | 0.74 |
| ASAT * | 32 (26–50.5) | 53.1 (32.9–97.6) | 0.11 |
| TB * | 0.7 (0.5–1.35) | 1.79 (0.88–2.85) | 0.04 |
| Albumin * | 4.33 (0.47) | 3.63 (0.61) | 0.001 |
| Platelet count * | 180.05 (84.54) | 142.75 (85.96) | 0.06 |
| AFP * | 5.27 (2.64–37.6) | 4.13 (2.30–6.39) | 0.34 |
| INR * | 1.15 (0.19) | 1.40 (0.28) | 0.03 |
| Triglycerides * | 100.80 (64.54) | 83.57 (37.18) | 0.12 |
| Cholesterol * | 155.78 (63.74) | 134.10 (43.42) | 0.14 |
| Ascites ** | 0.001 | ||
| Yes | 6 (14.6%) | 18 (45%) | |
| No | 35 (85.4%) | 22 (55%) | |
| Child-Pugh classification ** | 0.001 | ||
| A | 21 (51.2%) | 16 (40%) | |
| B | 6 (14.6%) | 24 (60%) | |
| without cirrhosis | 14 (34.1%) | 0 | |
| HVPG * | 8.72 (4.21) | 12.88 (3.27) | 0.02 |
| TE-LSM * | 14.49 (13.80) | 51.49 (25.63) | 0.001 |
| TAC score | N/A | N/A | |
| very low | 1 (2.4) | ||
| low | 9 (22) | ||
| medium | 27 (65.9) | ||
| high | 4 (9.8) | ||
| BCLC ** | N/A | N/A | |
| 0 | 4 (9.2%) | ||
| A | 34 (82.9%) | ||
| B | 3 (7.3%) | ||
| MELD score ** | 0.001 | ||
| ≤9 | 24 (58.5%) | 5 (12.5%) | |
| 10–19 | 17 (41.5%) | 35 (87.5%) | |
| Milan criteria ** | N/A | N/A | |
| In | 13 (31.7%) | ||
| Out | 28 (68.3%) |
| m/z | VIP | FC | log2FC | p-Value | MDA | Relative Variation | |
|---|---|---|---|---|---|---|---|
| 181.1081 | D-Glucose | 1.903 | 2.069 | 1.049 | 2.88 × 10−15 | <0.07 | HCC < CIR |
| 167.0921 | Methylxanthine | 1.762 | 2.008 | 1.01 | 1.65 × 10−12 | <0.07 | HCC < CIR |
| 143.0953 | 5-Hydroxymethyluracil | 1.967 | 1.863 | 0.898 | 2.00 × 10−19 | <0.07 | HCC < CIR |
| 595.3371 | LysoPI 18:3 | 1.781 | 0.715 | −0.484 | 8.77 × 10−9 | 0.0113 | HCC > CIR |
| 638.5631 | Cer(d16:2/24:0(2OH)) | 1.070 | 0.715 | −0.482 | 3.13 × 10−4 | 0.0074 | HCC > CIR |
| 600.4239 | DG(34:0) | 1.843 | 0.681 | −0.554 | 4.25 × 10−12 | <0.07 | HCC > CIR |
| 644.4472 | GlcCer(d18:1/12:0) | 1.857 | 0.624 | −0.680 | 9.47 × 10−13 | 0.0082 | HCC > CIR |
| 688.4706 | PE 32:2 | 1.756 | 0.568 | −0.815 | 2.41 × 10−11 | 0.0112 | HCC > CIR |
| 713.3923 | CerPE(d14:2/24:1) | 1.953 | 0.551 | −0.859 | 6.30 × 10−15 | 0.0111 | HCC > CIR |
| 628.4536 | DG 36:0 | 1.829 | 0.549 | −0.866 | 1.50 × 10−12 | 0.0095 | HCC > CIR |
| 581.3243 | LysoLPC 22:1 | 1.263 | 0.490 | −1.026 | 2.01 × 10−11 | 0.0073 | HCC > CIR |
| 589.3852 | LysoPI O-18:0 | 1.781 | 0.461 | −1.118 | 7.67 × 10−13 | 0.0078 | HCC > CIR |
| 669.3689 | DG 40:6 | 1.838 | 0.459 | −1.124 | 1.50 × 10−8 | 0.0131 | HCC > CIR |
| 649.4018 | PA(16:0/16:0) | 1.799 | 0.452 | −1.147 | 3.63 × 10−13 | 0.0105 | HCC > CIR |
| 732.4949 | PC(16:0/16:1) | 1.006 | 0.433 | −1.21 | 2.63 × 10−12 | 0.0083 | HCC > CIR |
| 757.4159 | CerPE(d16:2/24:1(2OH)) | 2.144 | 0.419 | −1.256 | 4.28 × 10−22 | 0.0130 | HCC > CIR |
| 677.4322 | SM(d18:0/14:0) | 1.678 | 0.402 | −1.31 | 4.08 × 10−11 | 0.0100 | HCC > CIR |
| 693.4258 | PA(36:6) | 1.866 | 0.377 | −1.409 | 1.80 × 10−14 | 0.0163 | HCC > CIR |
| 765.4801 | PS(18:0/16:0) | 1.786 | 0.326 | −1.615 | 6.27 × 10−13 | 0.0122 | HCC > CIR |
| 781.4709 | PA 42:4 | 1.774 | 0.279 | −1.840 | 1.01 × 10−12 | <0.07 | HCC > CIR |
| Identification | AUC | p-Value | Log2 FC | Relative Variation |
|---|---|---|---|---|
| CerPE(d16:2/24:1(2OH)) | 1 | 1.51 × 10−20 | −1.014 | HCC > CIR |
| Cer(t18:0/20:0(2OH)) | 0.991 | 1.01 × 10−12 | −0.636 | HCC > CIR |
| 5-Hydroxymethyluracil | 0.990 | 4.27 × 10−19 | 0.946 | HCC < CIR |
| PA(36:6) | 0.988 | 1.04 × 10−13 | −1.132 | HCC > CIR |
| DG(34:4) | 0.981 | 5.37 × 10−12 | −0.852 | HCC > CIR |
| PA(16:0/16:0) | 0.980 | 2.64 × 10−12 | −0.896 | HCC > CIR |
| D-Glucose | 0.976 | 2.07 × 10−16 | 1.127 | HCC < CIR |
| PA(34:4) | 0.975 | 8.97 × 10−13 | −0.564 | HCC > CIR |
| DG (44:12) | 0.973 | 4.60 × 10−15 | −0.671 | HCC > CIR |
| PS(18:0/16:0) | 0.969 | 2.50 × 10−12 | −1.340 | HCC > CIR |
| 5-Methoxytryptophan | 0.963 | 2.13 × 10−8 | 0.628 | HCC < CIR |
| SM(d18:0/14:0) | 0.962 | 1.51 × 10−10 | −1.047 | HCC > CIR |
| GlcCer(d18:1/12:0) | 0.962 | 8.46 × 10−13 | −0.502 | HCC > CIR |
| Palmitoleyl linolenate | 0.955 | 7.82 × 10−11 | −0.583 | HCC > CIR |
| Cer(t18:0/18:0(2OH)) | 0.955 | 3.36 × 10−12 | −0.385 | HCC > CIR |
| Metabolite Classes | AUC | HCC > CIR | HCC < CIR |
|---|---|---|---|
| Free fatty acids | 0.934–0.742 | C17:1; C38:0; C40:6; C20:5-O; C18:0; C14:0 | C22:5; C14:2; C30:3; C16:1; C22:6 |
| Fatty acid derivatives | 0.810–0.503 | Linoleyl arachidate; Linoleyl linoleate; Linoleyl arachidonate; Stearyl stearate; Oleyl palmitate | Stearamide; Docosenamide; Amino-octanoic acid; Myristyl palmitate; Myristoleyl arachidonate |
| Glycerophospholipids | 0.877–0.648 | PA 42:4; PS 34:0; PA 38:6; PG O-34:4; PA 36:6 | PC (23:2; O); PA 30:2; PA(O-18:0/16:0); PA 23:0; PE 30:3 |
| Lysophospholipids | 0.793–0.586 | LysoPC(20:3); LysoPC (22:1); LysoPC(22:6); LysoPI (18:3); LysoPA (18:1) | LysoPC (19:3); LysoPE (22:6); LysoPE 18:0); LysoPE (16:1) |
| Acylcarnitines | 0.777–0.516 | C18:1;O2; CAR 20:0; CAR 12:2; CAR 26:0; CAR 12:0;O; CAR 12:1;O; CAR 16:1;O; CAR 12:1; CAR 18:2; CAR 14:0 | CAR16:2; CAR 14:0; CAR 16:1; C16:0; CAR 18:3;O |
| Mono- and Diglycerides | 0.830–0.518 | DG40:7; MGDG (34:3); MGMG (16:2); DG(42:0); DG(34:1) | DG(33:4); DG(35:1); MG(20:4); DG (44:0); DG (40:1) |
| Sphingolipids | 0.911–0.666 | CerPE(d16:2/24:1(2OH)); SM(d18:1/18:1); SM(d18:0/14:0); CerPE(d16:2/20:1(2OH)); GlcCer(d18:1/14:0); CerPE(d16:1/16:0); Cer(t18:0/20:0(2OH)) | Sphingosine18:2; O2; CerPE(d14:2/16:0(2OH)); Cer(t18:1(6OH)/16:0(2OH)); C19 Sphingosine-1-phosphate; Cer(t18:0/19:0(2OH)); Cer(d18:2/20:1) |
| Sterol lipids | 0.771–0.580 | 25-Hydroxyvitamin D2; 3-Oxocholic acid; 21-hydroxypregnenolone; Dihomocholic acid; Deoxycholic acid; Ketodeoxycholic acid | Cortisol; Alpha-androstenol; Dihydrocorticosterone; Corticosterone; Hydroxycortisone; Dihydroxycholesterol; 18:0 Cholesterol ester; 12-Estrone 3-sulfate; Estradiol-17beta; Cortisol 21-sulfate |
| Antioxidants | 0.898–0.524 | Ascorbyl palmitate, all-trans-retinyl oleate; beta-carotene | Alpha-Tocotrienol |
| Oxylipins | 0.729–0.502 | PGF1a; Epoxy PGE1; Hydroxy-PGF1a; 15-HETE-GABA | HETE-Ethanolamine, PGE3; PGA2; 9-HODE; Lipoxin A4 |
| Polar molecules | 0.870–0.715 | N-Oleoylethanolamine; 5-Hydroxymethyluracil; Proline betaine; Phosphoserine; Trytptophan, N-stearoyl phenylalanine | N-Acetyl-D-glucosamine; N-Palmitoyltryptamine; D-Glucose; Taurine; L-Homocysteine sulfate 5-Hydroxymethyluracil |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ursu, C.-P.; Furcea, L.E.; Procopeț, B.; Ciocan, R.A.; Ursu, Ș.; Gherman, C.D.; Vălean, D.; Pop, R.S.; Moiș, E.I.; Ștefănescu, H.; et al. Differential Metabolic Dysregulations in Hepatocellular Carcinoma and Cirrhosis: Insights into Lipidomic Signatures. Biomolecules 2025, 15, 1575. https://doi.org/10.3390/biom15111575
Ursu C-P, Furcea LE, Procopeț B, Ciocan RA, Ursu Ș, Gherman CD, Vălean D, Pop RS, Moiș EI, Ștefănescu H, et al. Differential Metabolic Dysregulations in Hepatocellular Carcinoma and Cirrhosis: Insights into Lipidomic Signatures. Biomolecules. 2025; 15(11):1575. https://doi.org/10.3390/biom15111575
Chicago/Turabian StyleUrsu, Cristina-Paula, Luminița Elena Furcea, Bogdan Procopeț, Răzvan Alexandru Ciocan, Ștefan Ursu, Claudia Diana Gherman, Dan Vălean, Rodica Sorina Pop, Emil Ioan Moiș, Horia Ștefănescu, and et al. 2025. "Differential Metabolic Dysregulations in Hepatocellular Carcinoma and Cirrhosis: Insights into Lipidomic Signatures" Biomolecules 15, no. 11: 1575. https://doi.org/10.3390/biom15111575
APA StyleUrsu, C.-P., Furcea, L. E., Procopeț, B., Ciocan, R. A., Ursu, Ș., Gherman, C. D., Vălean, D., Pop, R. S., Moiș, E. I., Ștefănescu, H., Socaciu, C., Al Hajjar, N., & Graur, F. (2025). Differential Metabolic Dysregulations in Hepatocellular Carcinoma and Cirrhosis: Insights into Lipidomic Signatures. Biomolecules, 15(11), 1575. https://doi.org/10.3390/biom15111575








