Mining Thermophile Photosynthesis Genes: A Synthetic Operon Expressing Chloroflexota Species Reaction Center Genes in Rhodobacter sphaeroides
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Growth Conditions and Plasmids
2.2. Protein Purification, SDS-PAGE, and Spectroscopy
2.3. Bioinformatic Tools and Data
2.3.1. Curation of Hot Spring Metagenomes
2.3.2. Identification of Reaction Center Proteins PufL and PufM
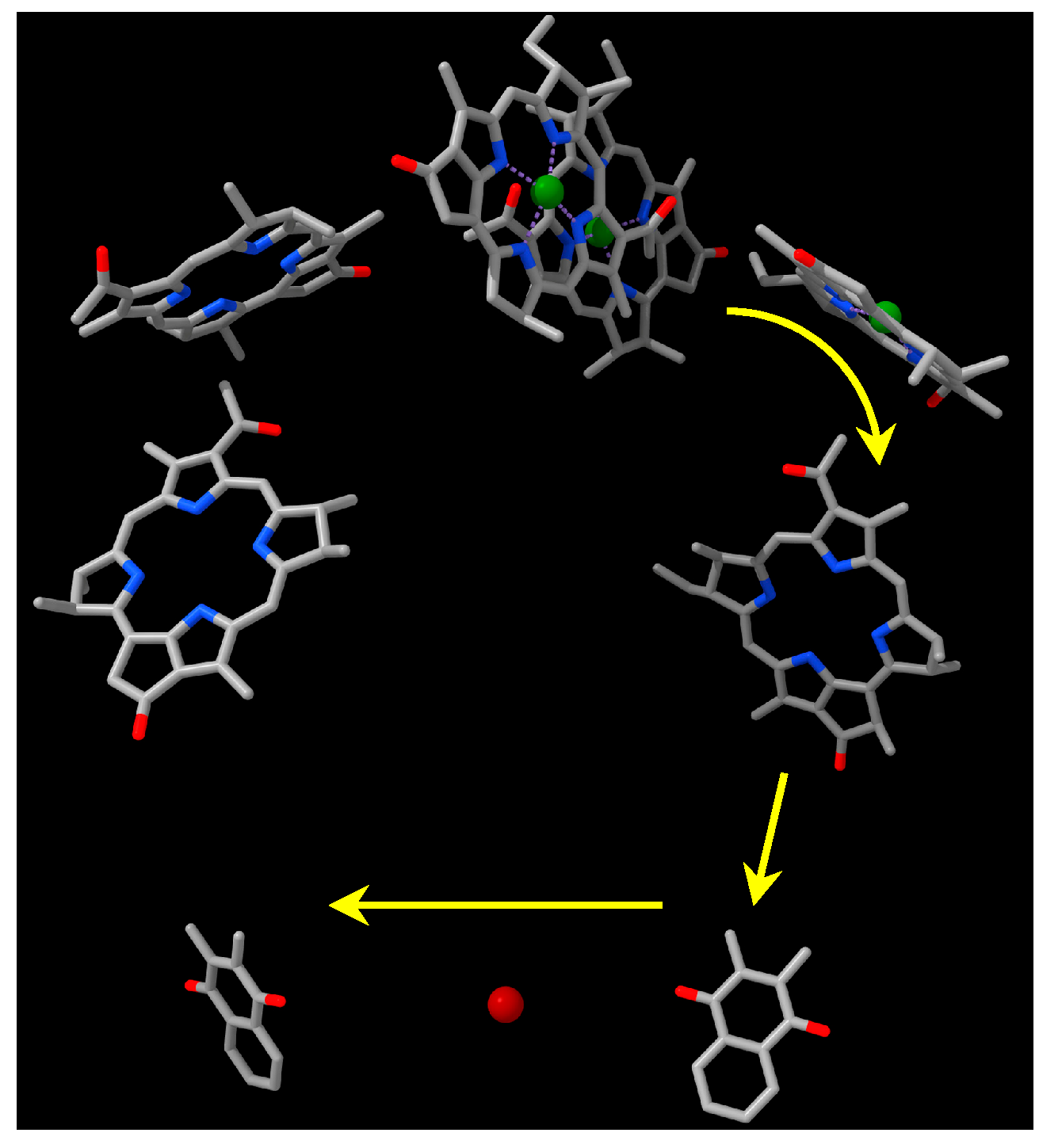
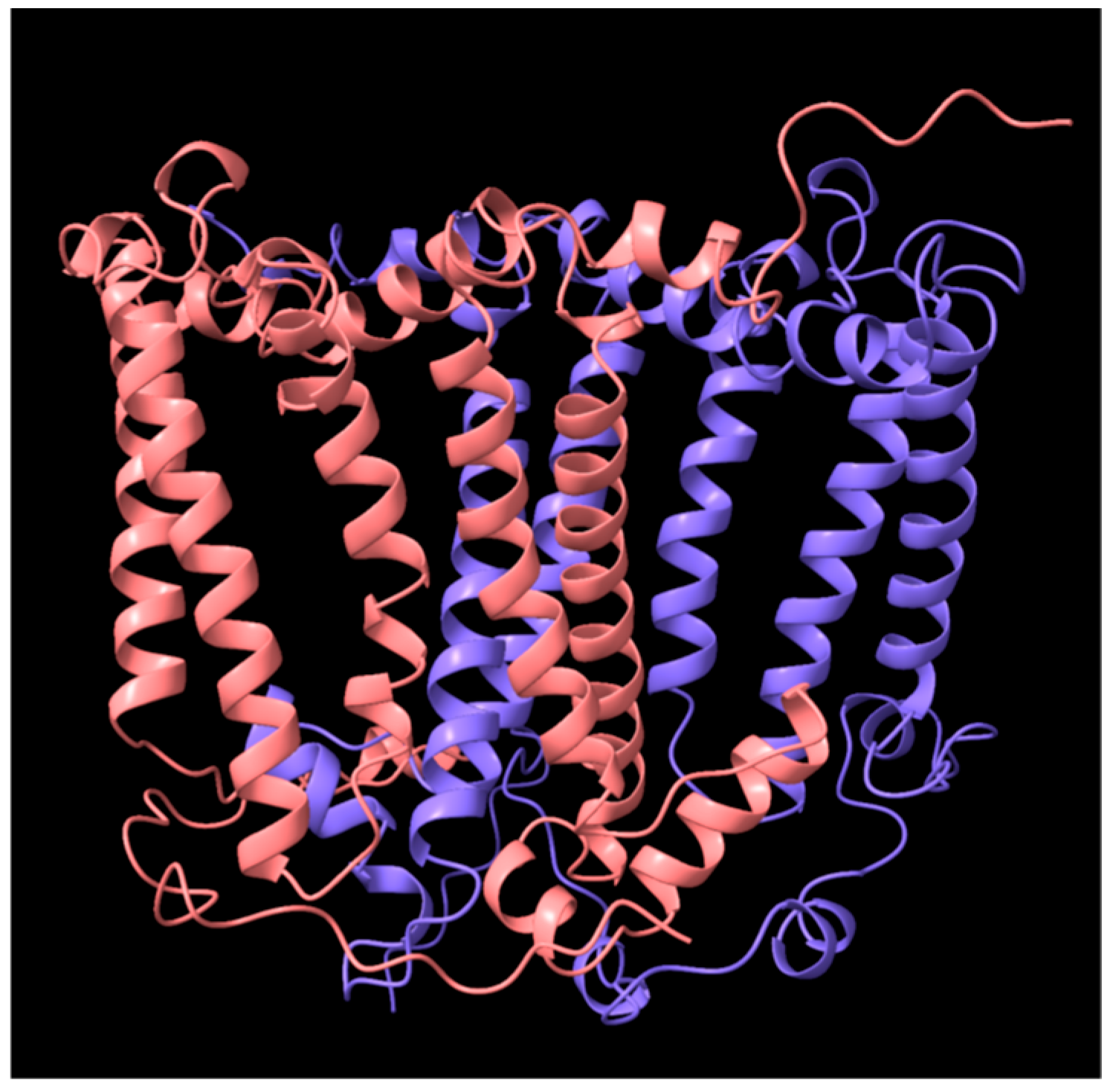
3. Results
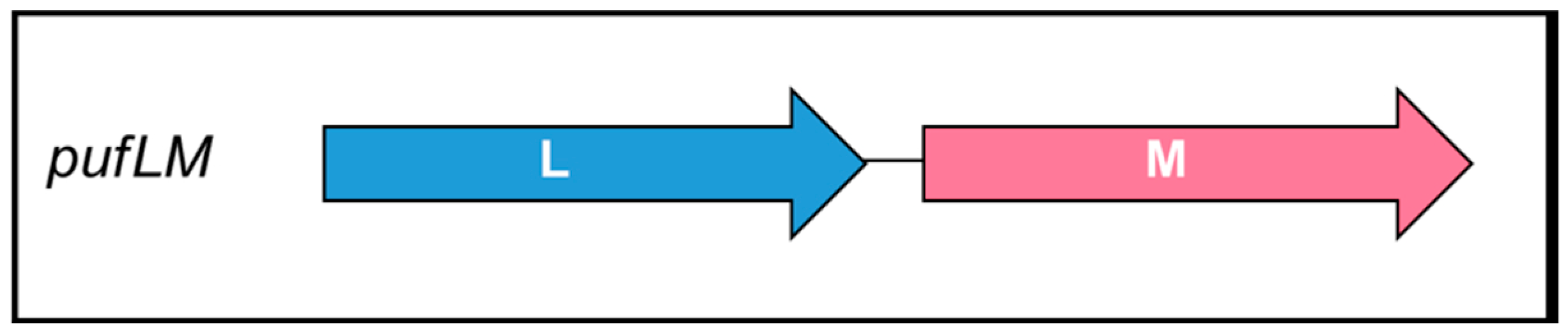
3.1. Design, Construction and Expression of Chloroflexota Synthetic pufLM Operon Encoding the RC
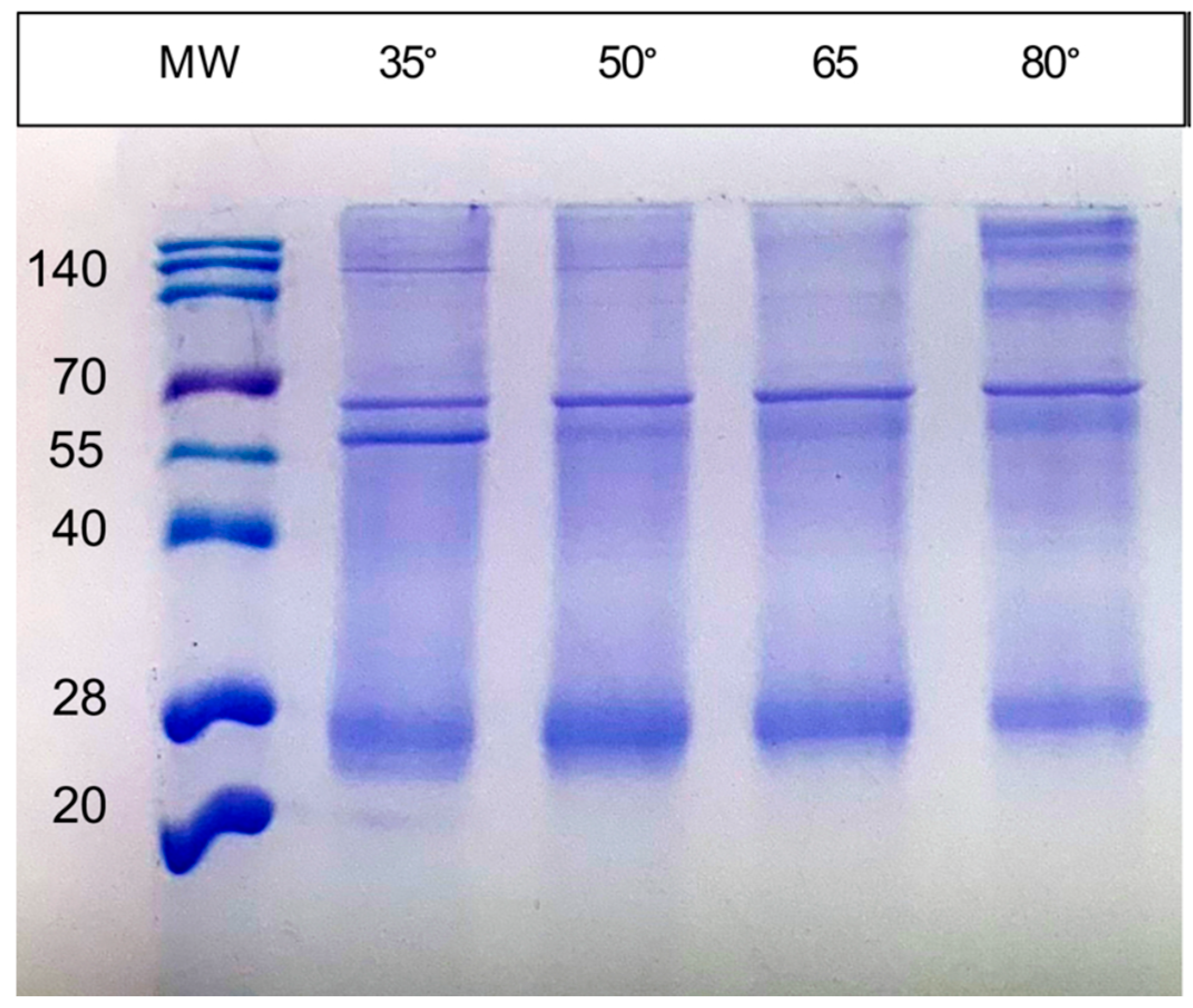
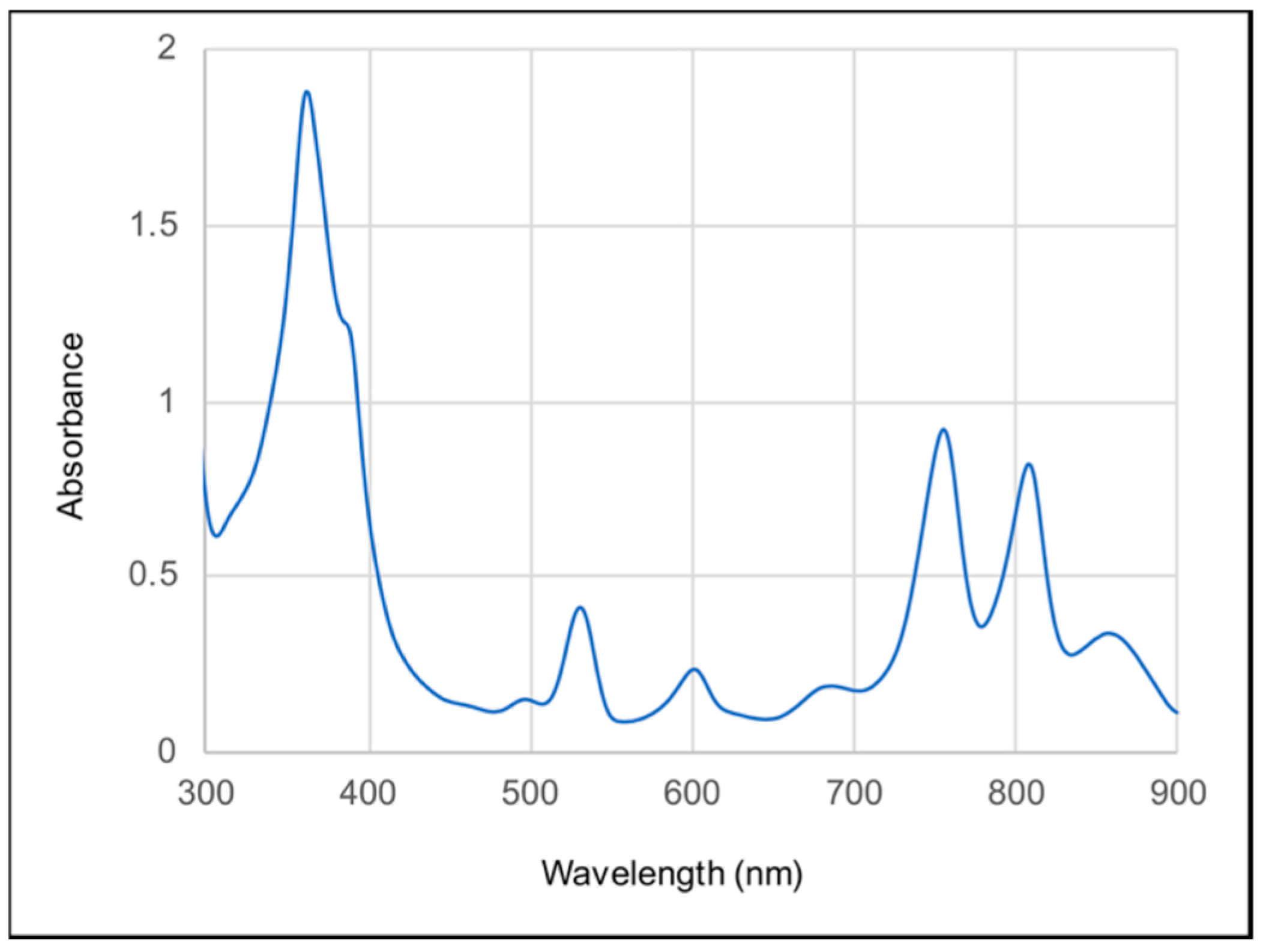
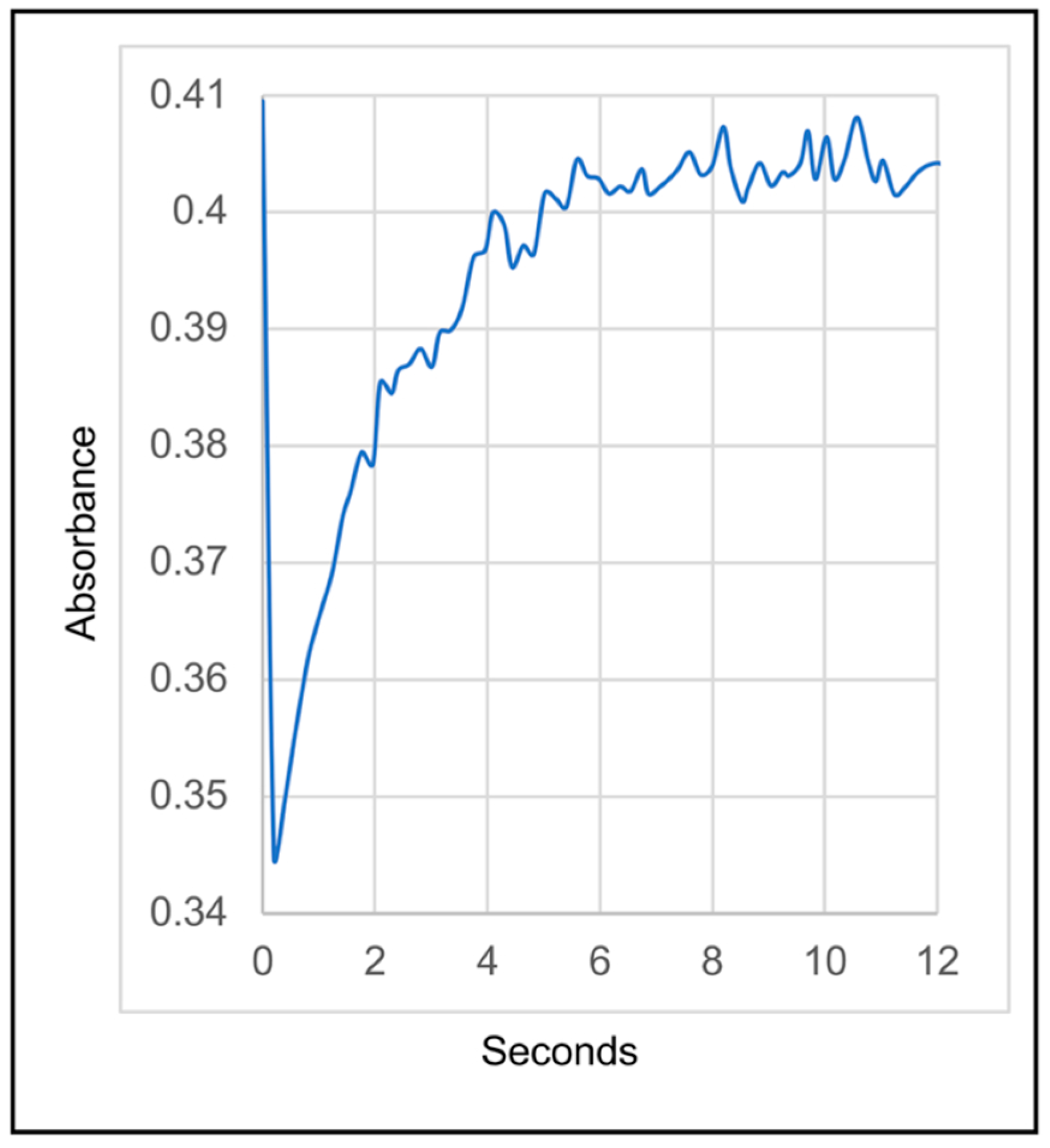
3.2. SDS-PAGE, Absorption Spectra and Catalytic Activity of the RC Complex Produced by the pufLM Operon
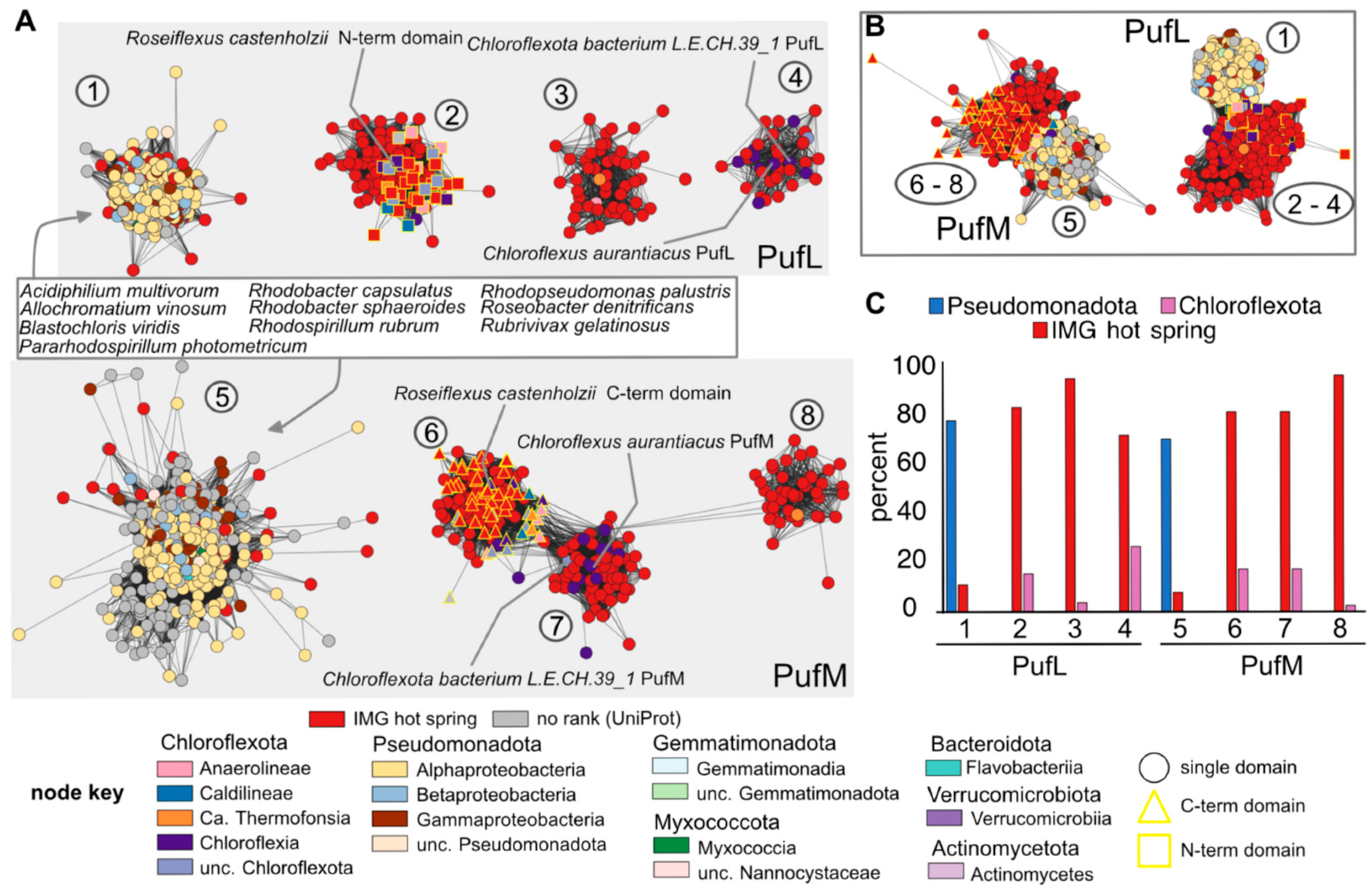
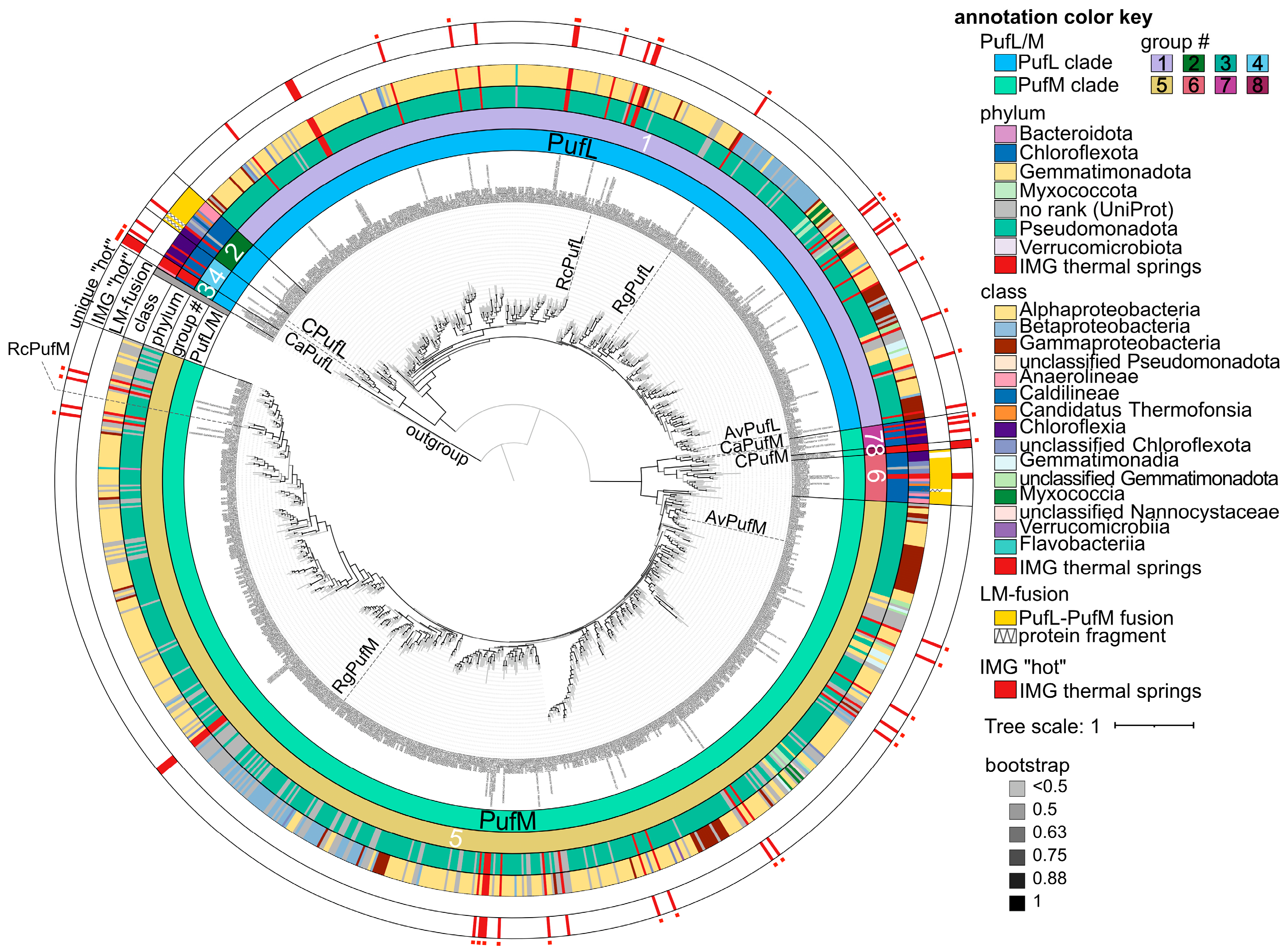
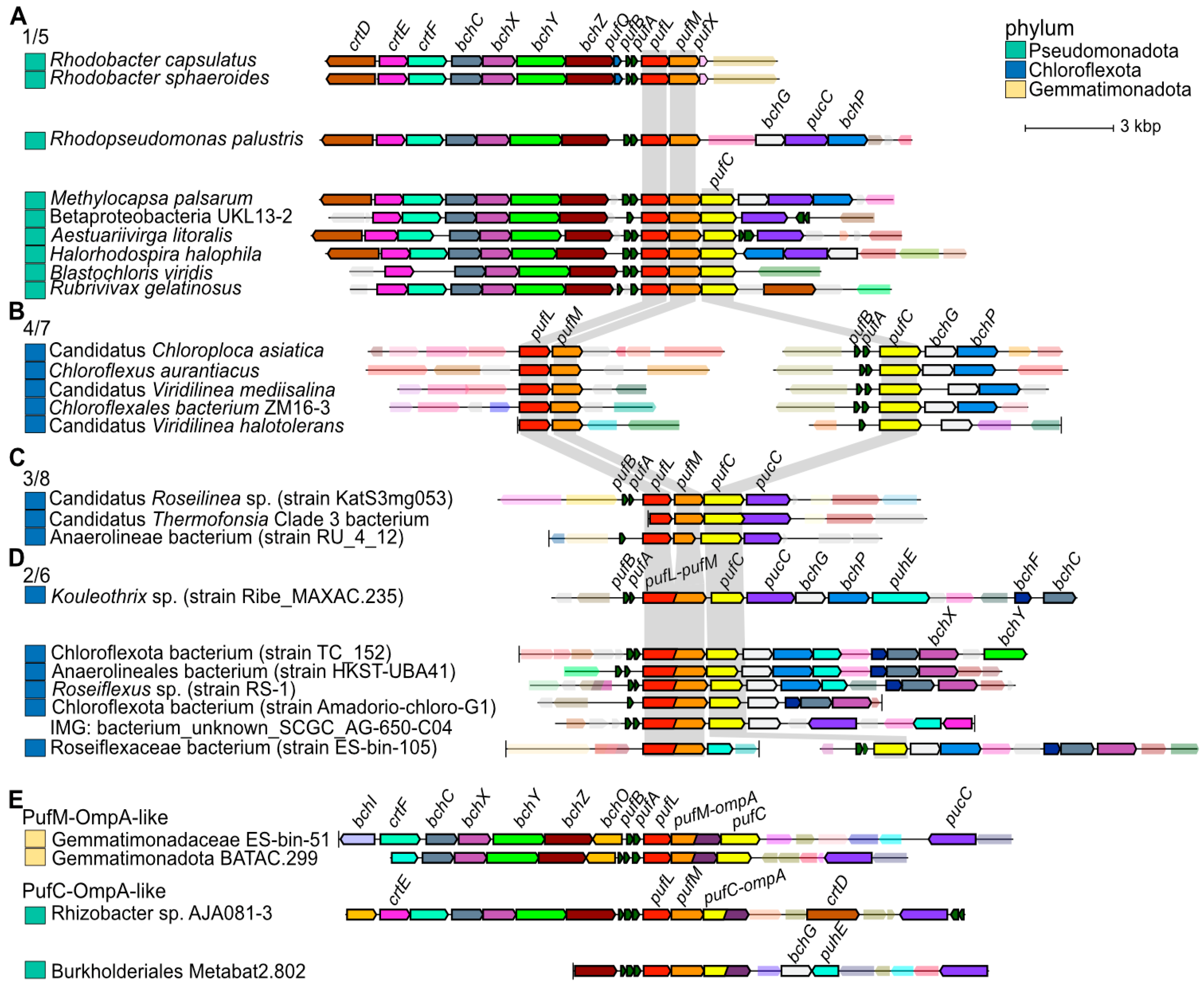
3.3. Bioinformatic Analyses of RC Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RC(s) | Reaction center(s) |
| BChl(s) | Bacteriochlorophyll(s) |
| BPhe(s) | Bacteriopheophytin(s) |
| P | Primary donor or special pair of BChls in the RC |
| QA | Quinone within the RC |
| QB | Quinone within the RC |
| sp. | Species |
| ms | millisecond |
| nm | nanometer |
| NTA | nitrilotriacetic acid |
| JGI | Joint Genome Institute |
| IMG/M | Integrated Microbial Genomes and Microbiomes |
| SSN(s) | Sequence similarity network(s) |
References
- Wiegand, S.; Sobol, M.; Schnepp-Pesch, L.K.; Yan, G.; Iqbal, S.; Vollmers, J.; Muller, J.A.; Kaster, A.K. Taxonomic re-classification and expansion of the phylum Chloroflexota based on over 5000 genomes and metagenome-assembled genomes. Microorganisms 2023, 11, 2612. [Google Scholar] [CrossRef]
- Freches, A.; Fradinho, J.C. The biotechnological potential of the Chloroflexota phylum. Appl. Environ. Microbiol. 2024, 90, e0175623. [Google Scholar] [CrossRef]
- Tsuji, J.M.; Shaw, N.A.; Nagashima, S.; Venkiteswaran, J.J.; Schiff, S.L.; Watanabe, T.; Fukui, M.; Hanada, S.; Tank, M.; Neufeld, J.D. Anoxygenic phototroph of the Chloroflexota uses a type I reaction centre. Nature 2024, 627, 915–922. [Google Scholar] [CrossRef]
- Blankenship, R.E. Molecular Mechanisms of Photosynthesis; Blackwell Science: Oxford, UK, 2002. [Google Scholar]
- Liu, L.N.; Bracun, L.; Li, M. Structural diversity and modularity of photosynthetic RC-LH1 complexes. Trends Microbiol. 2024, 32, 38–52. [Google Scholar] [CrossRef]
- Kirmaier, C.; Blankenship, R.E.; Holten, D. Formation and decay of radical-pair state P+I− in Chloroflexus aurantiacus reaction centers. Biochim. Biophys. Acta 1986, 850, 275–285. [Google Scholar] [CrossRef]
- Huang, G.Q.; Dong, S.S.; Ma, L.; Li, L.; Ju, J.X.; Wang, M.J.; Zhang, J.P.; Sui, S.F.; Qin, X.C. Cryo-EM structure of a minimal reaction center-light-harvesting complex from the phototrophic bacterium Chloroflexus aurantiacus. J. Integr. Plant Biol. 2025, 67, 967–978. [Google Scholar] [CrossRef]
- Gardiner, A.T.; Nguyen-Phan, T.C.; Cogdell, R.J. A comparative look at structural variation among RC-LH1 ‘Core’ complexes present in anoxygenic phototrophic bacteria. Photosynth. Res. 2020, 145, 83–96. [Google Scholar] [CrossRef]
- Xin, Y.; Shi, Y.; Niu, T.; Wang, Q.; Niu, W.; Huang, X.; Ding, W.; Yang, L.; Blankenship, R.E.; Xu, X.; et al. Cryo-EM structure of the RC-LH core complex from an early branching photosynthetic prokaryote. Nat. Commun. 2018, 9, 1568. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef] [PubMed]
- Pierson, B.K.; Thornber, J.P. Isolation and spectral characterization of photochemical reaction centers from the thermophilic green bacterium Chloroflexus aurantiacus strain J-10-f1. Proc. Natl. Acad. Sci. USA 1983, 80, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Bina, D.; Gardian, Z.; Vacha, F.; Litvin, R. Supramolecular organization of photosynthetic membrane proteins in the chlorosome-containing bacterium Chloroflexus aurantiacus. Photosynth. Res. 2014, 122, 13–21. [Google Scholar] [CrossRef]
- Swainsbury, D.J.K.; Qian, P.; Hitchcock, A.; Hunter, C.N. The structure and assembly of reaction centre-light-harvesting 1 complexes in photosynthetic bacteria. Biosci. Rep. 2023, 43, BSR20220089. [Google Scholar] [CrossRef]
- Qi, C.H.; Wang, G.L.; Wang, F.F.; Xin, Y.Y.; Zou, M.J.; Madigan, M.T.; Wang-Otomo, Z.Y.; Ma, F.; Yu, L.J. New insights on the photocomplex of revealed from comparisons of native and carotenoid-depleted complexes. J. Biol. Chem. 2023, 299, 105057. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.M.; Kirmaier, C.; Holten, D.; Blankenship, R.E. Kinetics and energetics of electron transfer in reaction centers of the photosynthetic bacterium Roseiflexus castenholzii. Biochim. Biophys. Acta 2011, 1807, 262–269. [Google Scholar] [CrossRef][Green Version]
- Wang, X.P.; Yu, B.Y.; Qi, C.H.; Wang, G.L.; Zou, M.J.; Zhang, C.F.; Yu, L.J.; Ma, F. Energy Transfer and Exciton Relaxation in B880-B800-RC Complex through Two-Dimensional Electronic Spectroscopy. J. Phys. Chem. Lett. 2024, 15, 3619–3626. [Google Scholar] [CrossRef]
- Xin, Y.Y.; Lin, S.; Blankenship, R.E. Femtosecond spectroscopy of the primary charge separation in reaction centers of Chloroflexus aurantiacus with selective excitation in the Q and soret bands. J. Phys. Chem. A 2007, 111, 9367–9373. [Google Scholar] [CrossRef]
- Zabelin, A.A.; Kovalev, V.B.; Khristin, A.M.; Khatypov, R.A.; Shkuropatov, A.Y. Primary charge separation in Chloroflexus aurantiacus reaction centers at room temperature: Ultrafast transient absorption measurements on QA-depleted preparations with native and chemically modified bacteriopheophytin composition. Photosynth. Res. 2025, 163, 1–15. [Google Scholar] [CrossRef]
- Kato, M.; Cardona, T.; Rutherford, A.W.; Reisner, E. Covalent immobilization of oriented photosystem II on a nanostructured electrode for solar water oxidation. J. Am. Chem. Soc. 2013, 135, 10610–10613. [Google Scholar] [CrossRef]
- Rasul, F.; You, D.W.; Jiang, Y.; Liu, X.J.; Daroch, M. Thermophilic cyanobacteria-exciting, yet challenging biotechnological chassis. Appl. Microbiol. Biotechnol. 2024, 108, 270. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.E.; Hunter, C.N. Engineering chlorophyll, bacteriochlorophyll, and carotenoid biosynthetic pathways in Escherichia coli. ACS Synth. Biol. 2023, 12, 2236–2244. [Google Scholar] [CrossRef] [PubMed]
- Levy-Booth, D.J.; Hashimi, A.; Roccor, R.; Liu, L.Y.; Renneckar, S.; Eltis, L.D.; Mohn, W.W. Genomics and metatranscriptomics of biogeochemical cycling and degradation of lignin-derived aromatic compounds in thermal swamp sediment. ISME J. 2021, 15, 879–893. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram-negative bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Jun, D.; Saer, R.G.; Madden, J.D.; Beatty, J.T. Use of new strains of Rhodobacter sphaeroides and a modified simple culture medium to increase yield and facilitate purification of the reaction centre. Photosynth. Res. 2014, 120, 197–205. [Google Scholar] [CrossRef]
- Jun, D.; Richardson-Sanchez, T.; Mahey, A.; Murphy, M.E.P.; Fernandez, R.C.; Beatty, J.T. Introduction of the menaquinone biosynthetic pathway into Rhodobacter sphaeroides and de novo synthesis of menaquinone for incorporation into heterologously expressed integral membrane proteins. ACS Synth. Biol. 2020, 9, 1190–1200. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 1989. [Google Scholar]
- Oberortner, E.; Cheng, J.F.; Hillson, N.J.; Deutsch, S. Streamlining the design-to-build transition with build-optimization software tools. ACS Synth. Biol. 2017, 6, 485–496. [Google Scholar] [CrossRef]
- Salis, H.M.; Mirsky, E.A.; Voigt, C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009, 27, 946–950. [Google Scholar] [CrossRef] [PubMed]
- Ind, A.C.; Porter, S.L.; Brown, M.T.; Byles, E.D.; de Beyer, J.A.; Godfrey, S.A.; Armitage, J.P. Inducible-expression plasmid for Rhodobacter sphaeroides and Paracoccus denitrificans. Appl. Environ. Microbiol. 2009, 75, 6613–6615. [Google Scholar] [CrossRef]
- Chen, I.M.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.H.; Huntemann, M.; Hajek, P.; Ritter, S.J.; Webb, C.; Wu, D.Y.; et al. The IMG/M data management and analysis system v.7: Content updates and new features. Nucleic Acids Res. 2023, 51, D723–D732. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Adesina, A.; Ahmad, S.; Bowler-Barnett, E.H.; Bye-A-Jee, H.; Carpentier, D.; Denny, P.; et al. UniProt: The universal protein knowledgebase in 2025. Nucleic Acids Res. 2024, 52, D609–D617. [Google Scholar] [CrossRef]
- Oberg, N.; Zallot, R.; Gerlt, J.A. EFI-EST, EFI-GNT, and EFI-CGFP: Enzyme function Iiitiative (EFI) web resource for genomic enzymology tools. J. Mol. Biol. 2023, 435, 168018. [Google Scholar] [CrossRef]
- Zallot, R.; Oberg, N.; Gerlt, J.A. The EFI web resource for genomic enzymology tools: Leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry 2019, 58, 4169–4182. [Google Scholar] [CrossRef]
- Suzek, B.E.; Wang, Y.Q.; Huang, H.Z.; McGarvey, P.B.; Wu, C.H.; Consortium, U. UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 2015, 31, 926–932. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.Z.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Andreeva, A.; Blum, M.; Chuguransky, S.R.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Llinares-López, F.; Meng-Papaxanthos, L.; et al. The Pfam protein families database: Embracing AI/ML. Nucleic Acids Res. 2024, 53, D523–D534. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Shelton, A.N.; Yu, F.B.; Grossman, A.R.; Bhaya, D. Abundant and active community members respond to diel cycles in hot spring phototrophic mats. ISME J. 2025, 19, wraf001. [Google Scholar] [CrossRef]
- Klatt, C.G.; Inskeep, W.P.; Herrgard, M.J.; Jay, Z.J.; Rusch, D.B.; Tringe, S.G.; Parenteau, M.N.; Ward, D.M.; Boomer, S.M.; Bryant, D.A.; et al. Community structure and function of high-temperature chlorophototrophic microbial mats inhabiting diverse geothermal environments. Front. Microbiol. 2013, 4, 106. [Google Scholar] [CrossRef]
- Klatt, C.G.; Liu, Z.F.; Ludwig, M.; Kühl, M.; Jensen, S.I.; Bryant, D.A.; Ward, D.M. Temporal metatranscriptomic patterning in phototrophic Chloroflexi inhabiting a microbial mat in a geothermal spring. ISME J. 2013, 7, 1775–1789. [Google Scholar] [CrossRef]
- Klatt, C.G.; Wood, J.M.; Rusch, D.B.; Bateson, M.M.; Hamamura, N.; Heidelberg, J.F.; Grossman, A.R.; Bhaya, D.; Cohan, F.M.; Kühl, M.; et al. Community ecology of hot spring cyanobacterial mats: Predominant populations and their functional potential. ISME J. 2011, 5, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Lynes, M.M.; Krukenberg, V.; Jay, Z.J.; Kohtz, A.J.; Gobrogge, C.A.; Spietz, R.L.; Hatzenpichler, R. Diversity and function of methyl-coenzyme M reductase-encoding archaea in Yellowstone hot springs revealed by metagenomics and mesocosm experiments. ISME Commun. 2023, 3, 22. [Google Scholar] [CrossRef]
- Jarett, J.K.; Dzunková, M.; Schulz, F.; Roux, S.; Paez-Espino, D.; Eloe-Fadrosh, E.; Jungbluth, S.P.; Ivanova, N.; Spear, J.R.; Carr, S.A.; et al. Insights into the dynamics between viruses and their hosts in a hot spring microbial mat. ISME J. 2020, 14, 2527–2541. [Google Scholar] [CrossRef]
- Inskeep, W.P.; Jay, Z.J.; Tringe, S.G.; Herrgård, M.J.; Rusch, D.B.; Co, Y.M.P.S. The YNP metagenome project: Environmental parameters responsible for microbial distribution in the Yellowstone geothermal ecosystem. Front. Microbiol. 2013, 4, 67. [Google Scholar] [CrossRef]
- Bowers, R.M.; Nayfach, S.; Schulz, F.; Jungbluth, S.P.; Ruhl, I.A.; Sheremet, A.; Lee, J.; Goudeau, D.; Eloe-Fadrosh, E.A.; Stepanauskas, R.; et al. Dissecting the dominant hot spring microbial populations based on community-wide sampling at single-cell genomic resolution. ISME J. 2022, 16, 1337–1347. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Zhang, H.; Hanada, S.; Nagashima, K.V.P.; Shimada, K.; Matsuura, K. Structural and spectroscopic properties of a reaction center complex from the chlorosome-lacking filamentous anoxygenic phototrophic bacterium. J. Bacteriol. 2005, 187, 1702–1709. [Google Scholar] [CrossRef]
- Yabe, S.; Muto, K.; Abe, K.; Yokota, A.; Staudigel, H.; Tebo, B.M. Vulcanimicrobium alpinus gen. nov. sp. nov., the first cultivated representative of the candidate phylum “Eremiobacterota”, is a metabolically versatile aerobic anoxygenic phototroph. ISME Commun. 2022, 2, 102. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv 2022. [CrossRef] [PubMed]
- Feher, G.; Allen, J.P.; Okamura, M.Y.; Rees, D.C. Structure and function of bacterial photosynthetic reaction centers. Nature 1989, 339, 111–116. [Google Scholar] [CrossRef]
- Venturoli, G.; Trotta, M.; Feick, R.; Melandri, B.A.; Zannoni, D. Temperature-dependence of charge recombination from the P+Qa- and P+Qb- states in photosynthetic reaction centers isolated from the thermophilic bacterium Chloroflexus-aurantiacus. Eur. J. Biochem. 1991, 202, 625–634. [Google Scholar] [CrossRef]
- Lips, S.; Schmitt-Jansen, M.; Borchert, E. Metagenomic analyses of the plastisphere reveals a common functional potential across oceans. bioRxiv 2024. [Google Scholar] [CrossRef]
- Pierson, B.K.; Thornber, J.P.; Seftor, R.E.B. Partial purification, subunit structure and thermal stability of the photochemical reaction center of the thermophilic green bacterium Chloroflexus aurantiacus. Biochim. Biophys. Acta 1983, 723, 322–326. [Google Scholar] [CrossRef]
- Nozawa, T.; Madigan, M.T. Temperature and solvent effects on reaction centers from Chloroflexus aurantiacus and Chromatium tepidum. J. Biochem. 1991, 110, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.J.; Hughes, A.V.; Fyfe, P.K.; Wakeham, M.C.; Holden-Dye, K.; Heathcote, P.; Jones, M.R. On the role of basic residues in adapting the reaction centre—LH1 complex for growth at elevated temperatures in purple bacteria. Photosynth. Res. 2005, 86, 81–100. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehman, Y.; Kim, Y.; Tong, M.; Blaby, I.K.; Blaby-Haas, C.E.; Beatty, J.T. Mining Thermophile Photosynthesis Genes: A Synthetic Operon Expressing Chloroflexota Species Reaction Center Genes in Rhodobacter sphaeroides. Biomolecules 2025, 15, 1529. https://doi.org/10.3390/biom15111529
Rehman Y, Kim Y, Tong M, Blaby IK, Blaby-Haas CE, Beatty JT. Mining Thermophile Photosynthesis Genes: A Synthetic Operon Expressing Chloroflexota Species Reaction Center Genes in Rhodobacter sphaeroides. Biomolecules. 2025; 15(11):1529. https://doi.org/10.3390/biom15111529
Chicago/Turabian StyleRehman, Yasir, Younghoon Kim, Michelle Tong, Ian K. Blaby, Crysten E. Blaby-Haas, and J. Thomas Beatty. 2025. "Mining Thermophile Photosynthesis Genes: A Synthetic Operon Expressing Chloroflexota Species Reaction Center Genes in Rhodobacter sphaeroides" Biomolecules 15, no. 11: 1529. https://doi.org/10.3390/biom15111529
APA StyleRehman, Y., Kim, Y., Tong, M., Blaby, I. K., Blaby-Haas, C. E., & Beatty, J. T. (2025). Mining Thermophile Photosynthesis Genes: A Synthetic Operon Expressing Chloroflexota Species Reaction Center Genes in Rhodobacter sphaeroides. Biomolecules, 15(11), 1529. https://doi.org/10.3390/biom15111529






