Beyond Processing: Furin as a Central Hub in Viral Pathogenesis and Genetic Susceptibility
Abstract
1. Introduction
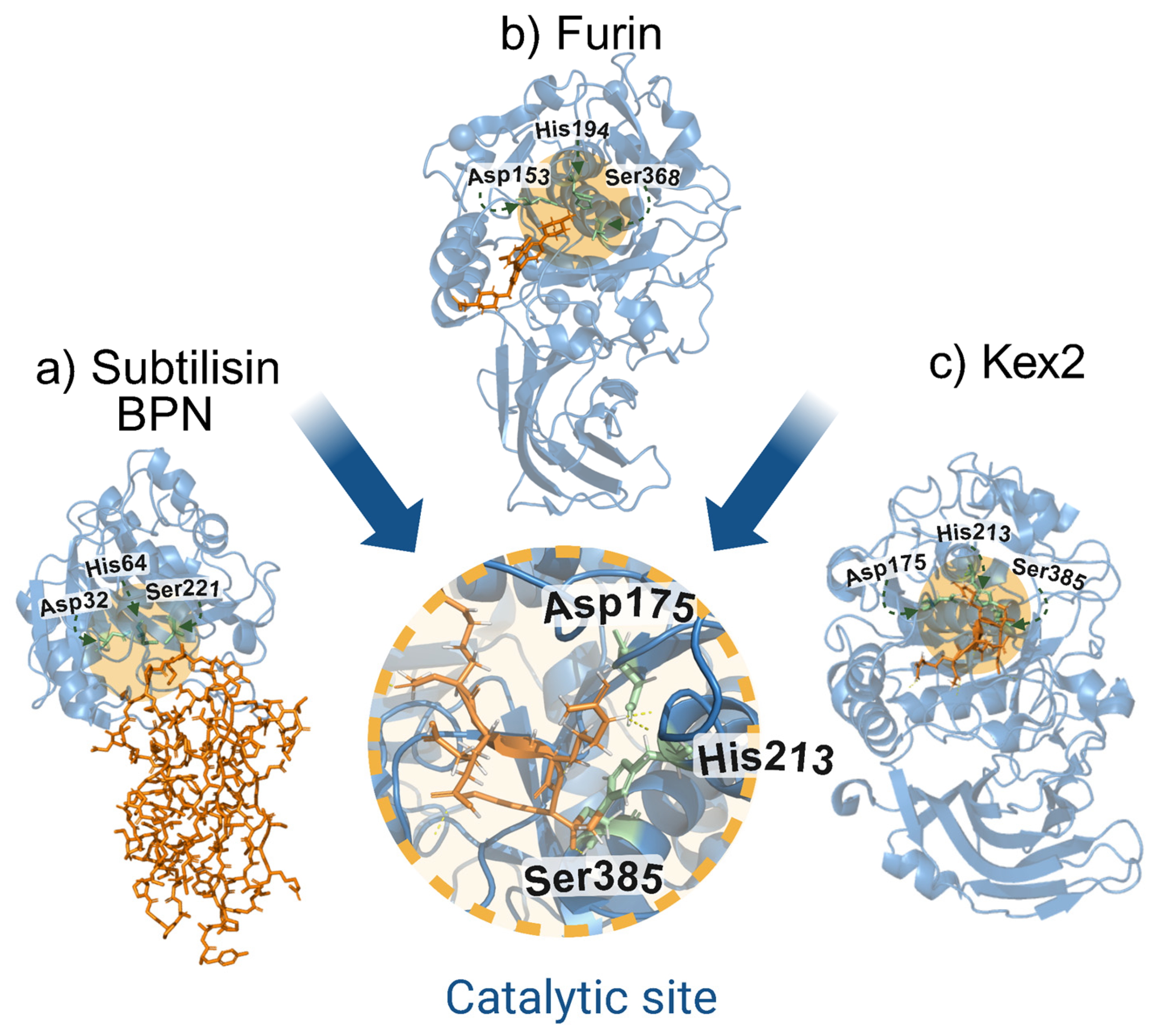
2. History of the Discovery of Furin
3. Furin
3.1. Encoding
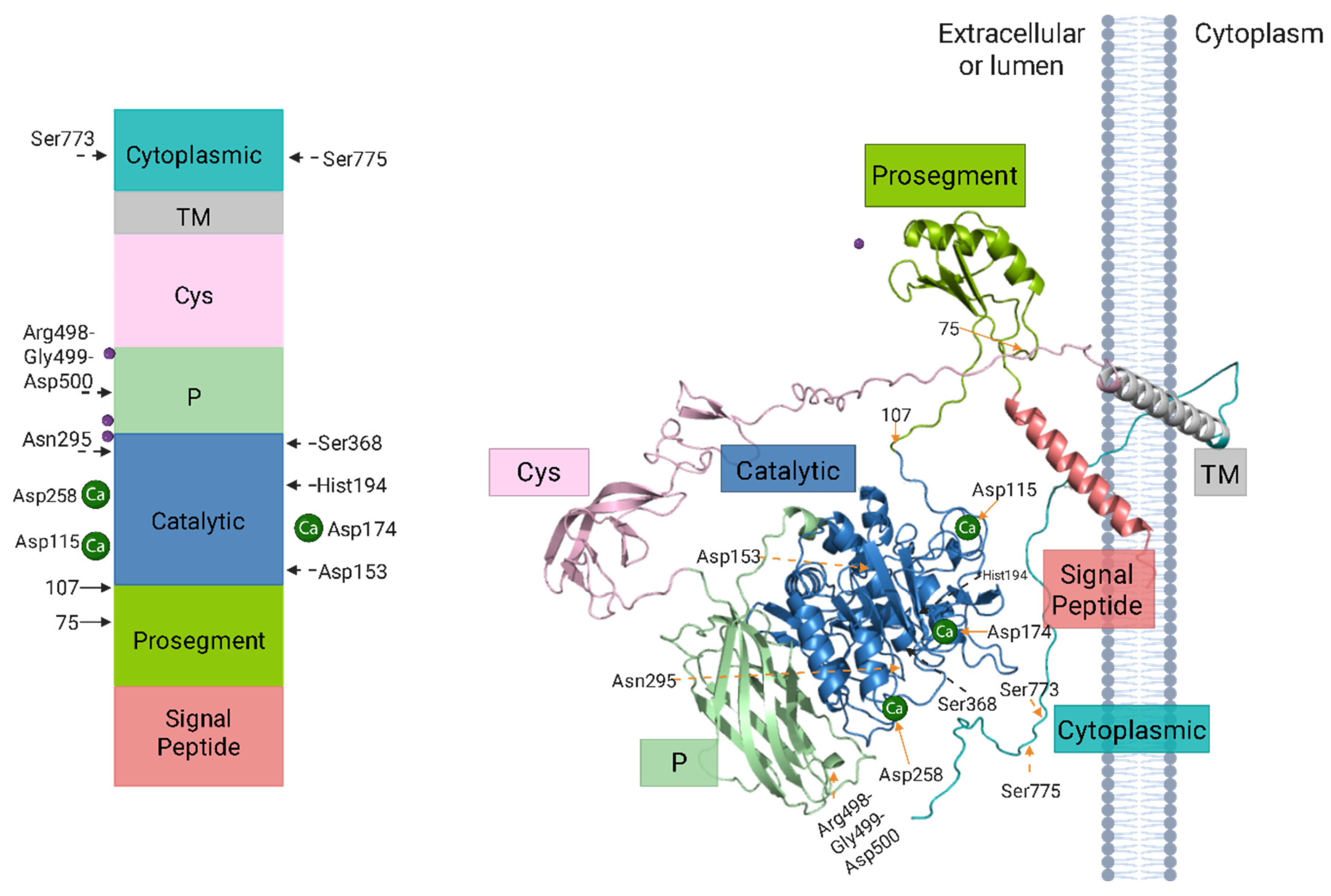
3.2. Molecular Structure and Functional Domains of Furin
3.3. Mechanisms of Furin Activation and Maturation
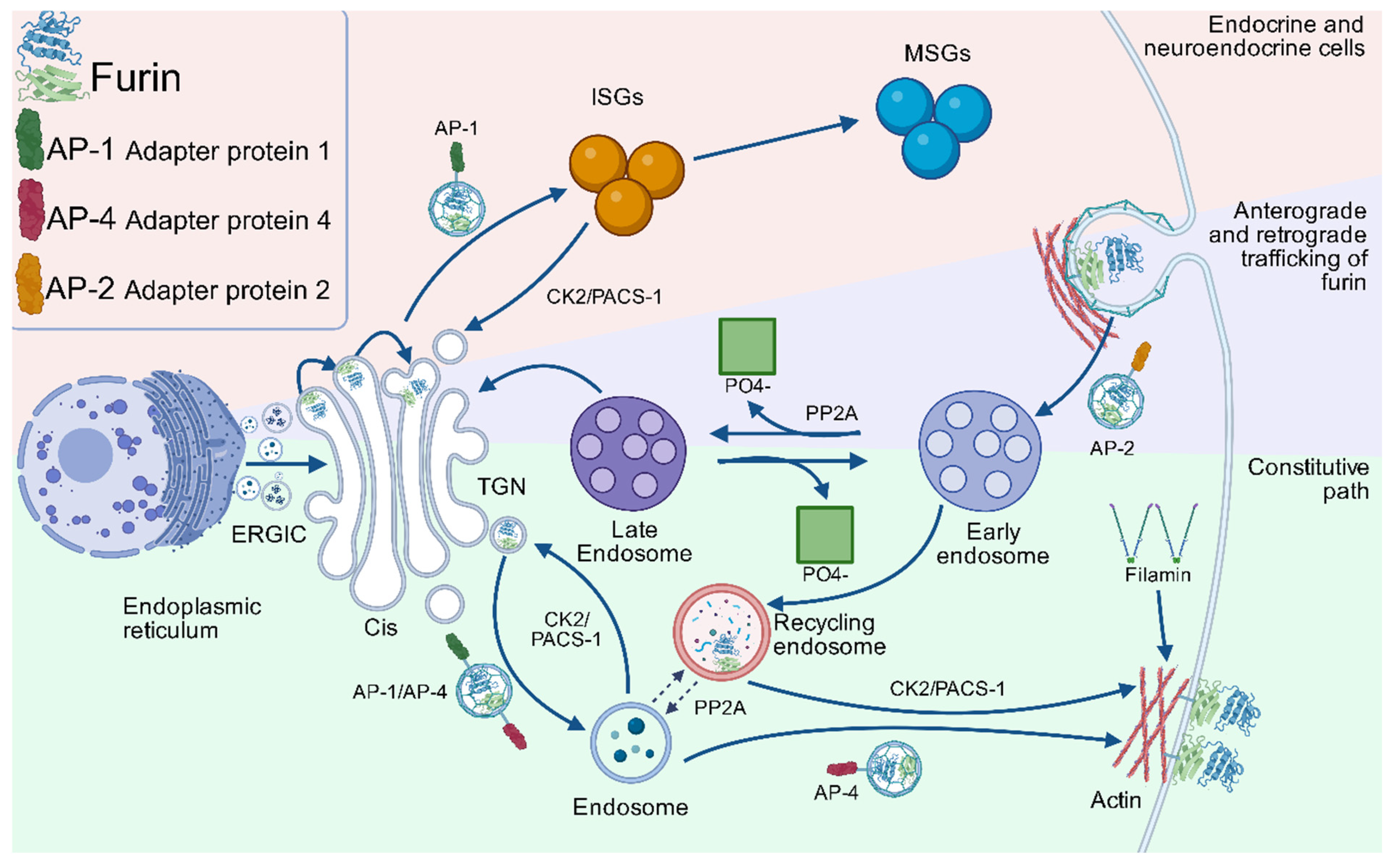
3.4. Intracellular Trafficking Dynamics and Recycling of Furin
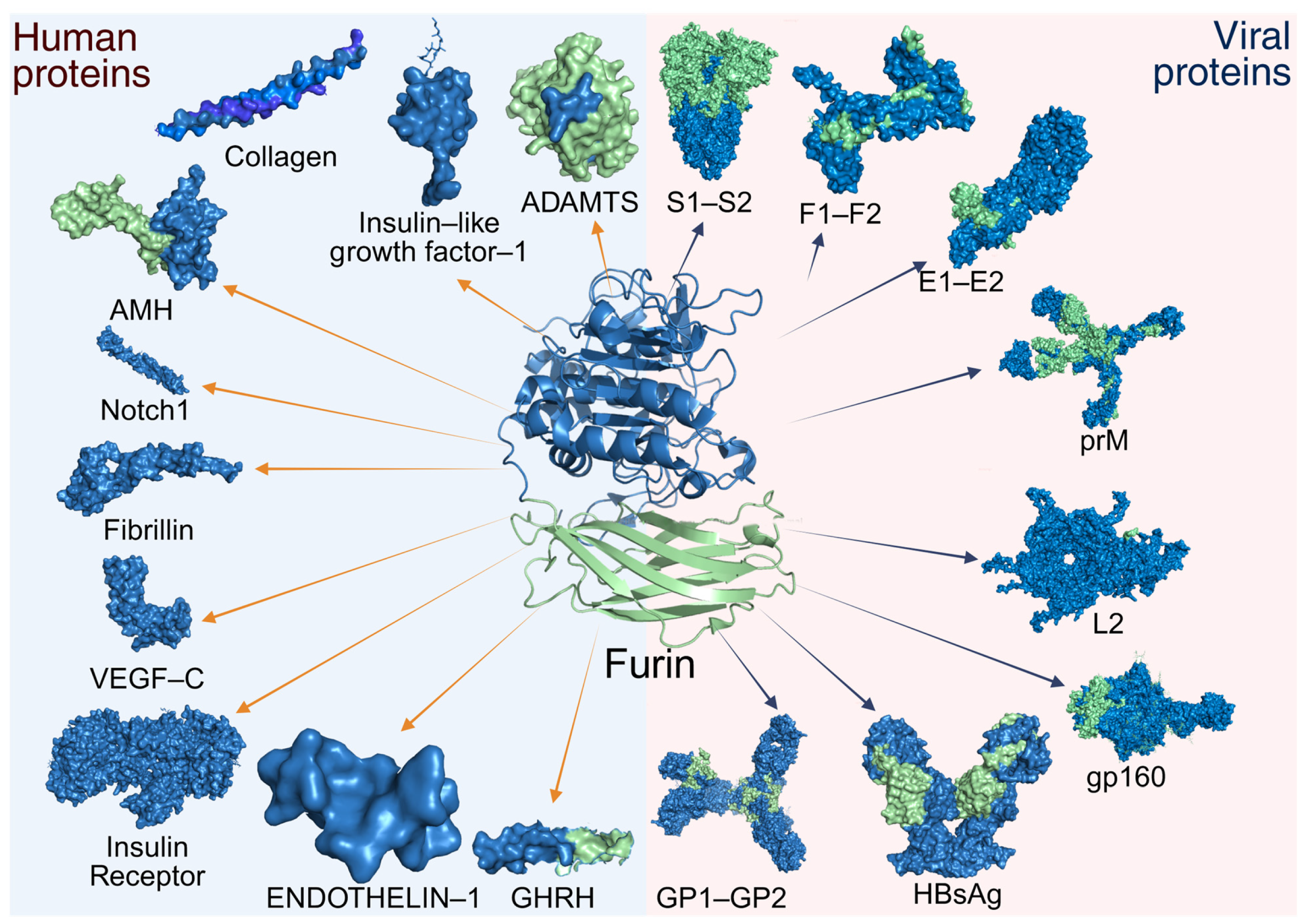
4. Functional Diversity of Furin Substrates
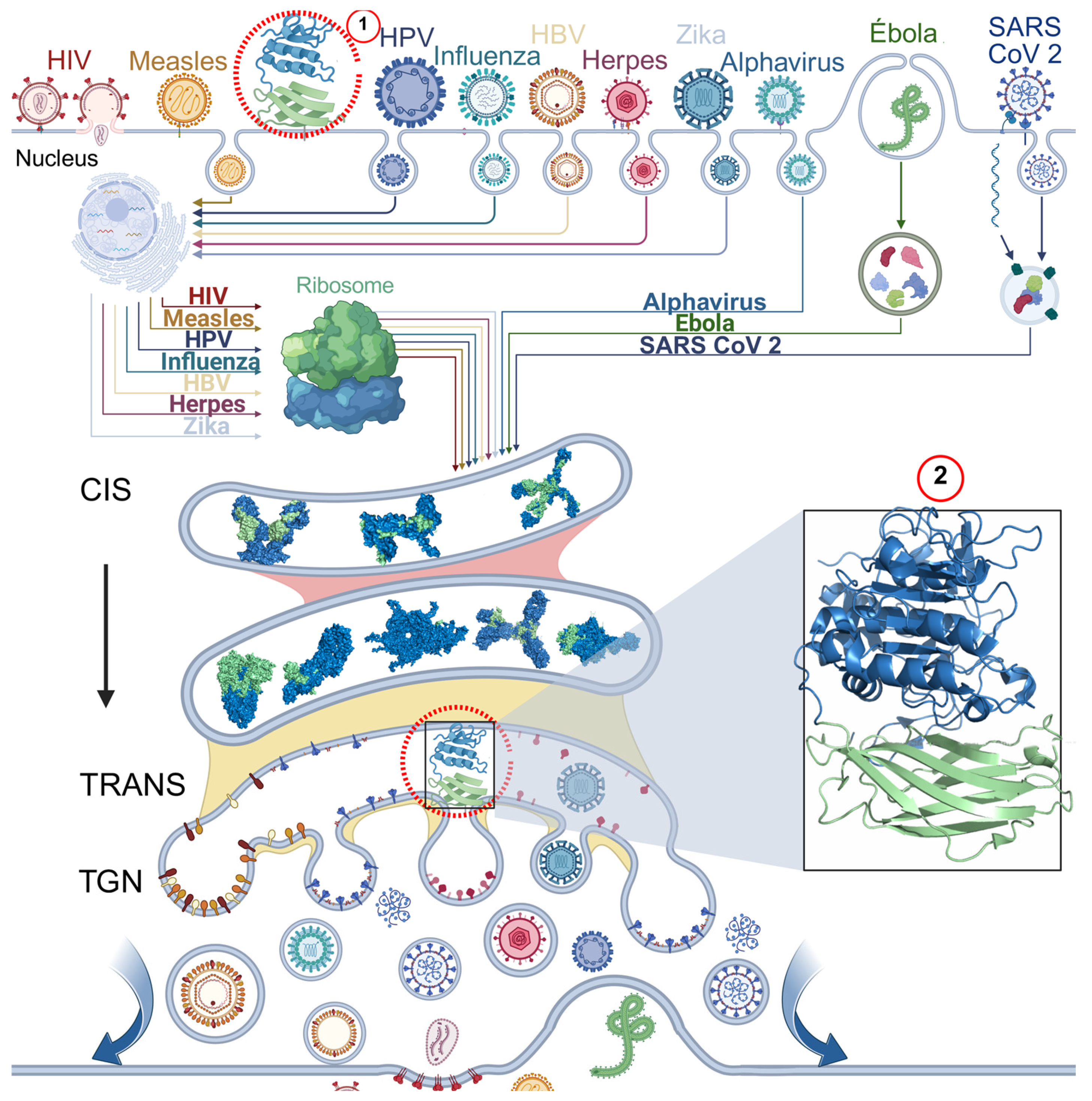
5. Furin-Dependent Viral Processing Mechanisms
5.1. Human Papillomavirus
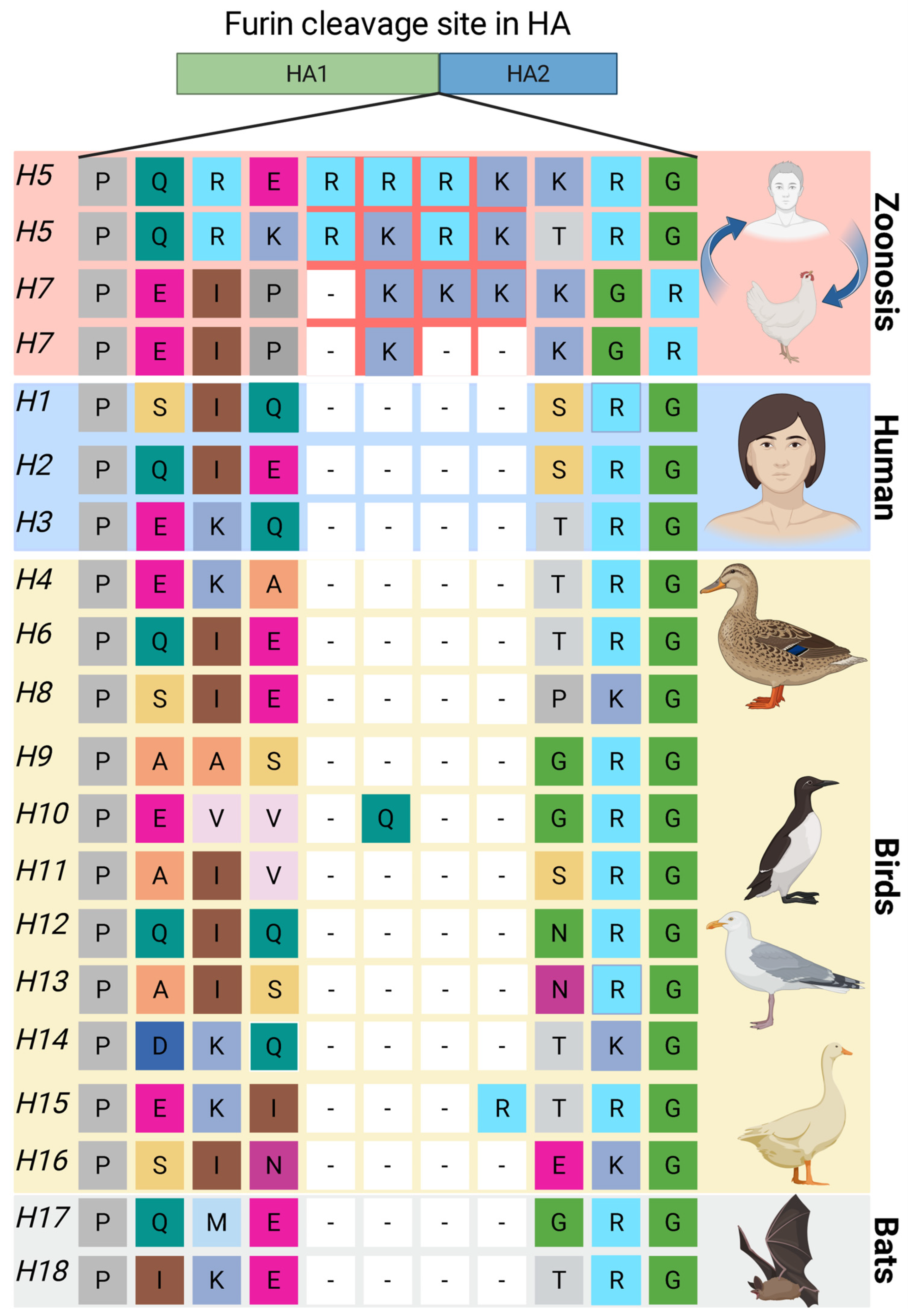
5.2. Influenza Virus
5.3. Herpesviridae Family
5.4. Togaviridae Family
5.5. Hepatitis B Virus
5.6. Filoviridae Family
5.7. Paramyxoviridae Family
5.8. Human Immunodeficiency Virus
5.9. Flavivirus Genus
5.10. Coronaviruses
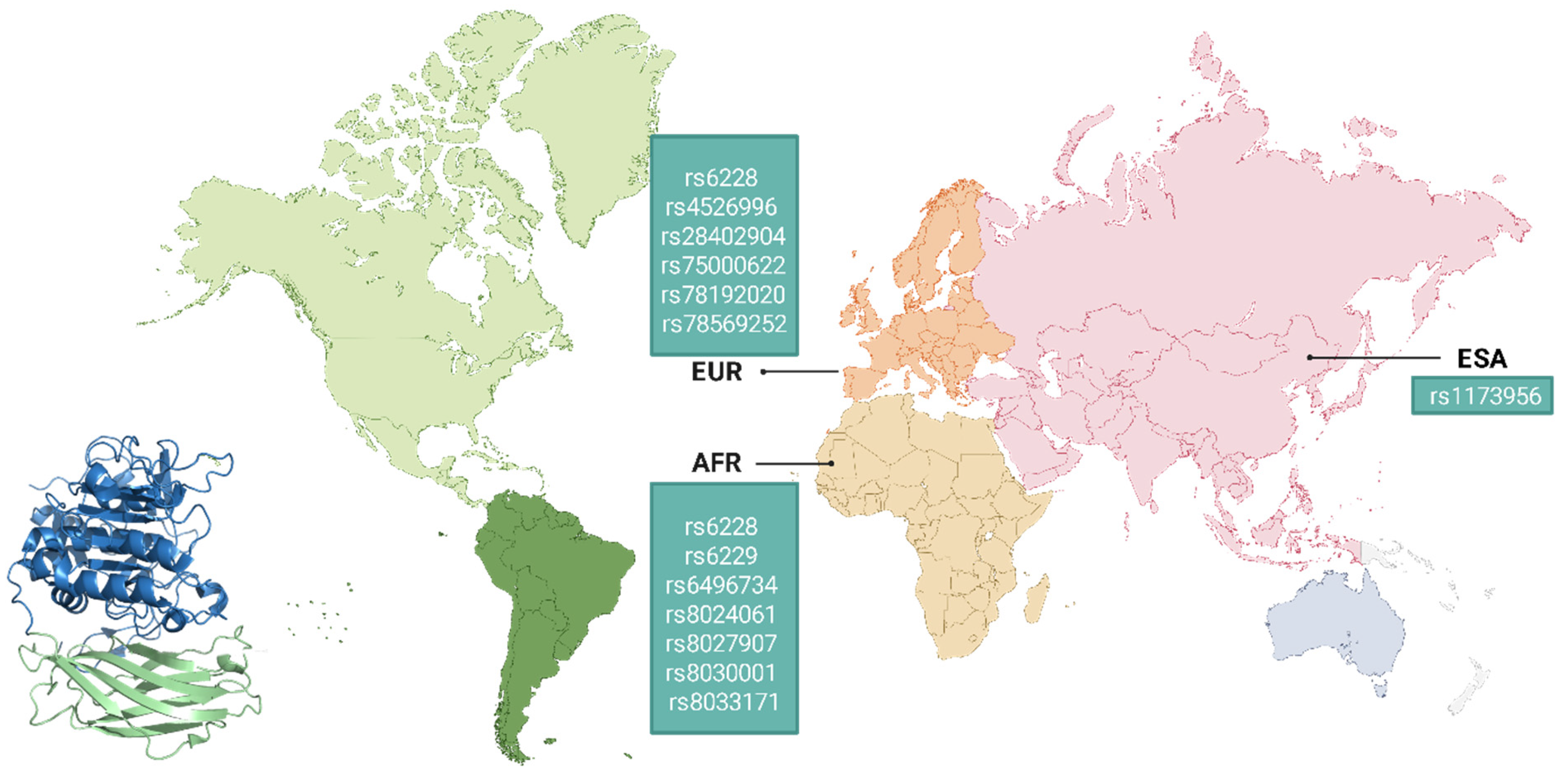
6. Genetic Variability of Furin and Its Implications in Viral Pathogenesis
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van de Ven, W.J.; Creemers, J.W.; Roebroek, A.J. Furin: The prototype mammalian subtilisin-like proprotein-processing enzyme. Endoproteolytic cleavage at paired basic residues of proproteins of the eukaryotic secretory pathway. Enzyme 1991, 45, 257–270. [Google Scholar] [CrossRef]
- Rozanov, A.S.; Shekhovtsov, S.V.; Bogacheva, N.V.; Pershina, E.G.; Ryapolova, A.V.; Bytyak, D.S.; Peltek, S.E. Production of subtilisin proteases in bacteria and yeast. Vavilovskii Zhurnal Genet. Sel. 2021, 25, 125–134. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Bateman, A. MEROPS: The peptidase database. Nucleic Acids Res. 2008, 36, D320–D325. [Google Scholar] [CrossRef] [PubMed]
- Thacker, C.; Rose, A.M. A look at the Caenorhabditis elegans Kex2/Subtilisin-like proprotein convertase family. BioEssays 2000, 22, 545–553. [Google Scholar] [CrossRef]
- Seidah, N.G.; Chrétien, M. Eukaryotic protein processing: Endoproteolysis of precursor proteins. Curr. Opin. Biotechnol. 1997, 8, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.; Jiang, T.; Zheng, X.L.; Zhao, G.J. Proprotein convertase furin/PCSK3 and atherosclerosis: New insights and potential therapeutic targets. Atherosclerosis 2017, 262, 163–170. [Google Scholar] [CrossRef]
- Declercq, J.; Creemers, J.W.M. Chapter 725—Furin. In Handbook of Proteolytic Enzymes, 3rd ed.; Rawlings, N.D., Salvesen, G., Eds.; Academic Press: Oxford, UK, 2013; pp. 3281–3285. [Google Scholar]
- Seidah, N.G.; Prat, A. The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discov. 2012, 11, 367–383. [Google Scholar] [CrossRef]
- Molloy, S.S.; Bresnahan, P.A.; Leppla, S.H.; Klimpel, K.R.; Thomas, G. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J. Biol. Chem. 1992, 267, 16396–16402. [Google Scholar] [CrossRef]
- Seidah, N.G.; Day, R.; Marcinkiewicz, M.; ChrÉtien, M. Precursor Convertases: An Evolutionary Ancient, Cell-Specific, Combinatorial Mechanism Yielding Diverse Bioactive Peptides and Proteins. Ann. N. Y. Acad. Sci. 1998, 839, 9–24. [Google Scholar] [CrossRef]
- Zhao, G.; Yang, W.; Wu, J.; Chen, B.; Yang, X.; Chen, J.; McVey, D.G.; Andreadi, C.; Gong, P.; Webb, T.R.; et al. Influence of a Coronary Artery Disease-Associated Genetic Variant on FURIN Expression and Effect of Furin on Macrophage Behavior. Arter. Thromb. Vasc. Biol. 2018, 38, 1837–1844. [Google Scholar] [CrossRef]
- Turpeinen, H.; Seppälä, I.; Lyytikäinen, L.P.; Raitoharju, E.; Hutri-Kähönen, N.; Levula, M.; Oksala, N.; Waldenberger, M.; Klopp, N.; Illig, T.; et al. A genome-wide expression quantitative trait loci analysis of proprotein convertase subtilisin/kexin enzymes identifies a novel regulatory gene variant for FURIN expression and blood pressure. Hum. Genet. 2015, 134, 627–636. [Google Scholar] [CrossRef]
- Marafie, S.K.; Al-Mulla, F. An Overview of the Role of Furin in Type 2 Diabetes. Cells 2023, 12, 2407. [Google Scholar] [CrossRef] [PubMed]
- Al Madhoun, A.; Mohammad, A.; Abu-Farha, M.; Hebbar, P.; Haddad, D.; Miranda, L.; Channanath, A.; Nizam, R.; Bourashed, M.; Ahmad, R.; et al. FURIN R81C variant: A link to type 2 diabetes via impaired enzymatic activity. Am. J. Physiol. Endocrinol. Metab. 2025, 329, E179–E189. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, Y.; Zhang, J.; Chen, L.; Li, J.; Zhang, M.; Zhang, Q.; Lu, Y.; Jiang, J.; Zhang, X.; et al. FURIN Promoter Methylation Predicts the Risk of Incident Diabetes: A Prospective Analysis in the Gusu Cohort. Front. Endocrinol. 2022, 13, 873012. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Fu, Z.Z.; Zhang, Y.Q.; Fu, B.H.; Dong, L. The G to A transformation of rs4702 polymorphism in 3′UTR of FURIN reduced the risk of radiotherapy-induced cognitive impairment in glioma patients. J. Cell Mol. Med. 2022, 26, 684–692. [Google Scholar] [CrossRef]
- Hou, Y.; Liang, W.; Zhang, J.; Li, Q.; Ou, H.; Wang, Z.; Li, S.; Huang, X.; Zhao, C. Schizophrenia-associated rs4702 G allele-specific downregulation of FURIN expression by miR-338-3p reduces BDNF production. Schizophr. Res. 2018, 199, 176–180. [Google Scholar] [CrossRef]
- Stieneke-Gröber, A.; Vey, M.; Angliker, H.; Shaw, E.; Thomas, G.; Roberts, C.; Klenk, H.D.; Garten, W. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992, 11, 2407–2414. [Google Scholar] [CrossRef]
- Hallenberger, S.; Bosch, V.; Angliker, H.; Shaw, E.; Klenk, H.D.; Garten, W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gpl60. Nature 1992, 360, 358–361. [Google Scholar] [CrossRef]
- Nakayama, K. Furin: A mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 1997, 327, 625–635. [Google Scholar] [CrossRef]
- Linderstrom-Land, K.; Ottesen, M. A new protein from ovalbumin. Nature 1947, 159, 807. [Google Scholar] [CrossRef]
- Guntelberg, A.V.; Ottesen, M. Preparation of crystals containing the plakalbumin-forming enzyme from Bacillus subtilis. Nature 1952, 170, 802. [Google Scholar] [CrossRef] [PubMed]
- Guntelberg, A.V.; Ottesen, M. Purification of the proteolytic enzyme from Bacillus subtilis. Comptes Rendus Trav. Lab. Carlsberg 1954, 29, 36–48. [Google Scholar]
- Smith, E.L.; Markland, F.S.; Kasper, C.B.; DeLange, R.J.; Landon, M.; Evans, W.H. The complete amino acid sequence of two types of subtilisin, BPN’ and Carlsberg. J. Biol. Chem. 1966, 241, 5974–5976. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.S.; Alden, R.A.; Kraut, J. Structure of subtilisin BPN’ at 2.5 angström resolution. Nature 1969, 221, 235–242. [Google Scholar] [CrossRef]
- Steiner, D.F.; Cunningham, D.; Spigelman, L.; Aten, B. Insulin biosynthesis: Evidence for a precursor. Science 1967, 157, 697–700. [Google Scholar] [CrossRef]
- Chance, R.E.; Ellis, R.M.; Bromer, W.W. Porcine proinsulin: Characterization and amino acid sequence. Science 1968, 161, 165–167. [Google Scholar] [CrossRef]
- Chrétien, M.; Li, C.H. Isolation, purification, and characterization of gamma-lipotropic hormone from sheep pituitary glands. Can. J. Biochem. 1967, 45, 1163–1174. [Google Scholar] [CrossRef]
- Seidah, N.G.; Mayer, G.; Zaid, A.; Rousselet, E.; Nassoury, N.; Poirier, S.; Essalmani, R.; Prat, A. The activation and physiological functions of the proprotein convertases. Int. J. Biochem. Cell Biol. 2008, 40, 1111–1125. [Google Scholar] [CrossRef]
- Alden, R.A.; Wright, C.S.; Kraut, J. A hydrogen-bond network at the active site of subtilisin BPN’. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1970, 257, 119–124. [Google Scholar]
- Figueiredo, J.; Sousa Silva, M.; Figueiredo, A. Subtilisin-like proteases in plant defence: The past, the present and beyond. Mol. Plant Pathol. 2018, 19, 1017–1028. [Google Scholar] [CrossRef]
- Valbuzzi, A.; Ferrari, E.; Albertini, A.M. A novel member of the subtilisin-like protease family from Bacillus subtilis. Microbiology 1999, 145 Pt 11, 3121–3127. [Google Scholar] [CrossRef] [PubMed]
- Leibowitz, M.J.; Wickner, R.B. A chromosomal gene required for killer plasmid expression, mating, and spore maturation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1976, 73, 2061–2065. [Google Scholar] [CrossRef] [PubMed]
- Fuller, R.S.; Sterne, R.E.; Thorner, J. Enzymes required for yeast prohormone processing. Annu. Rev. Physiol. 1988, 50, 345–362. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Nakamura, T.; Ohshima, T.; Tanaka, S.; Matsuo, H. Yeast KEX2 genes encodes an endopeptidase homologous to subtilisin-like serine proteases. Biochem. Biophys. Res. Commun. 1988, 156, 246–254. [Google Scholar] [CrossRef]
- Seidah, N.G.; Chrétien, M. Proprotein and prohormone convertases of the subtilisin family: Recent developments and future perspectives. Trends Endocrinol. Metab. 1992, 3, 133–140. [Google Scholar] [CrossRef]
- Bathurst, I.C.; Brennan, S.O.; Carrell, R.W.; Cousens, L.S.; Brake, A.J.; Barr, P.J. Yeast KEX2 protease has the properties of a human proalbumin converting enzyme. Science 1987, 235, 348–350. [Google Scholar] [CrossRef]
- Thomas, G.; Thorne, B.A.; Thomas, L.; Allen, R.G.; Hruby, D.E.; Fuller, R.; Thorner, J. Yeast KEX2 endopeptidase correctly cleaves a neuroendocrine prohormone in mammalian cells. Science 1988, 241, 226–230. [Google Scholar] [CrossRef]
- Roebroek, A.J.; Schalken, J.A.; Leunissen, J.A.; Onnekink, C.; Bloemers, H.P.; Van de Ven, W.J. Evolutionary conserved close linkage of the c-fes/fps proto-oncogene and genetic sequences encoding a receptor-like protein. EMBO J. 1986, 5, 2197–2202. [Google Scholar] [CrossRef]
- Schalken, J.A.; Roebroek, A.J.; Oomen, P.P.; Wagenaar, S.S.; Debruyne, F.M.; Bloemers, H.P.; Van de Ven, W.J. Fur gene expression as a discriminating marker for small cell and nonsmall cell lung carcinomas. J. Clin. Investig. 1987, 80, 1545–1549. [Google Scholar] [CrossRef]
- Fuller, R.S.; Brake, A.J.; Thorner, J. Intracellular targeting and structural conservation of a prohormone-processing endoprotease. Science 1989, 246, 482–486. [Google Scholar] [CrossRef]
- van den Ouweland, A.M.; van Duijnhoven, H.L.; Keizer, G.D.; Dorssers, L.C.; Van de Ven, W.J. Structural homology between the human fur gene product and the subtilisin-like protease encoded by yeast KEX2. Nucleic Acids Res. 1990, 18, 664. [Google Scholar] [CrossRef] [PubMed]
- Bresnahan, P.A.; Leduc, R.; Thomas, L.; Thorner, J.; Gibson, H.L.; Brake, A.J.; Barr, P.J.; Thomas, G. Human fur gene encodes a yeast KEX2-like endoprotease that cleaves pro-beta-NGF in vivo. J. Cell Biol. 1990, 111, 2851–2859. [Google Scholar] [CrossRef] [PubMed]
- van de Ven, W.J.; Voorberg, J.; Fontijn, R.; Pannekoek, H.; van den Ouweland, A.M.; van Duijnhoven, H.L.; Roebroek, A.J.; Siezen, R.J. Furin is a subtilisin-like proprotein processing enzyme in higher eukaryotes. Mol. Biol. Rep. 1990, 14, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Wise, R.J.; Barr, P.J.; Wong, P.A.; Kiefer, M.C.; Brake, A.J.; Kaufman, R.J. Expression of a human proprotein processing enzyme: Correct cleavage of the von Willebrand factor precursor at a paired basic amino acid site. Proc. Natl. Acad. Sci. USA 1990, 87, 9378–9382. [Google Scholar] [CrossRef]
- Cameron, A.; Appel, J.; Houghten, R.A.; Lindberg, I. Polyarginines are potent furin inhibitors. J. Biol. Chem. 2000, 275, 36741–36749. [Google Scholar] [CrossRef]
- Osman, E.E.A.; Rehemtulla, A.; Neamati, N. Why All the Fury over Furin? J. Med. Chem. 2022, 65, 2747–2784. [Google Scholar] [CrossRef]
- Laprise, M.-H.; Grondin, F.; Cayer, P.; McDonald, P.P.; Dubois, C.M. Furin gene (fur) regulation in differentiating human megakaryoblastic Dami cells: Involvement of the proximal GATA recognition motif in the P1 promoter and impact on the maturation of furin substrates. Blood 2002, 100, 3578–3587. [Google Scholar] [CrossRef]
- Ayoubi, T.A.; Creemers, J.W.; Roebroek, A.J.; Van de Ven, W.J. Expression of the dibasic proprotein processing enzyme furin is directed by multiple promoters. J. Biol. Chem. 1994, 269, 9298–9303. [Google Scholar] [CrossRef]
- Blanchette, F.; Rudd, P.; Grondin, F.; Attisano, L.; Dubois, C.M. Involvement of Smads in TGFbeta1-induced furin (fur) transcription. J. Cell. Physiol. 2001, 188, 264–273. [Google Scholar] [CrossRef]
- McMahon, S.; Grondin, F.; McDonald, P.P.; Richard, D.E.; Dubois, C.M. Hypoxia-enhanced expression of the proprotein convertase furin is mediated by hypoxia-inducible factor-1: Impact on the bioactivation of proproteins. J. Biol. Chem. 2005, 280, 6561–6569. [Google Scholar] [CrossRef]
- Creemers, J.W.; Siezen, R.J.; Roebroek, A.J.; Ayoubi, T.A.; Huylebroeck, D.; de Ven, W.J.V. Modulation of furin-mediated proprotein processing activity by site-directed mutagenesis. J. Biol. Chem. 1993, 268, 21826–21834. [Google Scholar] [CrossRef]
- Henrich, S.; Cameron, A.; Bourenkov, G.P.; Kiefersauer, R.; Huber, R.; Lindberg, I.; Bode, W.; Than, M.E. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat. Struct. Biol. 2003, 10, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, T.A.; Goder, V.; Heinrich, S.U.; Matlack, K.E.S. Membrane-protein integration and the role of the translocation channel. Trends Cell Biol. 2004, 14, 568–575. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 2002, 3, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Henrich, S.; Lindberg, I.; Bode, W.; Than, M.E. Proprotein convertase models based on the crystal structures of furin and kexin: Explanation of their specificity. J. Mol. Biol. 2005, 345, 211–227. [Google Scholar] [CrossRef]
- Dahms, S.O.; Arciniega, M.; Steinmetzer, T.; Huber, R.; Than, M.E. Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism. Proc. Natl. Acad. Sci. USA 2016, 113, 11196–11201. [Google Scholar] [CrossRef]
- Grossman, B.D.; Sanford, J.D.; Zhu, Y.; Zeller, C.B.; Weldon, J.E. Intracellular trafficking of furin enhances cellular intoxication by recombinant immunotoxins based on Pseudomonas exotoxin A. Biol. Open 2025, 14, bio061792. [Google Scholar] [CrossRef]
- Jones, B.G.; Thomas, L.; Molloy, S.S.; Thulin, C.D.; Fry, M.D.; Walsh, K.A.; Thomas, G. Intracellular trafficking of furin is modulated by the phosphorylation state of a casein kinase II site in its cytoplasmic tail. EMBO J. 1995, 14, 5869–5883. [Google Scholar] [CrossRef]
- Leduc, R.; Molloy, S.S.; Thorne, B.A.; Thomas, G. Activation of human furin precursor processing endoprotease occurs by an intramolecular autoproteolytic cleavage. J. Biol. Chem. 1992, 267, 14304–14308. [Google Scholar] [CrossRef]
- Creemers, J.W.M.; Khatib, A.-M. Knock-out mouse models of proprotein convertases: Unique functions or redundancy? Front. Biosci. 2008, 13, 4960–4971. [Google Scholar] [CrossRef]
- Anderson, E.D.; Molloy, S.S.; Jean, F.; Fei, H.; Shimamura, S.; Thomas, G. The Ordered and Compartment-specific Autoproteolytic Removal of the Furin Intramolecular Chaperone Is Required for Enzyme Activation. J. Biol. Chem. 2002, 277, 12879–12890. [Google Scholar] [CrossRef] [PubMed]
- Gawlik, K.; Shiryaev, S.A.; Zhu, W.; Motamedchaboki, K.; Desjardins, R.; Day, R.; Remacle, A.G.; Stec, B.; Strongin, A.Y. Autocatalytic activation of the furin zymogen requires removal of the emerging enzyme’s N-terminus from the active site. PLoS ONE 2009, 4, e5031. [Google Scholar] [CrossRef] [PubMed]
- Hatsuzawa, K.; Nagahama, M.; Takahashi, S.; Takada, K.; Murakami, K.; Nakayama, K. Purification and characterization of furin, a Kex2-like processing endoprotease, produced in Chinese hamster ovary cells. J. Biol. Chem. 1992, 267, 16094–16099. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Crump, C.M.; Thomas, G. Trans-Golgi network sorting. Cell Mol. Life Sci. 2001, 58, 1067–1084. [Google Scholar] [CrossRef]
- Molloy, S.S.; Anderson, E.D.; Jean, F.; Thomas, G. Bi-cycling the furin pathway: From TGN localization to pathogen activation and embryogenesis. Trends Cell Biol. 1999, 9, 28–35. [Google Scholar] [CrossRef]
- Tian, S.; Jianhua, W. Comparative study of the binding pockets of mammalian proprotein convertases and its implications for the design of specific small molecule inhibitors. Int. J. Biol. Sci. 2010, 6, 89–95. [Google Scholar] [CrossRef]
- Brennan, S.O.; Nakayama, K. Furin has the proalbumin substrate specificity and serpin inhibitory properties of an in situ hepatic convertase. FEBS Lett. 1994, 338, 147–151. [Google Scholar] [CrossRef]
- Misumi, Y.; Oda, K.; Fujiwara, T.; Takami, N.; Tashiro, K.; Ikehara, Y. Functional expression of furin demonstrating its intracellular localization and endoprotease activity for processing of proalbumin and complement pro-C3. J. Biol. Chem. 1991, 266, 16954–16959. [Google Scholar] [CrossRef]
- Wallin, R.; Stanton, C.; Ross, R.P. Intracellular proteolytic processing of the two-chain vitamin K-dependent coagulation factor X. Thromb. Res. 1994, 73, 395–403. [Google Scholar] [CrossRef]
- Wasley, L.C.; Rehemtulla, A.; Bristol, J.A.; Kaufman, R.J. PACE/furin can process the vitamin K-dependent pro-factor IX precursor within the secretory pathway. J. Biol. Chem. 1993, 268, 8458–8465. [Google Scholar] [CrossRef]
- Schlokat, U.; Preininger, A.; Himmelspach, M.; Mohr, G.; Fischer, B.; Dorner, F. High Yield Expression of Recombinant Plasma Factors: Use of Recombinant Endoprotease Derivatives In Vivo and In Vitro. In New Developments and New Applications in Animal Cell Technology: Proceedings of the 15th ESACT Meeting; Merten, O.-W., Perrin, P., Griffiths, B., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2002; pp. 69–76. [Google Scholar]
- Bendetowicz, A.V.; Morris, J.A.; Wise, R.J.; Gilbert, G.E.; Kaufman, R.J. Binding of factor VIII to von willebrand factor is enabled by cleavage of the von Willebrand factor propeptide and enhanced by formation of disulfide-linked multimers. Blood 1998, 92, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Drews, R.; Paleyanda, R.K.; Lee, T.K.; Chang, R.R.; Rehemtulla, A.; Kaufman, R.J.; Drohan, W.N.; Luboń, H. Proteolytic maturation of protein C upon engineering the mouse mammary gland to express furin. Proc. Natl. Acad. Sci. USA 1995, 92, 10462–10466. [Google Scholar] [CrossRef] [PubMed]
- Hendy, G.N.; Bennett, H.P.; Gibbs, B.F.; Lazure, C.; Day, R.; Seidah, N.G. Proparathyroid hormone is preferentially cleaved to parathyroid hormone by the prohormone convertase furin. A mass spectrometric study. J. Biol. Chem. 1995, 270, 9517–9525. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Goltzman, D.; Rabbani, S.A. Processing of pro-PTHRP by the prohormone convertase, furin: Effect on biological activity. Am. J. Physiol.-Endocrinol. Metab. 1995, 268, E832–E838. [Google Scholar] [CrossRef]
- Dey, A.; Norrbom, C.; Zhu, X.; Stein, J.; Zhang, C.; Ueda, K.; Steiner, D.F. Furin and prohormone convertase 1/3 are major convertases in the processing of mouse pro-growth hormone-releasing hormone. Endocrinology 2004, 145, 1961–1971. [Google Scholar] [CrossRef]
- Seidah, N.G.; Benjannet, S.; Pareek, S.; Chrétien, M.; Murphy, R.A. Cellular processing of the neurotrophin precursors of NT3 and BDNF by the mammalian proprotein convertases. FEBS Lett. 1996, 379, 247–250. [Google Scholar] [CrossRef]
- Denault, J.B.; Claing, A.; D’Orléans-Juste, P.; Sawamura, T.; Kido, T.; Masaki, T.; Leduc, R. Processing of proendothelin-1 by human furin convertase. FEBS Lett. 1995, 362, 276–280. [Google Scholar] [CrossRef]
- Nachtigal, M.W.; Ingraham, H.A. Bioactivation of Müllerian inhibiting substance during gonadal development by a kex2/subtilisin-like endoprotease. Proc. Natl. Acad. Sci. USA 1996, 93, 7711–7716. [Google Scholar] [CrossRef]
- Duguay, S.J.; Lai-Zhang, J.; Steiner, D.F. Mutational analysis of the insulin-like growth factor I prohormone processing site. J. Biol. Chem. 1995, 270, 17566–17574. [Google Scholar] [CrossRef]
- Dubois, C.M.; Laprise, M.H.; Blanchette, F.; Gentry, L.E.; Leduc, R. Processing of transforming growth factor beta 1 precursor by human furin convertase. J. Biol. Chem. 1995, 270, 10618–10624. [Google Scholar] [CrossRef]
- Siegfried, G.; Basak, A.; Cromlish, J.A.; Benjannet, S.; Marcinkiewicz, J.; Chrétien, M.; Seidah, N.G.; Khatib, A.-M. The secretory proprotein convertases furin, PC5, and PC7 activate VEGF-C to induce tumorigenesis. J. Clin. Investig. 2003, 111, 1723–1732. [Google Scholar] [CrossRef] [PubMed]
- McColl, B.K.; Paavonen, K.; Karnezis, T.; Harris, N.C.; Davydova, N.; Rothacker, J.; Nice, E.C.; Harder, K.W.; Roufail, S.; Hibbs, M.L.; et al. Proprotein convertases promote processing of VEGF-D, a critical step for binding the angiogenic receptor VEGFR-2. FASEB J. 2007, 21, 1088–1098. [Google Scholar] [CrossRef] [PubMed]
- Bravo, D.A.; Gleason, J.B.; Sanchez, R.I.; Roth, R.A.; Fuller, R.S. Accurate and efficient cleavage of the human insulin proreceptor by the human proprotein-processing protease furin. Characterization and kinetic parameters using the purified, secreted soluble protease expressed by a recombinant baculovirus. J. Biol. Chem. 1994, 269, 25830–25837. [Google Scholar] [CrossRef] [PubMed]
- Komada, M.; Hatsuzawa, K.; Shibamoto, S.; Ito, F.; Nakayama, K.; Kitamura, N. Proteolytic processing of the hepatocyte growth factor/scatter factor receptor by furin. FEBS Lett. 1993, 328, 25–29. [Google Scholar] [CrossRef]
- Willnow, T.E.; Moehring, J.M.; Inocencio, N.M.; Moehring, T.J.; Herz, J. The low-density-lipoprotein receptor-related protein (LRP) is processed by furin in vivo and in vitro. Biochem. J. 1996, 313 Pt 1, 71–76. [Google Scholar] [CrossRef]
- Lehmann, M.; Rigot, V.; Seidah, N.G.; Marvaldi, J.; Lissitzky, J.C. Lack of integrin alpha-chain endoproteolytic cleavage in furin-deficient human colon adenocarcinoma cells LoVo. Biochem. J. 1996, 317, 803–809. [Google Scholar] [CrossRef]
- Logeat, F.; Bessia, C.; Brou, C.; LeBail, O.; Jarriault, S.; Seidah, N.G.; Israël, A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 1998, 95, 8108–8112. [Google Scholar] [CrossRef]
- Sato, H.; Kinoshita, T.; Takino, T.; Nakayama, K.; Seiki, M. Activation of a recombinant membrane type 1-matrix metalloproteinase (MT1-MMP) by furin and its interaction with tissue inhibitor of metalloproteinases (TIMP)-2. FEBS Lett. 1996, 393, 101–104. [Google Scholar] [CrossRef]
- Murphy, G.; Nagase, H. Progress in matrix metalloproteinase research. Mol. Asp. Med. 2008, 29, 290–308. [Google Scholar] [CrossRef]
- Kang, T.; Zhao, Y.-G.; Pei, D.; Sucic, J.F.; Sang, Q.-X.A. Intracellular activation of human adamalysin 19/disintegrin and metalloproteinase 19 by furin occurs via one of the two consecutive recognition sites. J. Biol. Chem. 2002, 277, 25583–25591. [Google Scholar] [CrossRef]
- Kuno, K.; Terashima, Y.; Matsushima, K. ADAMTS-1 is an active metalloproteinase associated with the extracellular matrix. J. Biol. Chem. 1999, 274, 18821–18826. [Google Scholar] [CrossRef]
- Leighton, M.; Kadler, K.E. Paired basic/Furin-like proprotein convertase cleavage of Pro-BMP-1 in the trans-Golgi network. J. Biol. Chem. 2003, 278, 18478–18484. [Google Scholar] [CrossRef]
- Banyard, J.; Bao, L.; Zetter, B.R. Type XXIII collagen, a new transmembrane collagen identified in metastatic tumor cells. J. Biol. Chem. 2003, 278, 20989–20994. [Google Scholar] [CrossRef] [PubMed]
- Raghunath, M.; Putnam, E.A.; Ritty, T.; Hamstra, D.; Park, E.S.; Tschödrich-Rotter, M.; Peters, R.; Rehemtulla, A.; Milewicz, D.M. Carboxy-terminal conversion of profibrillin to fibrillin at a basic site by PACE/furin-like activity required for incorporation in the matrix. J. Cell Sci. 1999, 112 Pt 7, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, X.; Bai, X.; Yao, S.; Chang, Y.-Z.; Gao, G. The emerging role of furin in neurodegenerative and neuropsychiatric diseases. Transl. Neurodegener. 2022, 11, 39. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Luo, W.; Juhong, Z.; Yang, J.; Wang, H.; Zhou, L.; Chang, J. Associations between genetic variations in the FURIN gene and hypertension. BMC Med. Genet. 2010, 11, 124. [Google Scholar] [CrossRef]
- Jaaks, P.; Bernasconi, M. The proprotein convertase furin in tumour progression. Int. J. Cancer 2017, 141, 654–663. [Google Scholar] [CrossRef]
- Tsuneoka, M.; Nakayama, K.; Hatsuzawa, K.; Komada, M.; Kitamura, N.; Mekada, E. Evidence for involvement of furin in cleavage and activation of diphtheria toxin. J. Biol. Chem. 1993, 268, 26461–26465. [Google Scholar] [CrossRef]
- Klimpel, K.R.; Molloy, S.S.; Thomas, G.; Leppla, S.H. Anthrax toxin protective antigen is activated by a cell surface protease with the sequence specificity and catalytic properties of furin. Proc. Natl. Acad. Sci. USA 1992, 89, 10277–10281. [Google Scholar] [CrossRef]
- Inocencio, N.M.; Moehring, J.M.; Moehring, T.J. Furin activates Pseudomonas exotoxin A by specific cleavage in vivo and in vitro. J. Biol. Chem. 1994, 269, 31831–31835. [Google Scholar] [CrossRef]
- Garred, O.; van Deurs, B.; Sandvig, K. Furin-induced cleavage and activation of Shiga toxin. J. Biol. Chem. 1995, 270, 10817–10821. [Google Scholar] [CrossRef] [PubMed]
- Braun, E.; Sauter, D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunol. 2019, 8, e1073. [Google Scholar] [CrossRef] [PubMed]
- Bernard, H.-U.; Burk, R.D.; Chen, Z.; van Doorslaer, K.; zur Hausen, H.; de Villiers, E.-M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 2010, 401, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Mishra, V.; Sharma, N.; Khurana, N. Human Papillomavirus and Its Nature of Infection: An Overview. Asian J. Pharm. Clin. Res. 2018, 11, 12–16. [Google Scholar] [CrossRef]
- Cruz-Gregorio, A.; Aranda-Rivera, A.K. Human Papilloma Virus-Infected Cells. In Virus Infected Cells; Vijayakrishnan, S., Jiu, Y., Harris, J.R., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 213–226. [Google Scholar]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015, 25 (Suppl. S1), 2–23. [Google Scholar] [CrossRef]
- Day, P.M.; Schiller, J.T. The role of furin in papillomavirus infection. Future Microbiol. 2009, 4, 1255–1262. [Google Scholar] [CrossRef]
- Bronnimann, M.P.; Calton, C.M.; Chiquette, S.F.; Li, S.; Lu, M.; Chapman, J.A.; Bratton, K.N.; Schlegel, A.M.; Campos, S.K. Furin Cleavage of L2 during Papillomavirus Infection: Minimal Dependence on Cyclophilins. J. Virol. 2016, 90, 6224–6234. [Google Scholar] [CrossRef]
- Schiller, J.T.; Day, P.M.; Kines, R.C. Current understanding of the mechanism of HPV infection. Gynecol. Oncol. 2010, 118, S12–S17. [Google Scholar] [CrossRef]
- Cerqueira, C.; Samperio Ventayol, P.; Vogeley, C.; Schelhaas, M. Kallikrein-8 Proteolytically Processes Human Papillomaviruses in the Extracellular Space To Facilitate Entry into Host Cells. J. Virol. 2015, 89, 7038–7052. [Google Scholar] [CrossRef]
- Izaguirre, G.; Phan, L.M.U.; Asif, S.; Alam, S.; Meyers, C.; Rong, L. Diversity in Proprotein Convertase Reactivity among Human Papillomavirus Types. Viruses 2024, 16, 39. [Google Scholar] [CrossRef]
- Böttcher-Friebertshäuser, E.; Garten, W.; Matrosovich, M.; Klenk, H.D. The Hemagglutinin: A Determinant of Pathogenicity. In Influenza Pathogenesis and Control—Volume I; Compans, R.W., Oldstone, M.B.A., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 3–34. [Google Scholar]
- de Bruin, A.C.M.; Funk, M.; Spronken, M.I.; Gultyaev, A.P.; Fouchier, R.A.M.; Richard, M. Hemagglutinin Subtype Specificity and Mechanisms of Highly Pathogenic Avian Influenza Virus Genesis. Viruses 2022, 14, 1566. [Google Scholar] [CrossRef] [PubMed]
- Kido, H.; Okumura, Y.; Takahashi, E.; Pan, H.-Y.; Wang, S.; Yao, D.; Yao, M.; Chida, J.; Yano, M. Role of host cellular proteases in the pathogenesis of influenza and influenza-induced multiple organ failure. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2012, 1824, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lee, K.H.; Steinhauer, D.A.; Stevens, D.J.; Skehel, J.J.; Wiley, D.C. Structure of the Hemagglutinin Precursor Cleavage Site, a Determinant of Influenza Pathogenicity and the Origin of the Labile Conformation. Cell 1998, 95, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Franҫa, M.S.; Brown, J.D. Influenza Pathobiology and Pathogenesis in Avian Species. In Influenza Pathogenesis and Control—Volume I; Compans, R.W., Oldstone, M.B.A., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 221–242. [Google Scholar]
- Zambon, M.C. The pathogenesis of influenza in humans. Rev. Med. Virol. 2001, 11, 227–241. [Google Scholar] [CrossRef]
- Zhirnov, O.; Klenk, H.-D. Human influenza A viruses are proteolytically activated and do not induce apoptosis in CACO-2 cells. Virology 2003, 313, 198–212. [Google Scholar] [CrossRef]
- Tse, L.V.; Hamilton, A.M.; Friling, T.; Whittaker, G.R. A Novel Activation Mechanism of Avian Influenza Virus H9N2 by Furin. J. Virol. 2014, 88, 1673–1683. [Google Scholar] [CrossRef]
- Carneiro, V.C.; Pereira, J.G.; de Paula, V.S. Family Herpesviridae and neuroinfections: Current status and research in progress. Mem. Inst. Oswaldo Cruz 2022, 117, e220200. [Google Scholar] [CrossRef]
- Grünewald, K.; Desai, P.; Winkler, D.C.; Heymann, J.B.; Belnap, D.M.; Baumeister, W.; Steven, A.C. Three-Dimensional Structure of Herpes Simplex Virus from Cryo-Electron Tomography. Science 2003, 302, 1396–1398. [Google Scholar] [CrossRef]
- Roizmann, B.; Desrosiers, R.C.; Fleckenstein, B.; Lopez, C.; Minson, A.C.; Studdert, M.J. The family Herpesviridae: An update. Arch. Virol. 1992, 123, 425–449. [Google Scholar] [CrossRef]
- Gatherer, D.; Depledge, D.P.; Hartley, C.A.; Szpara, M.L.; Vaz, P.K.; Benkő, M.; Brandt, C.R.; Bryant, N.A.; Dastjerdi, A.; Doszpoly, A.; et al. ICTV Virus Taxonomy Profile: Herpesviridae 2021. J. Gen. Virol. 2021, 102, 001673. [Google Scholar] [CrossRef]
- Keil, G.M.; Klopfleisch, C.; Giesow, K.; Veits, J. Protein display by bovine herpesvirus type 1 glycoprotein B. Vet. Microbiol. 2010, 143, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Glauser, D.L.; Milho, R.; Frederico, B.; May, J.S.; Kratz, A.-S.; Gillet, L.; Stevenson, P.G. Glycoprotein B Cleavage Is Important for Murid Herpesvirus 4 To Infect Myeloid Cells. J. Virol. 2013, 87, 10828–10842. [Google Scholar] [CrossRef] [PubMed]
- Izaguirre, G. The Proteolytic Regulation of Virus Cell Entry by Furin and Other Proprotein Convertases. Viruses 2019, 11, 837. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, K. Proteolytic cleavage of glycoprotein B is dispensable for in vitro replication, but required for syncytium formation of pseudorabies virus. J. Gen. Virol. 2007, 88, 1859–1865. [Google Scholar] [CrossRef]
- Sorem, J.; Longnecker, R. Cleavage of Epstein–Barr virus glycoprotein B is required for full function in cell–cell fusion with both epithelial and B cells. J. Gen. Virol. 2009, 90, 591–595. [Google Scholar] [CrossRef]
- Backovic, M.; Leser, G.P.; Lamb, R.A.; Longnecker, R.; Jardetzky, T.S. Characterization of EBV gB indicates properties of both class I and class II viral fusion proteins. Virology 2007, 368, 102–113. [Google Scholar] [CrossRef]
- Oliver, S.L.; Sommer, M.; Zerboni, L.; Rajamani, J.; Grose, C.; Arvin, A.M. Mutagenesis of Varicella-Zoster Virus Glycoprotein B: Putative Fusion Loop Residues Are Essential for Viral Replication, and the Furin Cleavage Motif Contributes to Pathogenesis in Skin Tissue In Vivo. J. Virol. 2009, 83, 7495–7506. [Google Scholar] [CrossRef]
- Vey, M.; Schäfer, W.; Reis, B.; Ohuchi, R.; Britt, W.; Garten, W.; Klenk, H.-D.; Radsak, K. Proteolytic processing of human cytomegalovirus glycoprotein B (gpUL55) is mediatedby the human endoprotease furin. Virology 1995, 206, 746–749. [Google Scholar] [CrossRef]
- Griffiths, P.; Baraniak, I.; Reeves, M. The pathogenesis of human cytomegalovirus. J. Pathol. 2015, 235, 288–297. [Google Scholar] [CrossRef]
- Kropp, K.A.; Srivaratharajan, S.; Ritter, B.; Yu, P.; Krooss, S.; Polten, F.; Pich, A.; Alcami, A.; Viejo-Borbolla, A. Identification of the Cleavage Domain within Glycoprotein G of Herpes Simplex Virus Type 2. Viruses 2020, 12, 1428. [Google Scholar] [CrossRef]
- Connolly, S.A.; Jardetzky, T.S.; Longnecker, R. The structural basis of herpesvirus entry. Nat. Rev. Microbiol. 2021, 19, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Izaguirre, G.; Arciniega, M.; Quezada, A.G. Specific and Selective Inhibitors of Proprotein Convertases Engineered by Transferring Serpin B8 Reactive-Site and Exosite Determinants of Reactivity to the Serpin α1PDX. Biochemistry 2019, 58, 1679–1688. [Google Scholar] [CrossRef] [PubMed]
- Jean, F.; Thomas, L.; Molloy, S.S.; Liu, G.; Jarvis, M.A.; Nelson, J.A.; Thomas, G. A protein-based therapeutic for human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 2000, 97, 2864–2869. [Google Scholar] [CrossRef] [PubMed]
- Izaguirre, G.; Qi, L.; Lima, M.; Olson, S.T. Identification of serpin determinants of specificity and selectivity for furin inhibition through studies of α1PDX (α1-protease inhibitor Portland)-serpin B8 and furin active-site loop chimeras. J. Biol. Chem. 2013, 288, 21802–21814. [Google Scholar] [CrossRef]
- Kim, A.S.; Diamond, M.S. A molecular understanding of alphavirus entry and antibody protection. Nat. Rev. Microbiol. 2023, 21, 396–407. [Google Scholar] [CrossRef]
- Metz, S.W.; Geertsema, C.; Martina, B.E.; Andrade, P.; Heldens, J.G.; van Oers, M.M.; Goldbach, R.W.; Vlak, J.M.; Pijlman, G.P. Functional processing and secretion of Chikungunya virus E1 and E2 glycoproteins in insect cells. Virol. J. 2011, 8, 353. [Google Scholar] [CrossRef]
- Yap, M.L.; Klose, T.; Urakami, A.; Hasan, S.S.; Akahata, W.; Rossmann, M.G. Structural studies of Chikungunya virus maturation. Proc. Natl. Acad. Sci. USA 2017, 114, 13703–13707. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Zhang, W.; Gabler, S.; Chipman, P.R.; Strauss, E.G.; Strauss, J.H.; Baker, T.S.; Kuhn, R.J.; Rossmann, M.G. Mapping the Structure and Function of the E1 and E2 Glycoproteins in Alphaviruses. Structure 2006, 14, 63–73. [Google Scholar] [CrossRef]
- Gibbons, D.L.; Vaney, M.-C.; Roussel, A.; Vigouroux, A.; Reilly, B.; Lepault, J.; Kielian, M.; Rey, F.A. Conformational change and protein–protein interactions of the fusion protein of Semliki Forest virus. Nature 2004, 427, 320–325. [Google Scholar] [CrossRef]
- Uchime, O.; Fields, W.; Kielian, M. The Role of E3 in pH Protection during Alphavirus Assembly and Exit. J. Virol. 2013, 87, 10255–10262. [Google Scholar] [CrossRef]
- Zhang, X.; Fugère, M.; Day, R.; Kielian, M. Furin Processing and Proteolytic Activation of Semliki Forest Virus. J. Virol. 2003, 77, 2981–2989. [Google Scholar] [CrossRef]
- Ozden, S.; Lucas-Hourani, M.; Ceccaldi, P.-E.; Basak, A.; Valentine, M.; Benjannet, S.; Hamelin, J.; Jacob, Y.; Mamchaoui, K.; Mouly, V.; et al. Inhibition of Chikungunya Virus Infection in Cultured Human Muscle Cells by Furin Inhibitors: Impairment of the Maturation of the E2 Surface Glycoprotein *. J. Biol. Chem. 2008, 283, 21899–21908. [Google Scholar] [CrossRef]
- Chen, M.T.; Billaud, J.-N.; Sällberg, M.; Guidotti, L.G.; Chisari, F.V.; Jones, J.; Hughes, J.; Milich, D.R. A function of the hepatitis B virus precore protein is to regulate the immune response to the core antigen. Proc. Natl. Acad. Sci. USA 2004, 101, 14913–14918. [Google Scholar] [CrossRef] [PubMed]
- Liaw, Y.-F.; Lau, G.K.K.; Kao, J.-H.; Gane, E. Hepatitis B e Antigen Seroconversion: A Critical Event in Chronic Hepatitis B Virus Infection. Dig. Dis. Sci. 2010, 55, 2727–2734. [Google Scholar] [CrossRef] [PubMed]
- Messageot, F.; Salhi, S.; Eon, P.; Rossignol, J.-M. Proteolytic Processing of the Hepatitis B Virus e Antigen Precursor: Cleavage at Two Furin Consensus Sequences *. J. Biol. Chem. 2003, 278, 891–895. [Google Scholar] [CrossRef] [PubMed]
- Deroubaix, A.; Kramvis, A. In vitro expression of precore proteins of hepatitis B virus subgenotype A1 is affected by HBcAg, and can affect HBsAg secretion. Sci. Rep. 2021, 11, 8167. [Google Scholar] [CrossRef]
- Ito, K.; Kim, K.-H.; Lok, A.S.-F.; Tong, S. Characterization of Genotype-Specific Carboxyl-Terminal Cleavage Sites of Hepatitis B Virus e Antigen Precursor and Identification of Furin as the Candidate Enzyme. J. Virol. 2009, 83, 3507–3517. [Google Scholar] [CrossRef]
- Wu, J.-F.; Hsu, H.-Y.; Ni, Y.-H.; Chen, H.-L.; Wu, T.-C.; Chang, M.-H. Suppression of Furin by Interferon-γ and the Impact on Hepatitis B Virus Antigen Biosynthesis in Human Hepatocytes. Am. J. Pathol. 2012, 181, 19–25. [Google Scholar] [CrossRef]
- Yang, H.Y.; Zheng, N.Q.; Li, D.M.; Gu, L.; Peng, X.M. Entecavir combined with furin inhibitor simultaneously reduces hepatitis B virus replication and e antigen secretion. Virol. J. 2014, 11, 165. [Google Scholar] [CrossRef]
- Biedenkopf, N.; Bukreyev, A.; Chandran, K.; Di Paola, N.; Formenty, P.B.H.; Griffiths, A.; Hume, A.J.; Mühlberger, E.; Netesov, S.V.; Palacios, G.; et al. ICTV Virus Taxonomy Profile: Filoviridae 2024. J. Gen. Virol. 2024, 105, 001955. [Google Scholar] [CrossRef]
- Burgueño-Sosa, E.E.; Esquivel-Gómez, L.R.; Rivadeneyra-Gutiérrez, E.; León-López, A.A. Generalidades de la familia Filoviridae y el virus del Ébola: Una actualización de sus implicaciones en la población humana. Rev. Biomédica 2020, 31, 58–68. [Google Scholar] [CrossRef]
- Volchkov, V.E.; Feldmann, H.; Volchkova, V.A.; Klenk, H.-D. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. USA 1998, 95, 5762–5767. [Google Scholar] [CrossRef] [PubMed]
- Volchkov, V.E.; Volchkova, V.A.; Ströher, U.; Becker, S.; Dolnik, O.; Cieplik, M.; Garten, W.; Klenk, H.-D.; Feldmann, H. Proteolytic Processing of Marburg Virus Glycoprotein. Virology 2000, 268, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Feldmann, H.; Watanabe, S.; Lukashevich, I.; Kawaoka, Y. Reverse Genetics Demonstrates that Proteolytic Processing of the Ebola Virus Glycoprotein Is Not Essential for Replication in Cell Culture. J. Virol. 2002, 76, 406–410. [Google Scholar] [CrossRef]
- Neumann, G.; Geisbert, T.W.; Ebihara, H.; Geisbert, J.B.; Daddario-DiCaprio, K.M.; Feldmann, H.; Kawaoka, Y. Proteolytic Processing of the Ebola Virus Glycoprotein Is Not Critical for Ebola Virus Replication in Nonhuman Primates. J. Virol. 2007, 81, 2995–2998. [Google Scholar] [CrossRef]
- Zhu, W.; Banadyga, L.; Emeterio, K.; Wong, G.; Qiu, X. The Roles of Ebola Virus Soluble Glycoprotein in Replication, Pathogenesis, and Countermeasure Development. Viruses 2019, 11, 999. [Google Scholar] [CrossRef]
- Duprex, W.P.; Dutch, R.E. Paramyxoviruses: Pathogenesis, Vaccines, Antivirals, and Prototypes for Pandemic Preparedness. J. Infect. Dis. 2023, 228, S390–S397. [Google Scholar] [CrossRef]
- González-Reyes, L.; Ruiz-Argüello, M.B.; García-Barreno, B.; Calder, L.; López, J.A.; Albar, J.P.; Skehel, J.J.; Wiley, D.C.; Melero, J.A. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. USA 2001, 98, 9859–9864. [Google Scholar] [CrossRef]
- Plattet, P.; Alves, L.; Herren, M.; Aguilar, H.C. Measles Virus Fusion Protein: Structure, Function and Inhibition. Viruses 2016, 8, 112. [Google Scholar] [CrossRef]
- Panda, A.; Huang, Z.; Elankumaran, S.; Rockemann, D.D.; Samal, S.K. Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microb. Pathog. 2004, 36, 1–10. [Google Scholar] [CrossRef]
- Chang, A.; Dutch, R.E. Paramyxovirus Fusion and Entry: Multiple Paths to a Common End. Viruses 2012, 4, 613–636. [Google Scholar] [CrossRef] [PubMed]
- Ueo, A.; Kubota, M.; Shirogane, Y.; Ohno, S.; Hashiguchi, T.; Yanagi, Y. Lysosome-Associated Membrane Proteins Support the Furin-Mediated Processing of the Mumps Virus Fusion Protein. J. Virol. 2020, 94, e00050-20. [Google Scholar] [CrossRef] [PubMed]
- Hüttl, S.; Hoffmann, M.; Steinmetzer, T.; Sauder, C.; Krüger, N. The Amino Acid at Position 8 of the Proteolytic Cleavage Site of the Mumps Virus Fusion Protein Affects Viral Proteolysis and Fusogenicity. J. Virol. 2020, 94, e01732-20. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, T.; Fukuda, Y.; Matsuoka, R.; Kuroda, D.; Kubota, M.; Shirogane, Y.; Watanabe, S.; Tsumoto, K.; Kohda, D.; Plemper, R.K.; et al. Structures of the prefusion form of measles virus fusion protein in complex with inhibitors. Proc. Natl. Acad. Sci. USA 2018, 115, 2496–2501. [Google Scholar] [CrossRef]
- Diederich, S.; Dietzel, E.; Maisner, A. Nipah virus fusion protein: Influence of cleavage site mutations on the cleavability by cathepsin L, trypsin and furin. Virus Res. 2009, 145, 300–306. [Google Scholar] [CrossRef]
- Moll, M.; Diederich, S.; Klenk, H.-D.; Czub, M.; Maisner, A. Ubiquitous Activation of the Nipah Virus Fusion Protein Does Not Require a Basic Amino Acid at the Cleavage Site. J. Virol. 2004, 78, 9705–9712. [Google Scholar] [CrossRef]
- Weis, M.; Maisner, A. Nipah virus fusion protein: Importance of the cytoplasmic tail for endosomal trafficking and bioactivity. Eur. J. Cell Biol. 2015, 94, 316–322. [Google Scholar] [CrossRef]
- Pager, C.T.; Dutch, R.E. Cathepsin L Is Involved in Proteolytic Processing of the Hendra Virus Fusion Protein. J. Virol. 2005, 79, 12714–12720. [Google Scholar] [CrossRef]
- Liljeroos, L.; Krzyzaniak, M.A.; Helenius, A.; Butcher, S.J. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc. Natl. Acad. Sci. USA 2013, 110, 11133–11138. [Google Scholar] [CrossRef]
- Krzyzaniak, M.A.; Zumstein, M.T.; Gerez, J.A.; Picotti, P.; Helenius, A. Host Cell Entry of Respiratory Syncytial Virus Involves Macropinocytosis Followed by Proteolytic Activation of the F Protein. PLOS Pathog. 2013, 9, e1003309. [Google Scholar] [CrossRef]
- Bekker, L.-G.; Beyrer, C.; Mgodi, N.; Lewin, S.R.; Delany-Moretlwe, S.; Taiwo, B.; Masters, M.C.; Lazarus, J.V. HIV infection. Nat. Rev. Dis. Primers 2023, 9, 1–21. [Google Scholar] [CrossRef]
- Schneck, N.A.; Ivleva, V.B.; Cai, C.X.; Cooper, J.W.; Lei, Q.P. Characterization of the furin cleavage motif for HIV-1 trimeric envelope glycoprotein by intact LC-MS analysis. Analyst 2020, 145, 1636–1640. [Google Scholar] [CrossRef]
- Hallenberger, S.; Moulard, M.; Sordel, M.; Klenk, H.D.; Garten, W. The role of eukaryotic subtilisin-like endoproteases for the activation of human immunodeficiency virus glycoproteins in natural host cells. J. Virol. 1997, 71, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Decroly, E.; Benjannet, S.; Savaria, D.; Seidah, N.G. Comparative functional role of PC7 and furin in the processing of the HIV envelope glycoprotein gp160. FEBS Lett. 1997, 405, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Prévost, J.; Medjahed, H.; Vézina, D.; Chen, H.-C.; Hahn, B.H.; Smith, A.B.; Finzi, A. HIV-1 Envelope Glycoproteins Proteolytic Cleavage Protects Infected Cells from ADCC Mediated by Plasma from Infected Individuals. Viruses 2021, 13, 2236. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.; Xu, K.; Zhou, T.; Acharya, P.; Lemmin, T.; Liu, K.; Ozorowski, G.; Soto, C.; Taft, J.D.; Bailer, R.T.; et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 2016, 352, 828–833. [Google Scholar] [CrossRef]
- Caillat, C.; Guilligay, D.; Torralba, J.; Friedrich, N.; Nieva, J.L.; Trkola, A.; Chipot, C.J.; Dehez, F.L.; Weissenhorn, W. Structure of HIV-1 gp41 with its membrane anchors targeted by neutralizing antibodies. eLife 2021, 10, e65005. [Google Scholar] [CrossRef]
- Sarkar, A.; Bale, S.; Behrens, A.-J.; Kumar, S.; Sharma, S.K.; de Val, N.; Pallesen, J.; Irimia, A.; Diwanji, D.C.; Stanfield, R.L.; et al. Structure of a cleavage-independent HIV Env recapitulates the glycoprotein architecture of the native cleaved trimer. Nat. Commun. 2018, 9, 1956. [Google Scholar] [CrossRef]
- Sachan, V.; Lodge, R.; Mihara, K.; Hamelin, J.; Power, C.; Gelman, B.B.; Hollenberg, M.D.; Cohen, É.A.; Seidah, N.G. HIV-induced neuroinflammation: Impact of PAR1 and PAR2 processing by Furin. Cell Death Differ. 2019, 26, 1942–1954. [Google Scholar] [CrossRef]
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV Virus Taxonomy Profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar] [CrossRef]
- Zhao, R.; Wang, M.; Cao, J.; Shen, J.; Zhou, X.; Wang, D.; Cao, J. Flavivirus: From Structure to Therapeutics Development. Life 2021, 11, 615. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jia, R.; Shen, H.; Wang, M.; Yin, Z.; Cheng, A. Structures and Functions of the Envelope Glycoprotein in Flavivirus Infections. Viruses 2017, 9, 338. [Google Scholar] [CrossRef] [PubMed]
- Stadler, K.; Allison, S.L.; Schalich, J.; Heinz, F.X. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 1997, 71, 8475–8481. [Google Scholar] [CrossRef] [PubMed]
- Elshuber, S.; Allison, S.L.; Heinz, F.X.; Mandl, C.W. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virusFN1. J. Gen. Virol. 2003, 84, 183–191. [Google Scholar] [CrossRef]
- Stiasny, K.; Allison, S.L.; Mandl, C.W.; Heinz, F.X. Role of Metastability and Acidic pH in Membrane Fusion by Tick-Borne Encephalitis Virus. J. Virol. 2001, 75, 7392–7398. [Google Scholar] [CrossRef]
- Yu, I.-M.; Zhang, W.; Holdaway, H.A.; Li, L.; Kostyuchenko, V.A.; Chipman, P.R.; Kuhn, R.J.; Rossmann, M.G.; Chen, J. Structure of the Immature Dengue Virus at Low pH Primes Proteolytic Maturation. Science 2008, 319, 1834–1837. [Google Scholar] [CrossRef]
- Owczarek, K.; Chykunova, Y.; Jassoy, C.; Maksym, B.; Rajfur, Z.; Pyrc, K. Zika virus: Mapping and reprogramming the entry. Cell Commun. Signal. 2019, 17, 41. [Google Scholar] [CrossRef]
- Junjhon, J.; Edwards, T.J.; Utaipat, U.; Bowman, V.D.; Holdaway, H.A.; Zhang, W.; Keelapang, P.; Puttikhunt, C.; Perera, R.; Chipman, P.R.; et al. Influence of pr-M Cleavage on the Heterogeneity of Extracellular Dengue Virus Particles. J. Virol. 2010, 84, 8353–8358. [Google Scholar] [CrossRef]
- Keelapang, P.; Sriburi, R.; Supasa, S.; Panyadee, N.; Songjaeng, A.; Jairungsri, A.; Puttikhunt, C.; Kasinrerk, W.; Malasit, P.; Sittisombut, N. Alterations of pr-M Cleavage and Virus Export in pr-M Junction Chimeric Dengue Viruses. J. Virol. 2004, 78, 2367–2381. [Google Scholar] [CrossRef]
- Mukherjee, S.; Sirohi, D.; Dowd, K.A.; Chen, Z.; Diamond, M.S.; Kuhn, R.J.; Pierson, T.C. Enhancing dengue virus maturation using a stable furin over-expressing cell line. Virology 2016, 497, 33–40. [Google Scholar] [CrossRef]
- Kouretova, J.; Hammamy, M.Z.; Epp, A.; Hardes, K.; Kallis, S.; Zhang, L.; Hilgenfeld, R.; Bartenschlager, R.; Steinmetzer, T. Effects of NS2B-NS3 protease and furin inhibition on West Nile and Dengue virus replication. J. Enzym. Inhib. Med. Chem. 2017, 32, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Cao, S.; Wang, X.; Zhou, Y.; Yan, W.; Gu, X.; Wu, T.-C.; Pang, X. Generation and preliminary characterization of vertebrate-specific replication-defective Zika virus. Virology 2021, 552, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Zybert, I.A.; van der Ende-Metselaar, H.; Wilschut, J.; Smit, J.M. Functional importance of dengue virus maturation: Infectious properties of immature virions. J. Gen. Virol. 2008, 89, 3047–3051. [Google Scholar] [CrossRef] [PubMed]
- Moesker, B.; Rodenhuis-Zybert, I.A.; Meijerhof, T.; Wilschut, J.; Smit, J.M. Characterization of the functional requirements of West Nile virus membrane fusion. J. Gen. Virol. 2010, 91, 389–393. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; de Groot, R.J.; Haagmans, B.; Lau, S.K.P.; Neuman, B.W.; Perlman, S.; Sola, I.; van der Hoek, L.; Wong, A.C.P.; Yeh, S.-H. ICTV Virus Taxonomy Profile: Coronaviridae 2023. J. Gen. Virol. 2023, 104, 001843. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Al-Ahmed, S.H.; Haque, S.; Sah, R.; Tiwari, R.; Malik, Y.S.; Dhama, K.; Yatoo, M.I.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Infez. Med. 2020, 28, 174–184. [Google Scholar]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Lavie, M.; Dubuisson, J.; Belouzard, S. SARS-CoV-2 Spike Furin Cleavage Site and S2’ Basic Residues Modulate the Entry Process in a Host Cell-Dependent Manner. J. Virol. 2022, 96, e0047422. [Google Scholar] [CrossRef]
- Berche, P. Gain-of-function and origin of Covid19. Presse Med. 2023, 52, 104167. [Google Scholar] [CrossRef]
- Harrison, N.L.; Sachs, J.D. A call for an independent inquiry into the origin of the SARS-CoV-2 virus. Proc. Natl. Acad. Sci. USA 2022, 119, e2202769119. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, Q.; Zhang, Z. Global Diversification and Distribution of Coronaviruses With Furin Cleavage Sites. Front. Microbiol. 2021, 12, 649314. [Google Scholar] [CrossRef] [PubMed]
- Mousavizadeh, L.; Ghasemi, S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2021, 54, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Millet, J.K.; Whittaker, G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA 2014, 111, 15214–15219. [Google Scholar] [CrossRef]
- Kleine-Weber, H.; Elzayat, M.T.; Hoffmann, M.; Pöhlmann, S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci. Rep. 2018, 8, 16597. [Google Scholar] [CrossRef]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. [Google Scholar] [CrossRef]
- Gui, M.; Song, W.; Zhou, H.; Xu, J.; Chen, S.; Xiang, Y.; Wang, X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017, 27, 119–129. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef]
- Papa, G.; Mallery, D.L.; Albecka, A.; Welch, L.G.; Cattin-Ortolá, J.; Luptak, J.; Paul, D.; McMahon, H.T.; Goodfellow, I.G.; Carter, A.; et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLOS Pathog. 2021, 17, e1009246. [Google Scholar] [CrossRef]
- Song, W.; Gui, M.; Wang, X.; Xiang, Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLOS Pathog. 2018, 14, e1007236. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Johnson, B.A.; Xie, X.; Bailey, A.L.; Kalveram, B.; Lokugamage, K.G.; Muruato, A.; Zou, J.; Zhang, X.; Juelich, T.; Smith, J.K.; et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 2021, 591, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Essalmani, R.; Jain, J.; Susan-Resiga, D.; Andréo, U.; Evagelidis, A.; Derbali, R.M.; Huynh, D.N.; Dallaire, F.; Laporte, M.; Delpal, A.; et al. Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity. J. Virol. 2022, 96, e00128-22. [Google Scholar] [CrossRef] [PubMed]
- Kocyigit, A.; Sogut, O.; Durmus, E.; Kanimdan, E.; Guler, E.M.; Kaplan, O.; Yenigun, V.B.; Eren, C.; Ozman, Z.; Yasar, O. Circulating furin, IL-6, and presepsin levels and disease severity in SARS-CoV-2–infected patients. Sci. Prog. 2021, 104, 00368504211026119. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I.; et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022, 602, 300–306. [Google Scholar] [CrossRef]
- Zhang, L.; Mann, M.; Syed, Z.A.; Reynolds, H.M.; Tian, E.; Samara, N.L.; Zeldin, D.C.; Tabak, L.A.; Ten Hagen, K.G. Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation. Proc. Natl. Acad. Sci. USA 2021, 118, e2109905118. [Google Scholar] [CrossRef]
- Cherian, S.; Potdar, V.; Jadhav, S.; Yadav, P.; Gupta, N.; Das, M.; Rakshit, P.; Singh, S.; Abraham, P.; Panda, S.; et al. SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms 2021, 9, 1542. [Google Scholar] [CrossRef]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.T.M.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef]
- Jawaid, M.Z.; Baidya, A.; Mahboubi-Ardakani, R.; Davis, R.L.; Cox, D.L. SARS-CoV-2 omicron spike simulations: Broad antibody escape, weakened ACE2 binding, and modest furin cleavage. Microbiol. Spectr. 2023, 11, e01213-22. [Google Scholar] [CrossRef]
- Schubert, M.; Bertoglio, F.; Steinke, S.; Heine, P.A.; Ynga-Durand, M.A.; Maass, H.; Sammartino, J.C.; Cassaniti, I.; Zuo, F.; Du, L.; et al. Human serum from SARS-CoV-2-vaccinated and COVID-19 patients shows reduced binding to the RBD of SARS-CoV-2 Omicron variant. BMC Med. 2022, 20, 102. [Google Scholar] [CrossRef]
- Ueyama, C.; Horibe, H.; Yamase, Y.; Fujimaki, T.; Oguri, M.; Kato, K.; Arai, M.; Watanabe, S.; Murohara, T.; Yamada, Y. Association of FURIN and ZPR1 polymorphisms with metabolic syndrome. Biomed. Rep. 2015, 3, 641–647. [Google Scholar] [CrossRef]
- Fry, H.; Mazidi, M.; Kartsonaki, C.; Clarke, R.; Walters, R.G.; Chen, Z.; Millwood, I.Y. The Role of Furin and Its Therapeutic Potential in Cardiovascular Disease Risk. Int. J. Mol. Sci. 2024, 25, 9237. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.N.; Mia, M.A.; Akter, Y.; Chowdhury, M.A.; Rahman, M.H.; Siddiqua, H.; Shathi, U.S.; Al-Mamun, A.; Siddika, F.; Marzan, L.W. Variations in Furin SNPs, a Major Concern of SARS-CoV-2 Susceptibility Among Different Populations: An In-Silico Approach. Bioinform. Biol. Insights 2024, 18, 11779322241306388. [Google Scholar] [CrossRef] [PubMed]
- Seidah, N.G.; Pasquato, A.; Andréo, U. How Do Enveloped Viruses Exploit the Secretory Proprotein Convertases to Regulate Infectivity and Spread? Viruses 2021, 13, 1229. [Google Scholar] [CrossRef] [PubMed]
- Al-Mulla, F.; Mohammad, A.; Al Madhoun, A.; Haddad, D.; Ali, H.; Eaaswarkhanth, M.; John, S.E.; Nizam, R.; Channanath, A.; Abu-Farha, M.; et al. ACE2 and FURIN variants are potential predictors of SARS-CoV-2 outcome: A time to implement precision medicine against COVID-19. Heliyon 2021, 7, e06133. [Google Scholar] [CrossRef]
- Pandey, R.K.; Srivastava, A.; Mishra, R.K.; Singh, P.P.; Chaubey, G. Novel genetic association of the Furin gene polymorphism rs1981458 with COVID-19 severity among Indian populations. Sci. Rep. 2024, 14, 7822. [Google Scholar]
- Elgedawy, G.A.; Elabd, N.S.; Salem, R.H.; Awad, S.M.; Amer, A.A.; Torayah, M.M.; El-Koa, A.A.; Abozeid, M.; Montaser, B.A.; Aboshabaan, H.S.; et al. FURIN, IFNL4, and TLR2 gene polymorphisms in relation to COVID-19 severity: A case-control study in Egyptian patients. Infection 2024, 52, 2213–2229. [Google Scholar] [CrossRef]
- Torre-Fuentes, L.; Matías-Guiu, J.; Hernández-Lorenzo, L.; Montero-Escribano, P.; Pytel, V.; Porta-Etessam, J.; Gómez-Pinedo, U.; Matías-Guiu, J.A. ACE2, TMPRSS2, and Furin variants and SARS-CoV-2 infection in Madrid, Spain. J. Med. Virol. 2021, 93, 863–869. [Google Scholar] [CrossRef]
- Ji, H.-L.; Zhao, R.; Matalon, S.; Matthay, M.A. Elevated Plasmin(ogen) as a Common Risk Factor for COVID-19 Susceptibility. Physiol. Rev. 2020, 100, 1065–1075. [Google Scholar] [CrossRef]
- Lei, R.X.; Shi, H.; Peng, X.M.; Zhu, Y.H.; Cheng, J.; Chen, G.H. Influence of a single nucleotide polymorphism in the P1 promoter of the furin gene on transcription activity and hepatitis B virus infection#. Hepatology 2009, 50, 763. [Google Scholar] [CrossRef]
- Zou, J.; Cao, Z.; Zhang, J.; Chen, T.; Yang, S.; Huang, Y.; Hong, D.; Li, Y.; Chen, X.; Wang, X.; et al. Variants in human papillomavirus receptor and associated genes are associated with type-specific HPV infection and lesion progression of the cervix. Oncotarget 2016, 7, 40135–40147. [Google Scholar] [CrossRef]
- Coto, E.; Albaiceta, G.M.; Amado-Rodríguez, L.; García-Clemente, M.; Cuesta-Llavona, E.; Vázquez-Coto, D.; Alonso, B.; Iglesias, S.; Melón, S.; Alvarez-Argüelles, M.E.; et al. FURIN gene variants (rs6224/rs4702) as potential markers of death and cardiovascular traits in severe COVID-19. J. Med. Virol. 2022, 94, 3589–3595. [Google Scholar] [CrossRef]
- Cilhoroz, B.T.; Schifano, E.D.; Panza, G.A.; Ash, G.I.; Corso, L.; Chen, M.-H.; Deshpande, V.; Zaleski, A.; Farinatti, P.; Santos, L.P. FURIN variant associations with postexercise hypotension are intensity and race dependent. Physiol. Rep. 2019, 7, e13952. [Google Scholar] [CrossRef]

| Polymorphism | Protector/Pathogenic | Direct or Indirect | Virus or Comorbidity | Bibliographic Source |
|---|---|---|---|---|
| SNP229 | Pathogenic | Direct | HBV | Lei et al., 2009 [235] |
| rs17514846 | Protective | Direct | VPH | Zou et al., 2016 [236] |
| rs2575712 | Protective | Direct | VPH | Zou et al., 2016 [236] |
| rs6226 | Mixed | Direct | SARS-CoV-2 | Uddin et al., 2024; Al-Mulla et al., 2021; (Elgedawy et al., 2024) [228,230,232] |
| rs150925934 | Pathogenic | Direct | SARS-CoV-2 | Uddin et al., 2024; Al-Mulla et al., 2021;(Elgedawy et al., 2024) [228,230,232] |
| rs201172453 | Protective | Direct | SARS-CoV-2 | Al-Mulla et al., 2021 [230] |
| rs8039305 | Protective | Direct | SARS-CoV-2 | Al-Mulla et al., 2021 [230] |
| rs1981458 | Pathogenic | Direct | SARS-CoV-2 | Pandey et al., 2024 [231] |
| rs6224 | Pathogenic | Indirect | Hypercholesterolemia | Coto et al., 2022 [237] |
| rs4702 | Pathogenic | Indirect | Hypercholesterolemia | Coto et al., 2022 [237] |
| rs17514846 | Pathogenic | Indirect | Coronary heart disease | Zhao et al., 2018 [11] |
| rs753334944 | N | N | N | Torre--Fuentes et al., 2021 [233] |
| rs16944971 | N | N | N | Torre--Fuentes et al., 2021 [233] |
| rs73489557 | N | N | N | Torre--Fuentes et al., 2021 [233] |
| rs6225 | N | N | N | Torre--Fuentes et al., 2021 [233] |
| rs12917264 | N | N | Hypotension | Cilhoroz et al., 2019 [238] |
| rs75493298 | N | N | Hypotension | Cilhoroz et al., 2019 [238] |
| rs74037507 | N | N | Increased systolic pressure | Cilhoroz et al., 2019 [238] |
| rs4702 | N | N | Less cognitive impairment induced by radiotherapy | Yang et al., 2022 [16] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva-Ríos, A.A.; Mora-Ornelas, C.E.; Flores-Medina, L.G.; Muñoz-Valle, J.F.; Díaz-Palomera, C.D.; García-Chagollan, M.; Vizcaíno-Quirarte, A.M.; Viera-Segura, O. Beyond Processing: Furin as a Central Hub in Viral Pathogenesis and Genetic Susceptibility. Biomolecules 2025, 15, 1530. https://doi.org/10.3390/biom15111530
Silva-Ríos AA, Mora-Ornelas CE, Flores-Medina LG, Muñoz-Valle JF, Díaz-Palomera CD, García-Chagollan M, Vizcaíno-Quirarte AM, Viera-Segura O. Beyond Processing: Furin as a Central Hub in Viral Pathogenesis and Genetic Susceptibility. Biomolecules. 2025; 15(11):1530. https://doi.org/10.3390/biom15111530
Chicago/Turabian StyleSilva-Ríos, Adrián Alejandro, Carlos Ernesto Mora-Ornelas, Luna Galilea Flores-Medina, José Francisco Muñoz-Valle, Carlos Daniel Díaz-Palomera, Mariel García-Chagollan, Alexis Missael Vizcaíno-Quirarte, and Oliver Viera-Segura. 2025. "Beyond Processing: Furin as a Central Hub in Viral Pathogenesis and Genetic Susceptibility" Biomolecules 15, no. 11: 1530. https://doi.org/10.3390/biom15111530
APA StyleSilva-Ríos, A. A., Mora-Ornelas, C. E., Flores-Medina, L. G., Muñoz-Valle, J. F., Díaz-Palomera, C. D., García-Chagollan, M., Vizcaíno-Quirarte, A. M., & Viera-Segura, O. (2025). Beyond Processing: Furin as a Central Hub in Viral Pathogenesis and Genetic Susceptibility. Biomolecules, 15(11), 1530. https://doi.org/10.3390/biom15111530








