Mitochondrial Genome Features and Phylogenetic Analyses of Four Chrysochroinae Species (Coleoptera: Buprestidae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling and DNA Extraction
2.2. Mitogenome Sequencing, Annotation and Analysis
2.3. Phylogenetic Aanlysis
3. Results
3.1. Genome Organization and Base Composition
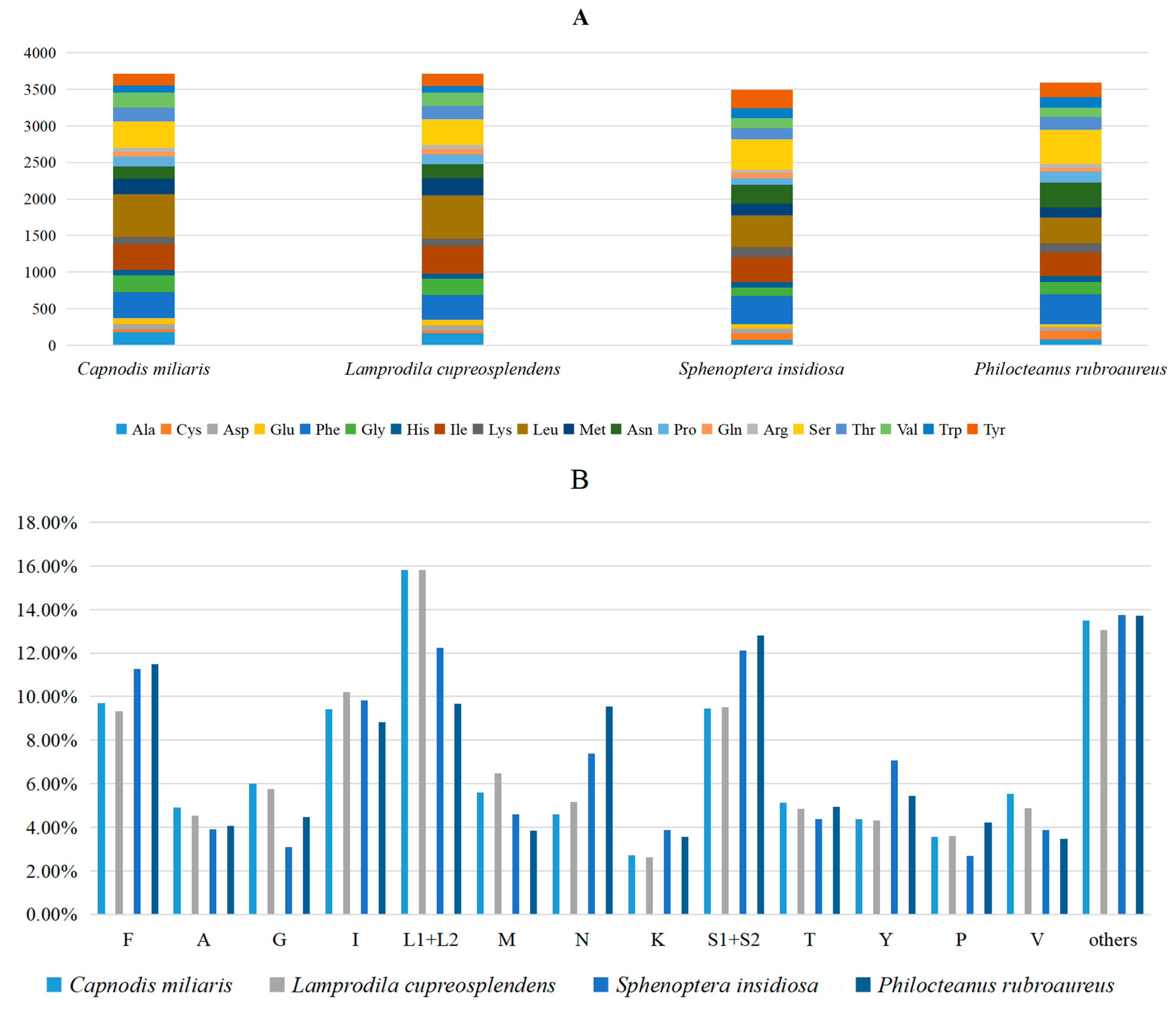
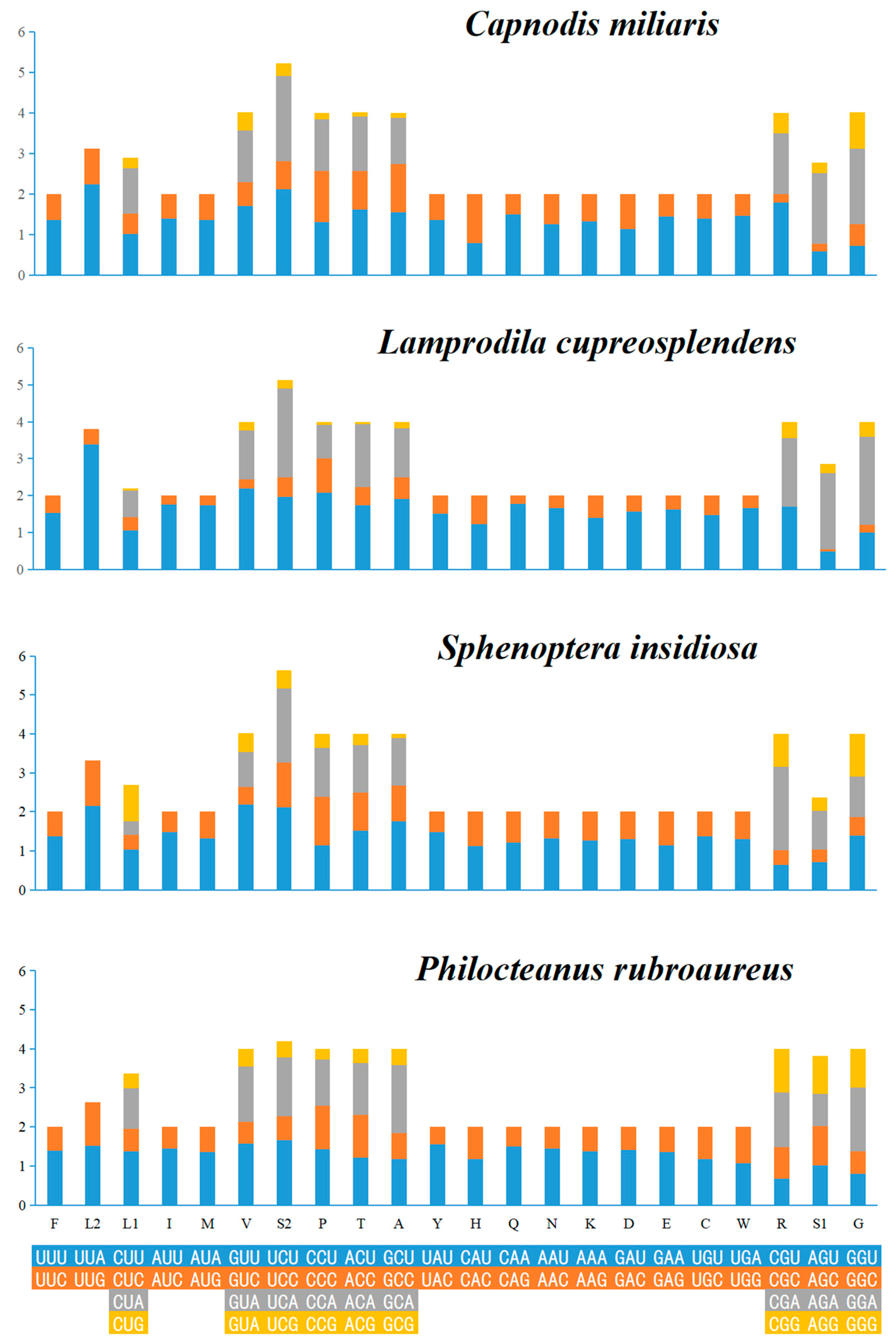
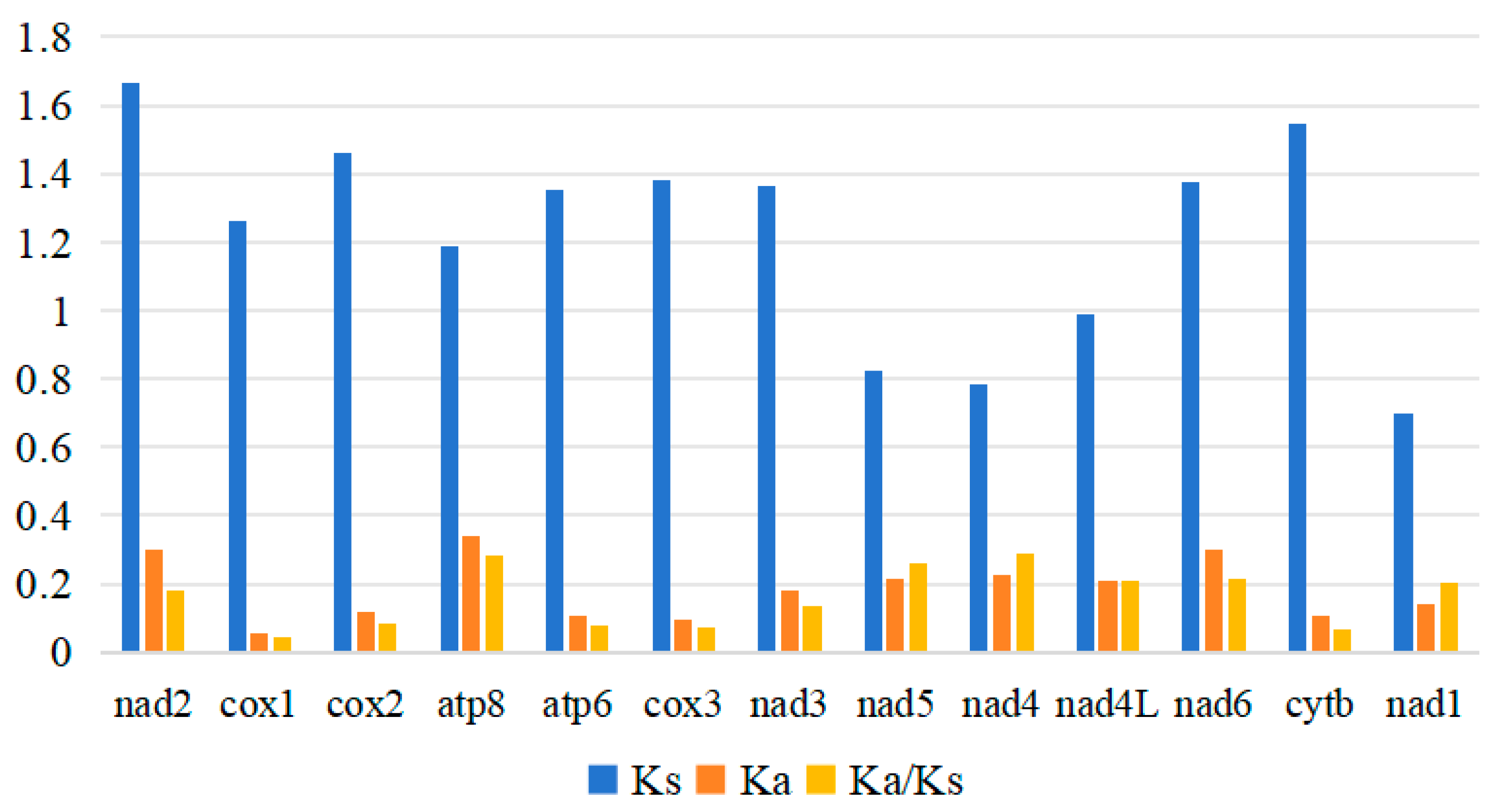
3.2. Protein-Coding Regions, Codon Usage and Nucleotide Diversity
3.3. Ribosomal and Transfer RNA Genes
3.4. Control Region and Gene Arrangement
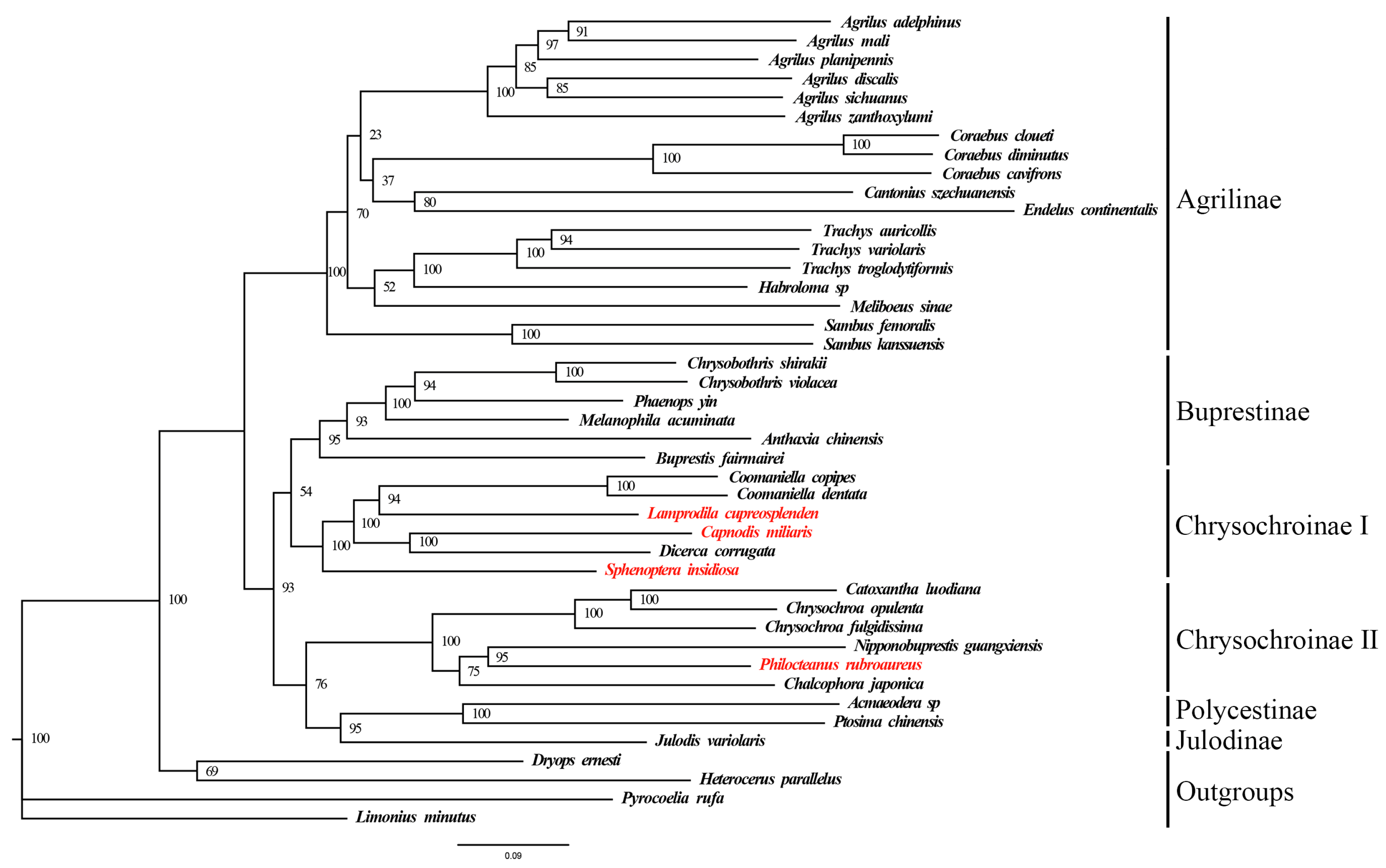
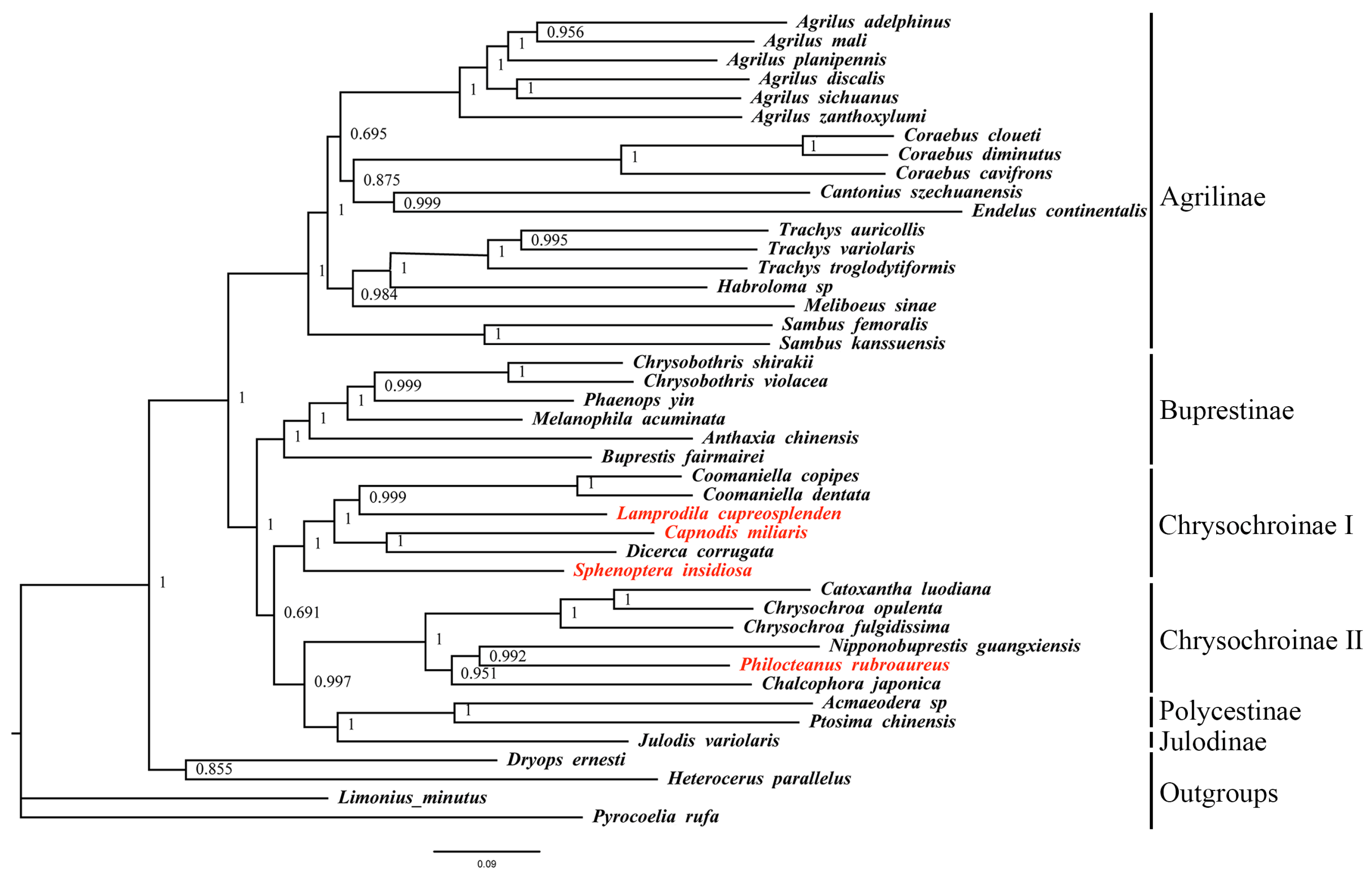
3.5. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bellamy, C.L. A World Catalogue and Bibliography of the Jewel Beetles (Coleoptera: Buprestoidea), Pensoft Series Faunistica; Pensoft Publishers: Sofia, Bulgaria; Moscow, Russia, 2008; Volume 1–4, pp. 1–2684. [Google Scholar]
- Bellamy, C.L.; Volkovitsh, M.G. 18 Buprestoidea Crowson, 1955. In Handbook of Zoology, Arthropoda: Insecta, Coleoptera, Beetles, 2nd ed.; Beutel, R.G., Kristensen, N.P., Eds.; Walter de Gruyter GmbH: Berlin, Germany; Boston, MA, USA, 2016; Volume 1, pp. 543–552. [Google Scholar]
- Herms, D.A.; McCullough, D.G. Emerald ash borer invasion of North America: History, biology, ecology, impact and management. Annu. Rev. Entomol. 2014, 59, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Bozorov, T.A.; Luo, Z.H.; Li, X.S.; Zhang, D.Y. Agrilus mali Matsumara (Coleoptera: Buprestidae), a new invasive pest of wild apple in western China: DNA barcoding and life cycle. Ecol. Evol. 2018, 9, 1160–1172. [Google Scholar] [CrossRef] [PubMed]
- Herms, D.A. Strategies for deployment of insect resistant ornamental plants. In Mechanisms and Deployment of Resistance in Trees to Insects; Wagner, M.R., Clancy, K.M., Lieutier, F., Paine, T.D., Eds.; Kluwer Academic Publishing: Dordrecht, The Netherlands, 2002; pp. 217–237. [Google Scholar]
- Apel, K.H.; Katzel, R.; Luttschwager, D.; Schmitz, H.; Schutz, S. Investigations on possible mechanisms of the host finding by Phaenops cyanea F. (Col., Buprestidae). Mitt. Dtsch. Ges. Allg. Angew. Ent. 2000, 12, 23–27. [Google Scholar]
- Xu, G.X.; Liu, Y.M.; Wu, W. Occurrence rhythm and control of Chrysobothris igai at Orange Stand. J. Zhejiang For. Sci. Technol. 2006, 26, 49–52. [Google Scholar]
- Åström, M.; Pettersson, L.B.; Öckinger, E.; Hedin, J. Habitat preferences and conservation of the marbled jewel beetle Poecilonota variolosa (Buprestidae). J. Insect Conserv. 2013, 17, 1145–1154. [Google Scholar] [CrossRef]
- Ohmomo, S.; Fukutomi, H. The Buprestid Beetles of Japan; Mushi-Sha: Tokyo, Japan, 2013. [Google Scholar]
- Ong, U.; Hattori, T. Jewel Beetles of Taiwan; Ministry of Beetles: Tainan, China, 2019; Volume 1. [Google Scholar]
- Volkovitsh, M.G.; Karpun, N.N. A new invasive species of buprestid beetles in the Russian fauna: Lamprodila (Palmar) festiva (L.) (Coleoptera, Buprestidae), a pest of Cupressaceae. Entomol. Rev. 2017, 97, 425–437. [Google Scholar] [CrossRef]
- Ruicănescu, A.; Stoica, A.I. The distribution and behaviour studies on a new invasive Buprestid species, Lamprodila festiva (Coleoptera: Buprestidae) in Romania. Trav. Mus. Natn. Hist. Nat. “Grigore Antipa” 2019, 62, 43–56. [Google Scholar]
- Zeng, F.Y.; Wang, T.; Zong, S.X. Damage characteristics and spatial distribution of Sphenoptera sp. larvae. Forest Res. 2012, 25, 223–226. [Google Scholar]
- Wang, J.W.; Li, Y.H.; Han, W.D.; Luo, Y.Q.; Zong, S.X. Biological characteristics of Sphenoptera sp. on Artemisia ordosoca. Chin. J. Appl. Entomol. 2012, 48, 141–146. [Google Scholar]
- Hong, M.Y.; Jeong, H.C.; Kim, M.J.; Jeong, H.U.; Lee, S.H.; Kim, I. Complete mitogenome sequence of the jewel beetle, Chrysochroa fulgidissima (Coleoptera: Buprestidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2009, 20, 46–60. [Google Scholar] [CrossRef]
- Xiao, L.F.; Zhang, S.D.; Long, C.P.; Guo, Q.Y.; Xu, J.S.; Dai, X.H.; Wang, J.G. Complete mitogenome of a leaf-mining buprestid beetle, Trachys auricollis, and its phylogenetic implications. Genes 2019, 10, 992. [Google Scholar] [CrossRef] [PubMed]
- Hołyński, R.B. Taxonomic Structure of the Subtribe Chrysochroina Cast with Review of the Genus Chrysochroa Dej. (Coleoptera: Buprestidae); Gondwana: Warszawa, Poland, 2009; pp. 1–379. [Google Scholar]
- Evans, A.M.; Mckenna, D.D.; Bellamy, C.L.; Farrell, B.D. Large-scale molecular phylogeny of metallic wood-boring beetles (Coleoptera: Buprestoidea) provides new insights into relationships and reveals multiple evolutionary origins of the larval leaf-mining habit. Syst. Entomol. 2015, 40, 385–400. [Google Scholar] [CrossRef]
- Kolibááč, J. Classification and phylogeny of the Buprestoidea (Insecta: Coleoptera). Acta Mus. Moraviae Sci. biol. 2000, 85, 113–184. [Google Scholar]
- Volkovitsh, M.G. The comparative morphology of antennal structures in Buprestidae (Coleoptera): Evolutionary trends, taxonomic and phylogenetic implications. Part 1. Acta Mus. Morav. Sci. Biol. 2001, 86, 43–169. [Google Scholar]
- Li, H.; Shao, R.F.; Song, N.; Song, F.; Jiang, P.; Li, Z.H.; Cai, W.Z. Higher-level phylogeny of paraneopteran insects inferred from mitochondrial genome sequences. Sci. Rep. 2015, 5, 8527. [Google Scholar] [CrossRef]
- Qin, J.; Zhang, Y.Z.; Zhou, X.; Kong, X.B.; Wei, S.J.; Ward, R.D.; Zhang, A.B. Mitochondrial phylogenomics and genetic relationships of closely related pine moth (Lasiocampidae: Dendrolimus) species in China, using whole mitochondrial genomes. BMC Genom. 2015, 16, 428. [Google Scholar] [CrossRef]
- Tian, T.; Yuan, H.; Chen, B. Phylogeny of hydradephagan water beetles (Coleoptera: Adephaga) inferred with mitochondrial genome sequences. Acta Entomol. Sin. 2020, 63, 1016–1027. [Google Scholar]
- Krzywinski, J.; Li, C.; Morris, M.; Conn, J.E.; Lima, J.B.; Povoa, M.M.; Wilkerson, R.C. Analysis of the evolutionary forces shaping mitochondrial genomes of a Neotropical malaria vector complex. Mol. Phylogenet. Evol. 2011, 58, 469–477. [Google Scholar] [CrossRef]
- Yan, L.P.; Pape, T.; Elgar, M.A.; Gao, Y.Y.; Zhang, D. Evolutionary history of stomach bot flies in the light of mitogenomics. Syst. Entomol. 2019, 44, 797–809. [Google Scholar] [CrossRef]
- Motyka, M.; Kusy, D.; Háva, J.; Jahodářová, E.; Bílková, R.; Vogler, A.P.; Bocak, L. Mitogenomic data elucidate the phylogeny and evolution of life strategies in Dermestidae (Coleoptera). Syst. Entomol. 2022, 47, 82–93. [Google Scholar] [CrossRef]
- Nie, R.E.; Wei, J.; Zhang, S.K.; Vogler, A.P.; Wu, L.; Konstantinov, A.S.; Li, W.Z.; Yang, X.K.; Xue, H.J. Diversification of mitogenomes in three sympatric Altica flea beetles (Insecta, Chrysomelidae). Zool. Scr. 2019, 48, 657–666. [Google Scholar] [CrossRef]
- Park, J.S.; Cho, Y.; Kim, M.J.; Nam, S.H.; Kim, I. Description of complete mitochondrial genome of the black-veined white, Aporia crataegi (Lepidoptera: Papilionoidea), and comparison to papilionoid species. J. Asia-Pac. Entomol. 2012, 15, 331–341. [Google Scholar] [CrossRef]
- Hahn, C.; Bachmann, L.; Chevreux, B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—A baiting and iterative mapping approach. Nucleic Acids Res. 2013, 41, e129. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Peng, Z.L. A study on the genus Nipponobuprestis Obenberger (Coleoptera: Buprestidae). Insect Sci. 1995, 2, 95–103. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Phylogenetics Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weight, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijin, M.H.L.; Droujn, A.R.J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; Schreier, P.H.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. TRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Shen, X.; Li, X.; Sha, Z.; Yan, B.; Xu, Q. Complete mitochondrial genome of the Japanese snapping shrimp Alpheus japonicus (Crustacea: Decapoda: Caridea): Gene rearrangement and phylogeny within Caridea. Sci. China Life Sci. 2012, 55, 591–598. [Google Scholar] [CrossRef]
- Chen, X.Q. Phylogenetic Relationships of Buprestidae (Coleoptera) Based on Molecular and Morphological Data. J. Insect Syst. Evol. 2024, 52, 189–205. [Google Scholar]
- Zhang, Y.J.; Jiang, N.; Liu, Q.; Zhu, Y.; Huang, X. The role of mitochondrial damage in cadmium-induced hepatocyte apoptosis and DNA damage. J. Hygiene Res. 2020, 49, 290–297. [Google Scholar]
- Huang, X.Y.; Wei, Z.H.; Lu, J.W.; Shi, A.M. Mitogenomic analysis and phylogenetic relationships of Agrilinae: Insights into the evolutionary patterns of a diverse buprestid subfamily. PLoS ONE 2023, 18, e0291820. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhnam, S.; Largetm, B.; Lium, L.; Suchardm, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Wei, Z.H. The complete mitochondrial genomes of five Agrilinae (Coleoptera, Buprestidae) species and phylogenetic implications. ZooKeys 2022, 1092, 195–212. [Google Scholar] [CrossRef]
- Ouyang, B.W.; Huang, X.Y.; Gan, Y.J.; Wei, Z.H.; Shi, A.M. Three mitochondrial genomes of Chrysochroinae (Coleoptera, Buprestidae) and phylogenetic analyses. Genes 2024, 15, 1336. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Ouyang, B.; Wei, Z.; Shi, A. Four Mitochondrial Genomes of Buprestinae (Coleoptera: Buprestidae) and Phylogenetic Analyses. Genes 2025, 16, 828. [Google Scholar] [CrossRef]
- Huang, X.Y.; Chen, B.; Wei, Z.H.; Shi, A.M. First report of complete mitochondrial genome in the tribes Coomaniellini and Dicercini (Coleoptera: Buprestidae) and phylogenetic implications. Genes 2022, 13, 1074. [Google Scholar] [CrossRef]
- Li, H.; Liu, H.Y.; Song, F.; Shi, A.M.; Zhou, X.G.; Cai, W.Z. Comparative mitogenomic analysis of damsel bugs representing three tribes in the family Nabidae (Insecta: Hemiptera). PLoS ONE 2012, 7, e45925. [Google Scholar] [CrossRef]
- Song, F.; Li, H.; Jiang, P.; Zhou, X.; Liu, J.P.; Sun, C.H.; Vogler, A.P.; Cai, W.Z. Capturing the phylogeny of holometabola with mitochondrial genome data and Bayesian site-heterogeneous mixture models. Genome Biol. Evol. 2016, 8, 1411–1426. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.H.; Huang, X.Y.; Shi, A.M. First mitochondrial genome of subfamily Julodinae (Coleoptera, Buprestidae) with its phylogenetic implications. ZooKeys 2023, 1139, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Tôyama, M. The systematic positions of some buprestid genera (Coleoptera, Buprestidae). Elytra 1987, 15, 1–11. [Google Scholar]
- Hołyński, R.B. Remarks on the general classification of Buprestidae Leach as applied to Maoraxiina. Folia Entomol. Hung. 1988, 49, 49–54. [Google Scholar]
- Hołyński, R.B. A reassessment of the internal classification of the Buprestidae Leach (Coleoptera). Cryst. Ser. Zool. 1993, 1, 1–42. [Google Scholar]
- Kubáň, V. New nomenclatorial and taxonomic acts, and comments. Buprestidae: Various groups. In Catalogue of Palaearctic Coleoptera; Löbl, I., Löbl, D., Eds.; Brill: Leiden, The Netherlands; Boston, MA, USA, 2016; Volume 3, pp. 19–27, 432–438, 456–470, 494–523, 554–574, 983. [Google Scholar]
| Gene | Strand | Position | Codons | Intergenic Nucleotides | ||
|---|---|---|---|---|---|---|
| From | To | Start | Stop | |||
| trnI | J | 1/1/1/1 | 64/64/64/66 | 0/0/0/0 | ||
| trnQ | N | 77/62/62/64 | 145/130/130/132 | 12/−3/−3/−3 | ||
| trnM | J | 145/130/130/132 | 214/198/198/200 | −1/−1/−1/−1 | ||
| nad2 | J | 215/199/253/201 | 1240/1221/1224/1220 | ATT/ATT/ATT/ATA | TAA/TAA/TAA/TAG | 0/0/54/0 |
| trnW | J | 1239/1220/1223/1222 | 1305/1286/1288/1293 | −2/−2/−2/1 | ||
| trnC | N | 1298/1279/1281/1286 | 1363/1340/1341/1346 | −8/−8/−8/−8 | ||
| trnY | N | 1364/1341/1342/1347 | 1427/1404/1404/1411 | 0/0/0/0 | ||
| cox1 | J | 1429/1406/1403/1404 | 2959/2936/2941/2944 | ?/?/ACG/ATT | T/T/TAA/T | 1/1/−2/−8 |
| trnL2 | J | 2960/2937/2937/2945 | 3024/3001/3002/3010 | 0/0/−5/0 | ||
| cox2 | J | 3025/3002/3051/3011 | 3712/3683/3725/3698 | ATA/ATA/ATA/ATT | T/T/TAA/T | 0/0/48/0 |
| trnK | J | 3713/3684/3691/3699 | 3783/3753/3761/3768 | 0/0/−35/0 | ||
| trnD | J | 3783/3754/3761/3769 | 3847/3818/3826/3832 | −1/0/−1/0 | ||
| atp8 | J | 3848/3819/3827/3961 | 4003/3974/3985/4137 | ATC/ATA/ATT/ATC | TAA/TAA/TAA/TAA | 0/0/0/128 |
| atp6 | J | 3997/3968/3979/4131 | 4671/4642/4653/4805 | ATG/ATG/ATG/ATG | TAA/TAA/TAA/TAA | −7/−7/−7/−7 |
| cox3 | J | 4671/4642/4653/4805 | 5457/5428/5440/5591 | ATG/ATG/ATG/ATG | T/T/T/T | −1/−1/−1/−1 |
| trnG | J | 5458/5429/5440/5592 | 5521/5490/5503/5654 | 0/0/−1/0 | ||
| nad3 | J | 5522/5491/5504/5655 | 5875/5844/5857/6008 | ATT/ATT/ATA/ATA | TAG/TAG/TAA/TAG | 0/0/0/0 |
| trnA | J | 5874/5843/5863/6007 | 5935/5905/5928/6070 | −2/−2/5/−2 | ||
| trnR | J | 5935/5905/5928/6071 | 6001/5968/5994/6132 | −1/−1/−1/0 | ||
| trnN | J | 6001/5968/5994/6132 | 6065/6033/6058/6195 | −1/−1/−1/−1 | ||
| trnS1 | J | 6066/6034/6059/6196 | 6132/6100/6125/6262 | 0/0/0/−1 | ||
| trnE | J | 6133/6101/6126/6265 | 6195/6162/6190/6326 | 0/0/0/2 | ||
| trnF | N | 6195/6162/6190/6326 | 6258/6225/6255/6389 | −1/−1/−1/−1 | ||
| nad5 | N | 6259/6226/6255/6399 | 7978/7945/7967/8106 | ATT/ATT/ATT/ATT | T/T/TAA/T | 0/0/−1/9 |
| trnH | N | 7979/7946/7977/8110 | 8042/8009/8040/8171 | 0/0/9/3 | ||
| nad4 | N | 8043/8010/8095/8149 | 9375/9342/9376/9507 | ATG/ATG/ATG/ATG | T/T/T/TAA | 0/0/54/−24 |
| nad4L | N | 9369/9336/9370/9501 | 9656/9626/9660/9770 | ATG/ATG/ATG/ATG | TAA/TAA/TAA/TAA | −7/−7/−7/−7 |
| trnT | J | 9659/9629/9663/9791 | 9724/9692/9727/9853 | 2/2/2/20 | ||
| trnP | N | 9725/9693/9727/9854 | 9789/97,575/9792/9918 | 0/0/0/0 | ||
| nad6 | J | 9791/9759/9794/9923 | 10,294/10,262/10,303/10,423 | ATA/ATA/ATT/ATA | TAA/TAA/TAA/TAA | 1/1/1/4 |
| cytb | J | 10,298/10,262/10,303/10,423 | 11,440/11,404/11,445/11,562 | ATG/ATG/ATG/ATG | TAG/TAG/TAG/TAG | 3/−1/−1/−1 |
| trnS2 | J | 11,439/11,403/11,444/11,561 | 11,506/11,470/11,510/11,628 | −2/−2/−2/−2 | ||
| nad1 | N | 11,543/11,489/11,528/11,646 | 12,487/12,439/12,477/12,596 | TTG/TTG/ATA/TTG | TAA/TAA/TAA/TAG | 36/18/16/17 |
| trnL1 | N | 12,489/12,441/12,483/12,597 | 12,552/12,504/12,542/12,660 | 1/1/22/0 | ||
| rrnL | N | 12,553/12,505/12,533/12,694 | 13,842/13,790/13,759/13,878 | 0/0/−15/33 | ||
| trnV | N | 13,843/13,791/13,845/13,962 | 13,912/13,860/13,914/14,031 | 0/0/17/83 | ||
| rrnS | N | 13,913/13,861/13,914/14,033 | 14,649/14,587/14,538/14,639 | 0/0/−1/1 | ||
| CR | 14,650/14,588/14,695/14,640 | 16,230/16,222/16,183/15,778 | 0/0/0/0 | |||
| Species | PCGs | rRNAs | tRNAs | A + T-Rich Region | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size (bp) | A + T (%) | A + T Skew | Size (bp) | A + T (%) | A + T Skew | Size (bp) | A + T (%) | A + T Skew | Size (bp) | A + T (%) | A + T Skew | |
| C. miliaris | 11,150 | 71.93 | −0.15 | 2013 | 76.66 | −0.03 | 1438 | 74.40 | 0.02 | 1635 | 78.58 | −0.01 |
| L. cupreosplendens | 11,159 | 66.16 | −0.16 | 2027 | 73.31 | −0.07 | 1450 | 73.75 | 0.02 | 1581 | 76.58 | 0.03 |
| S.insidiosa | 11,034 | 71.46 | −0.14 | 2076 | 76.74 | −0.09 | 1452 | 73.77 | 0.01 | 1488 | 82.00 | 0.07 |
| P. rubroaureus | 11,140 | 67.92 | −0.12 | 1790 | 72.18 | −0.16 | 1410 | 70.85 | 0.03 | 1138 | 75.66 | 0.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Li, Y.; Wei, Z.; Shi, A. Mitochondrial Genome Features and Phylogenetic Analyses of Four Chrysochroinae Species (Coleoptera: Buprestidae). Biomolecules 2025, 15, 1531. https://doi.org/10.3390/biom15111531
Wang J, Li Y, Wei Z, Shi A. Mitochondrial Genome Features and Phylogenetic Analyses of Four Chrysochroinae Species (Coleoptera: Buprestidae). Biomolecules. 2025; 15(11):1531. https://doi.org/10.3390/biom15111531
Chicago/Turabian StyleWang, Jieqiong, Yingying Li, Zhonghua Wei, and Aimin Shi. 2025. "Mitochondrial Genome Features and Phylogenetic Analyses of Four Chrysochroinae Species (Coleoptera: Buprestidae)" Biomolecules 15, no. 11: 1531. https://doi.org/10.3390/biom15111531
APA StyleWang, J., Li, Y., Wei, Z., & Shi, A. (2025). Mitochondrial Genome Features and Phylogenetic Analyses of Four Chrysochroinae Species (Coleoptera: Buprestidae). Biomolecules, 15(11), 1531. https://doi.org/10.3390/biom15111531





