Efficient Mining and Characterization of Two Novel Keratinases from Metagenomic Database
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis
2.2. Cloning and Heterologous Expression of Keratinase Genes
2.2.1. Plasmid Construction and Cloning of Keratinase Genes
2.2.2. Transformation and Co-Expression of Recombinant Keratinase Genes
2.2.3. Co-Expression with Molecular Chaperone
2.2.4. Optimization of Expression Conditions
- Induction Temperature:Protein expression was induced at temperatures of 16 °C, 20 °C, 28 °C, and 37 °C to determine the optimal conditions for soluble protein expression. Soluble expression was monitored using SDS-PAGE (Bio-Rad, Hercules, CA, USA).
- IPTG Concentration: The effect of IPTG concentration on protein expression was evaluated by varying the IPTG concentration from 0.02 mM to 0.5 mM. Optimal conditions were selected based on the level of soluble protein expression.
2.2.5. Protein Purification
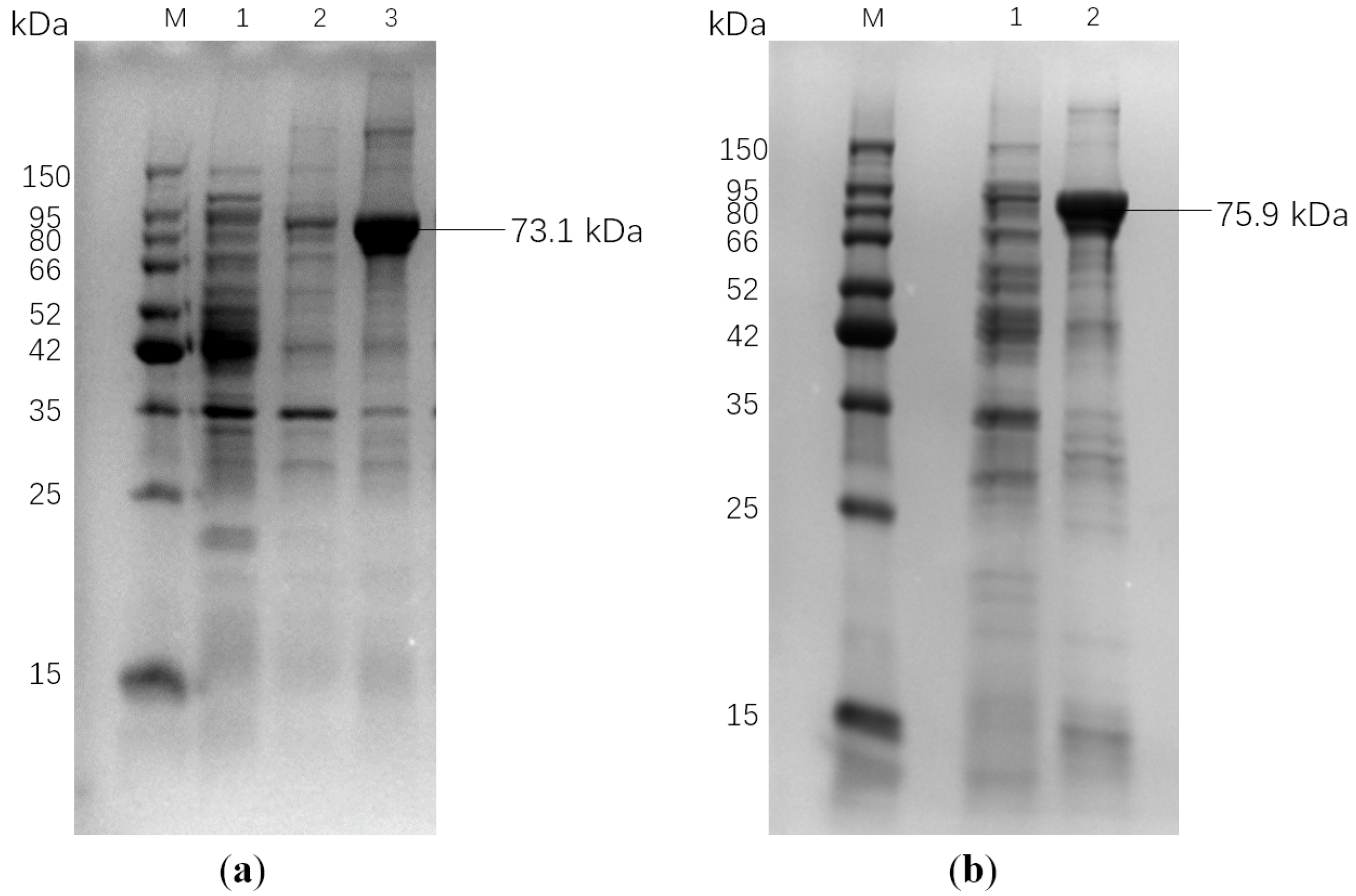
2.2.6. Evaluation of Expression and Purity
2.3. Enzymatic Characterization
2.3.1. Enzymatic Activity Assays
2.3.2. Optimal Temperature and pH Determination
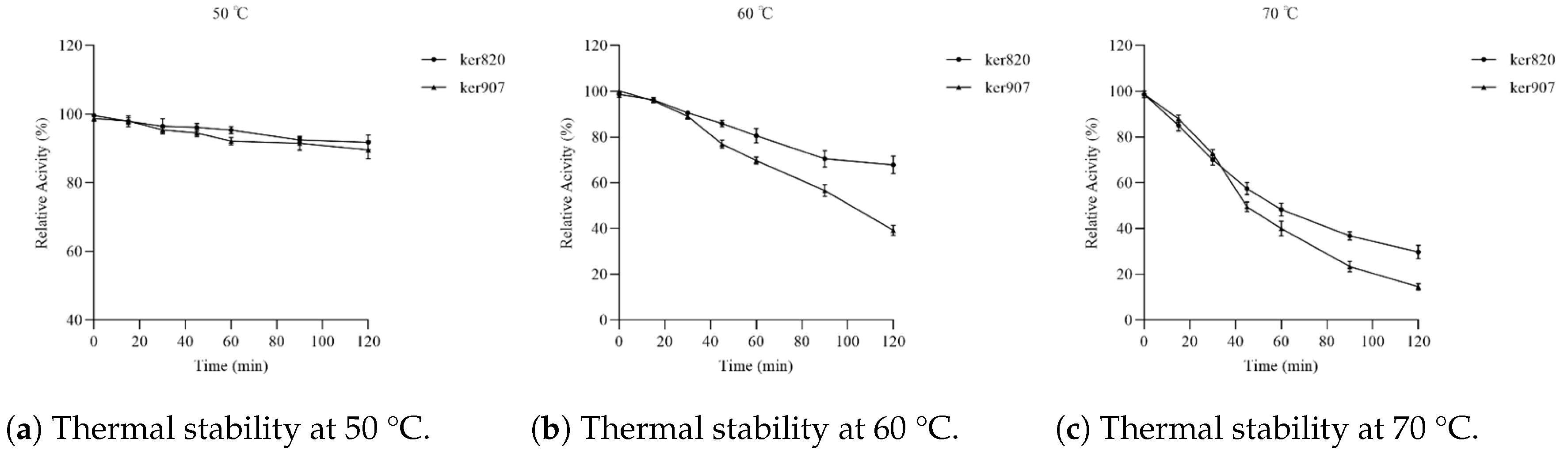
2.3.3. Thermal Stability Assay
2.3.4. Effect of Metal Ions and Additives
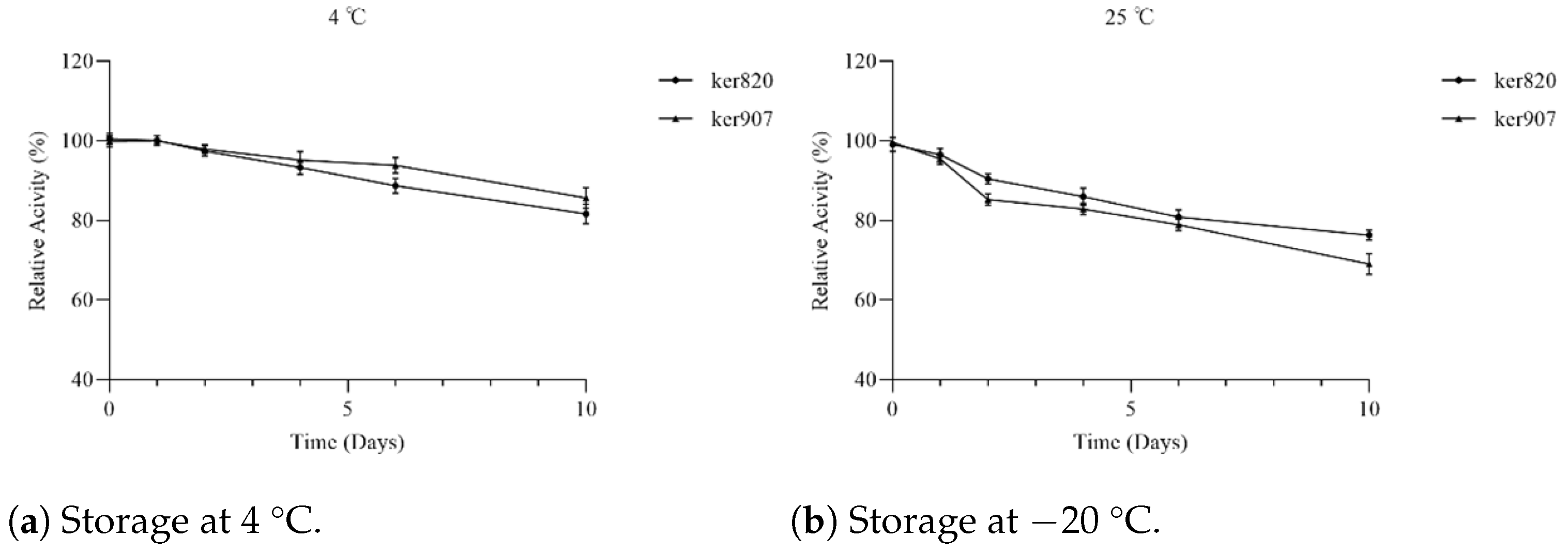
2.3.5. Storage Stability Assay
2.3.6. Enzyme Kinetics and Lineweaver–Burk Plot
2.4. In Vitro Degradation of Keratin Substrates
2.5. Amino Acid Analysis by Hplc
2.6. Statistical Processing of Results
3. Results
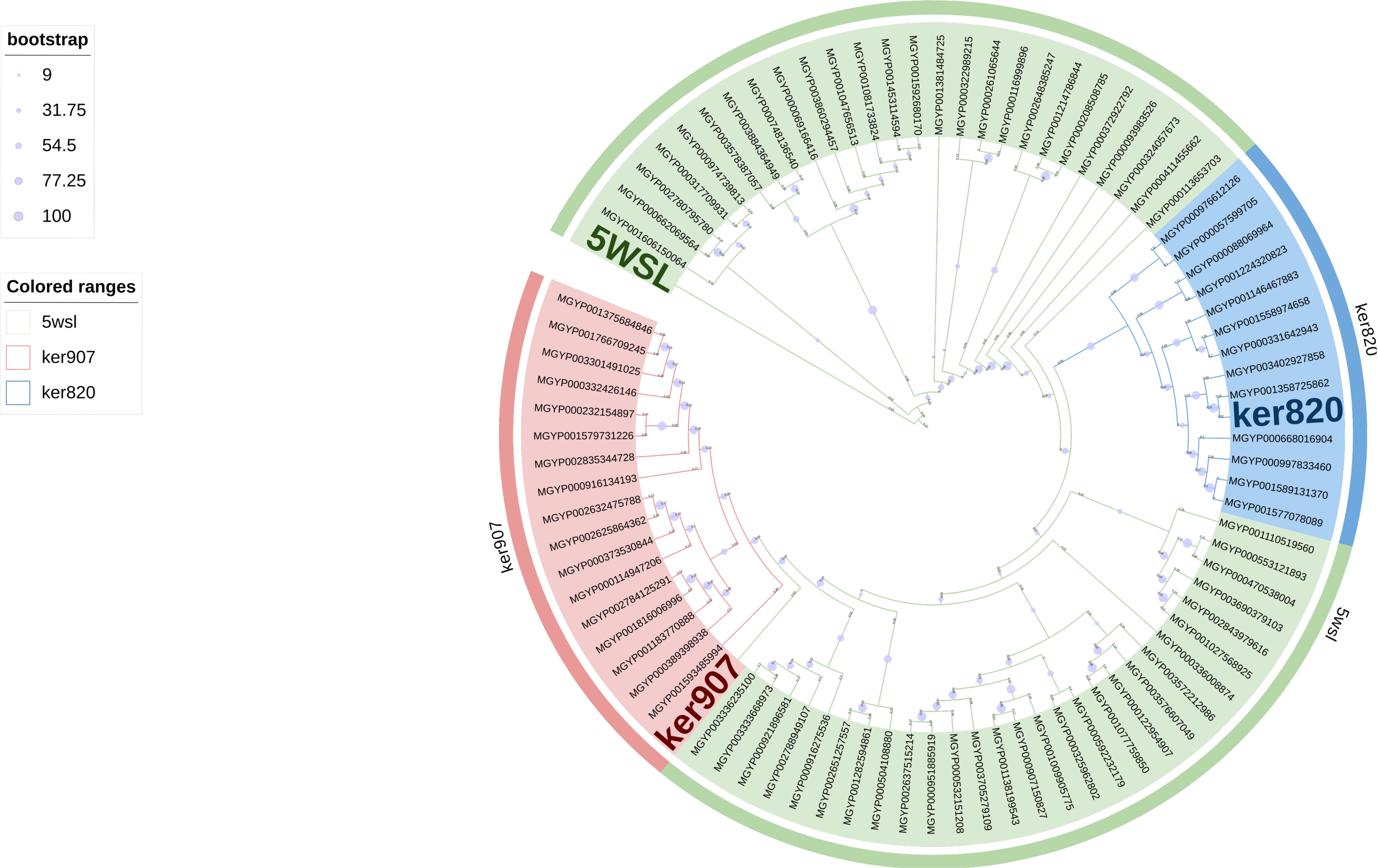
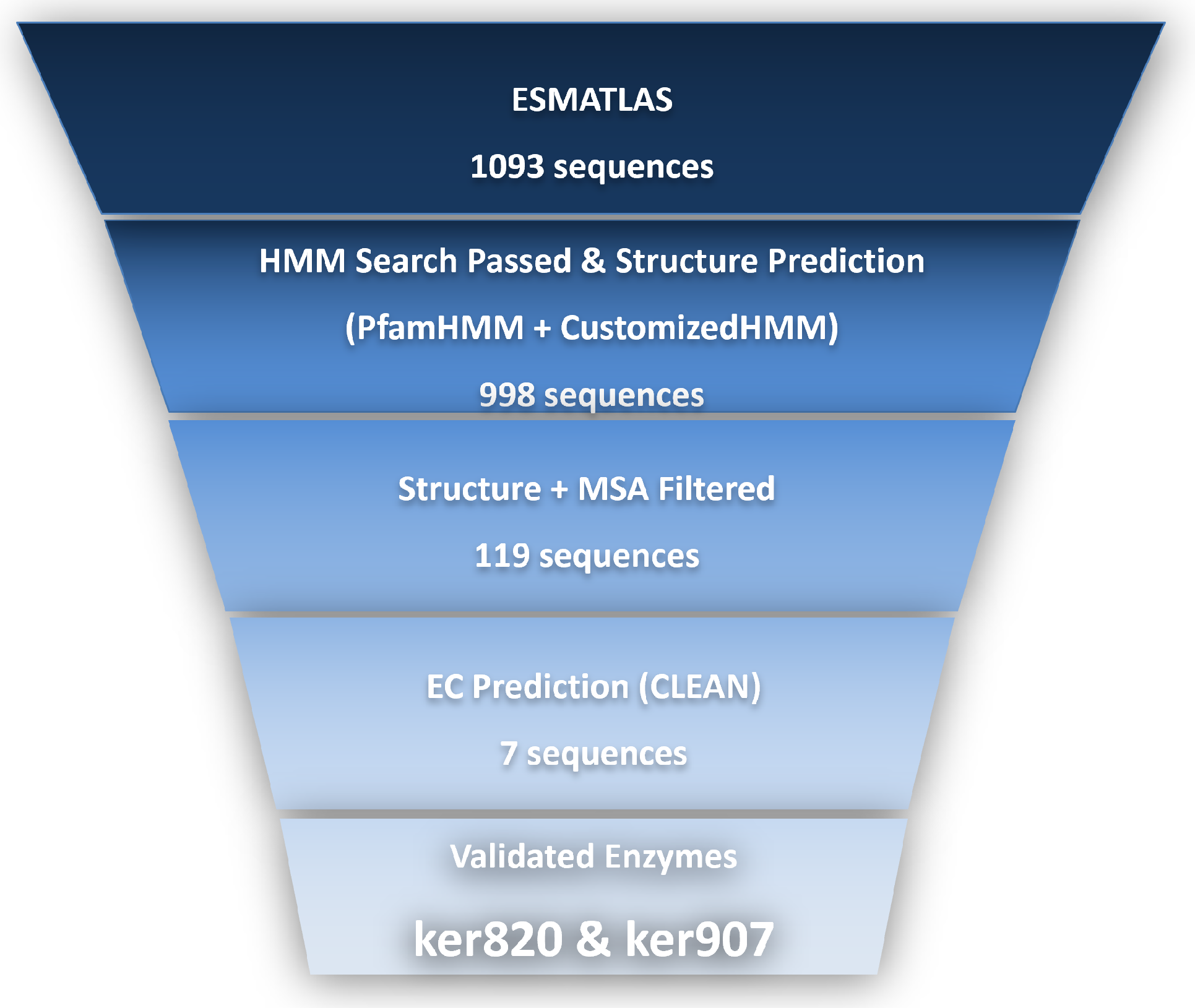
3.1. Identification of Novel Keratinase Genes
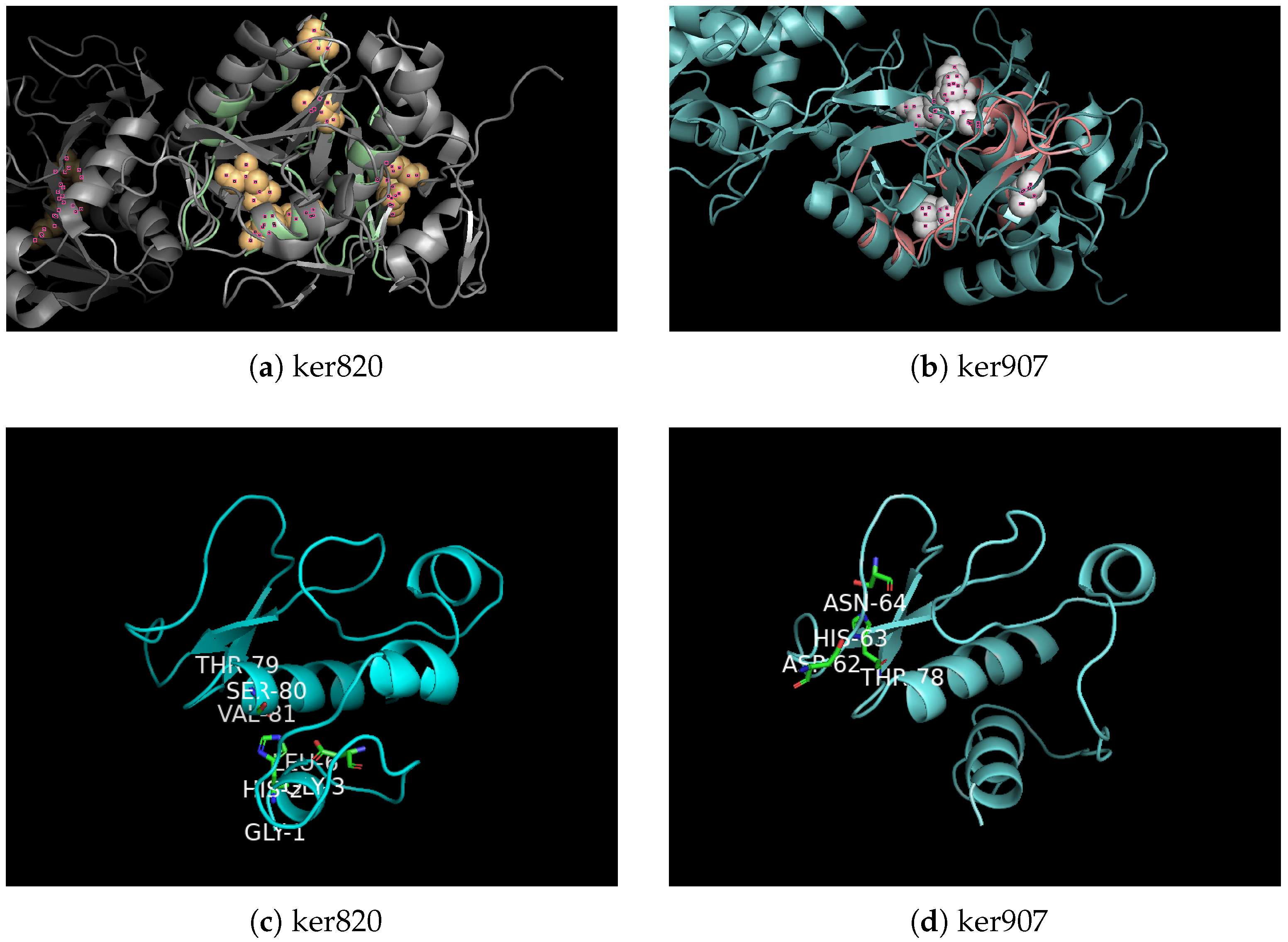
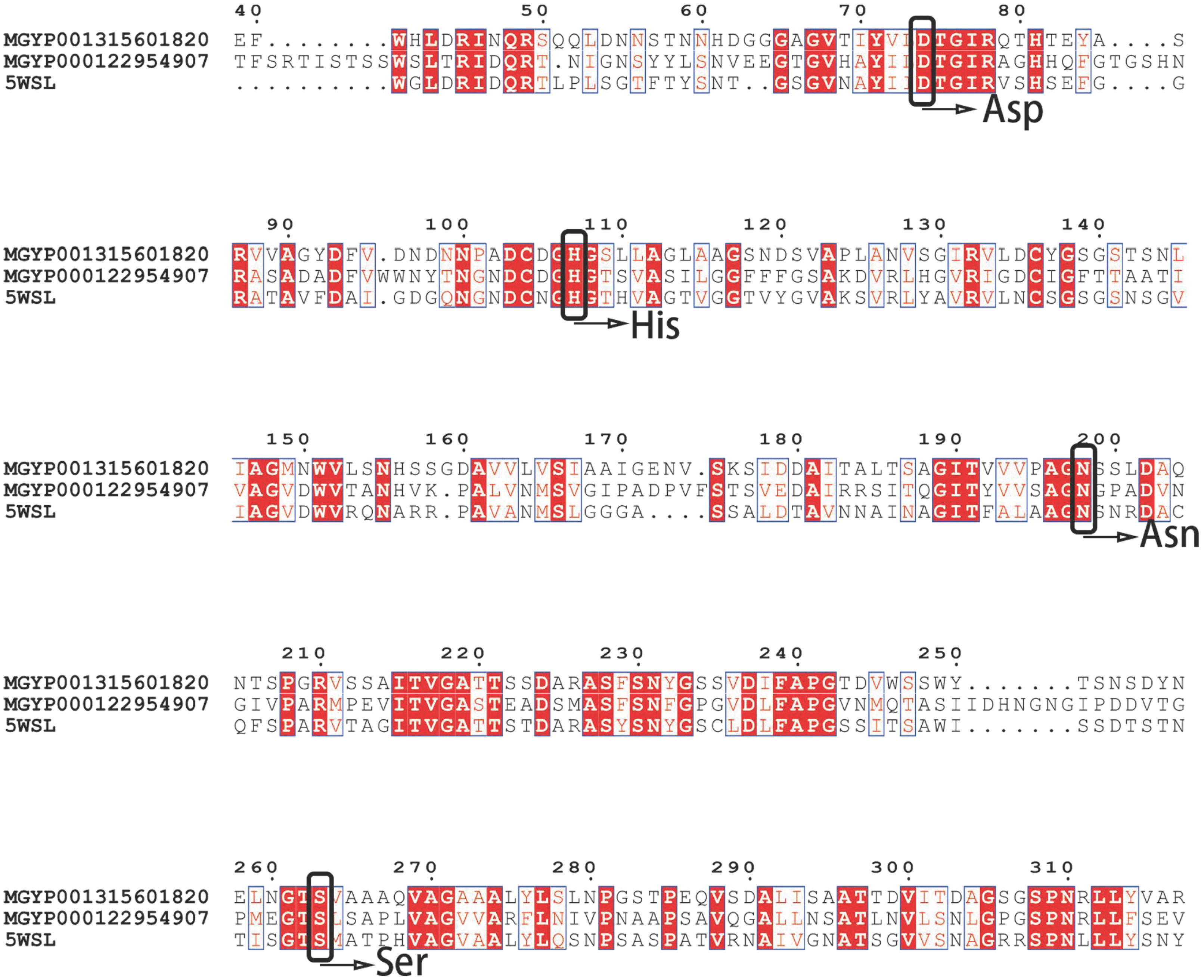
3.2. Sequence Analysis and Structural Prediction
3.3. Cloning and Expression of Keratinase Genes
3.4. Biochemical and Kinetic Properties of New Keratinases
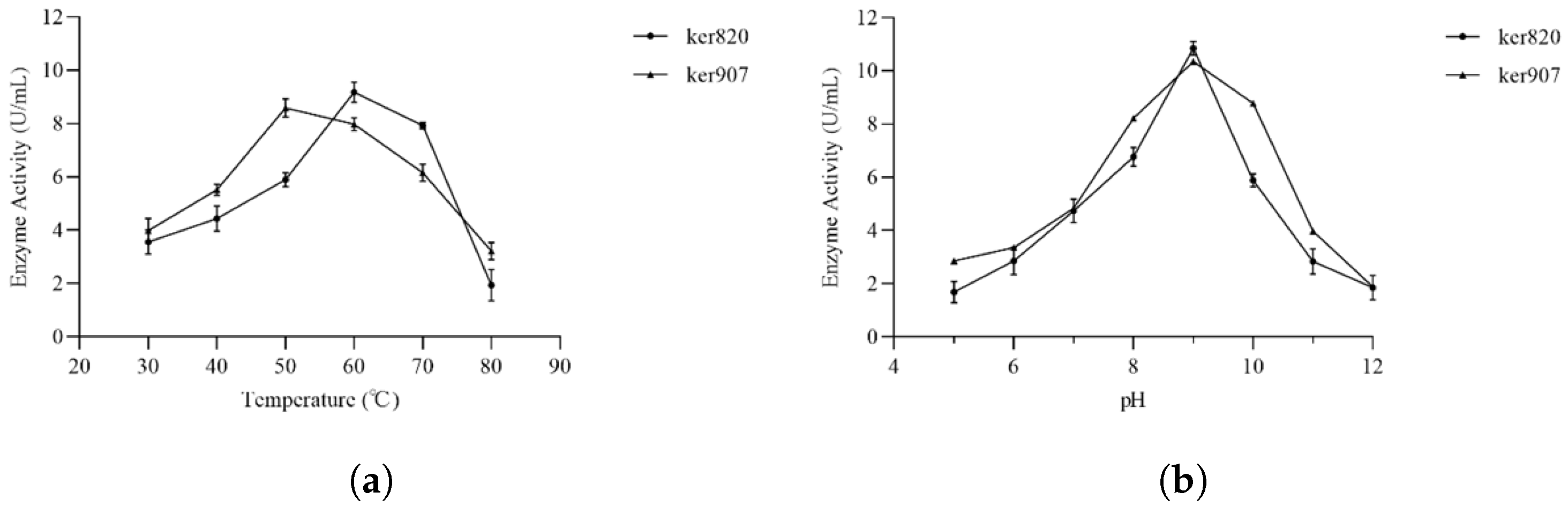
3.4.1. Optimal pH and Temperature
3.4.2. Kinetic Parameters
3.5. Substrate Specificity
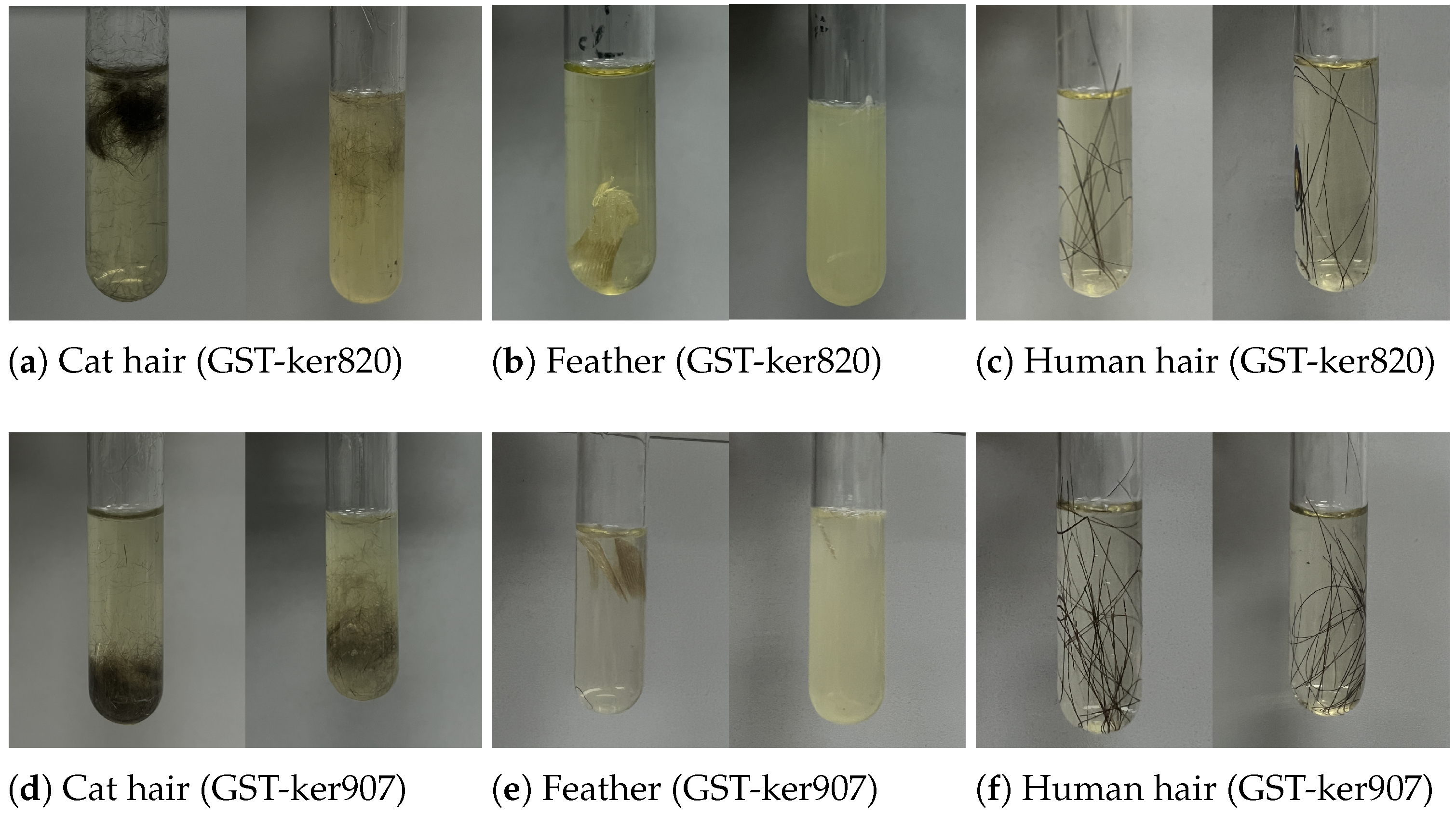
3.5.1. Substrate Degradation
3.5.2. Hplc Analysis of Hydrolysis Products
3.6. Stability of Recombinant Keratinases
Thermal Stability
3.7. Storage Stability
3.8. Effect of Metal Ions and Chemical Additives
3.9. Inhibition Studies
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BLAST | Basic Local Alignment Search Tool |
| HMM | Hidden Markov Model |
| EC | Enzyme Commission (number) |
| RMSD | Root Mean Square Deviation |
| MAFFT | Multiple Alignment using Fast Fourier Transform |
| MEGA X | Molecular Evolutionary Genetics Analysis X |
| CLEAN | Contrastive Learning–Enabled Enzyme Annotation |
Appendix A
Appendix A.1. Seuqneces of Novel Keratinases
Appendix A.2. Primers Designed for Seamless Cloning
- ker820-F: 5’-CGCGTGGATCCCCGGAATTCATGAACCACAAAGTACATCATCACCAT-3’
- ker820-R: 5’-TCACGATGCGGCCGCTCGAGCTCGAGTTATTCAGCGGT-3’
- ker907-F: 5’-CGCGTGGATCCCCGGAATTCATGAACCACAAA-3’
- ker907-R: 5’-TCACGATGCGGCCGCTCGAGCTCGAGTTATTT-3’
Appendix A.3. Phylogenetic Tree
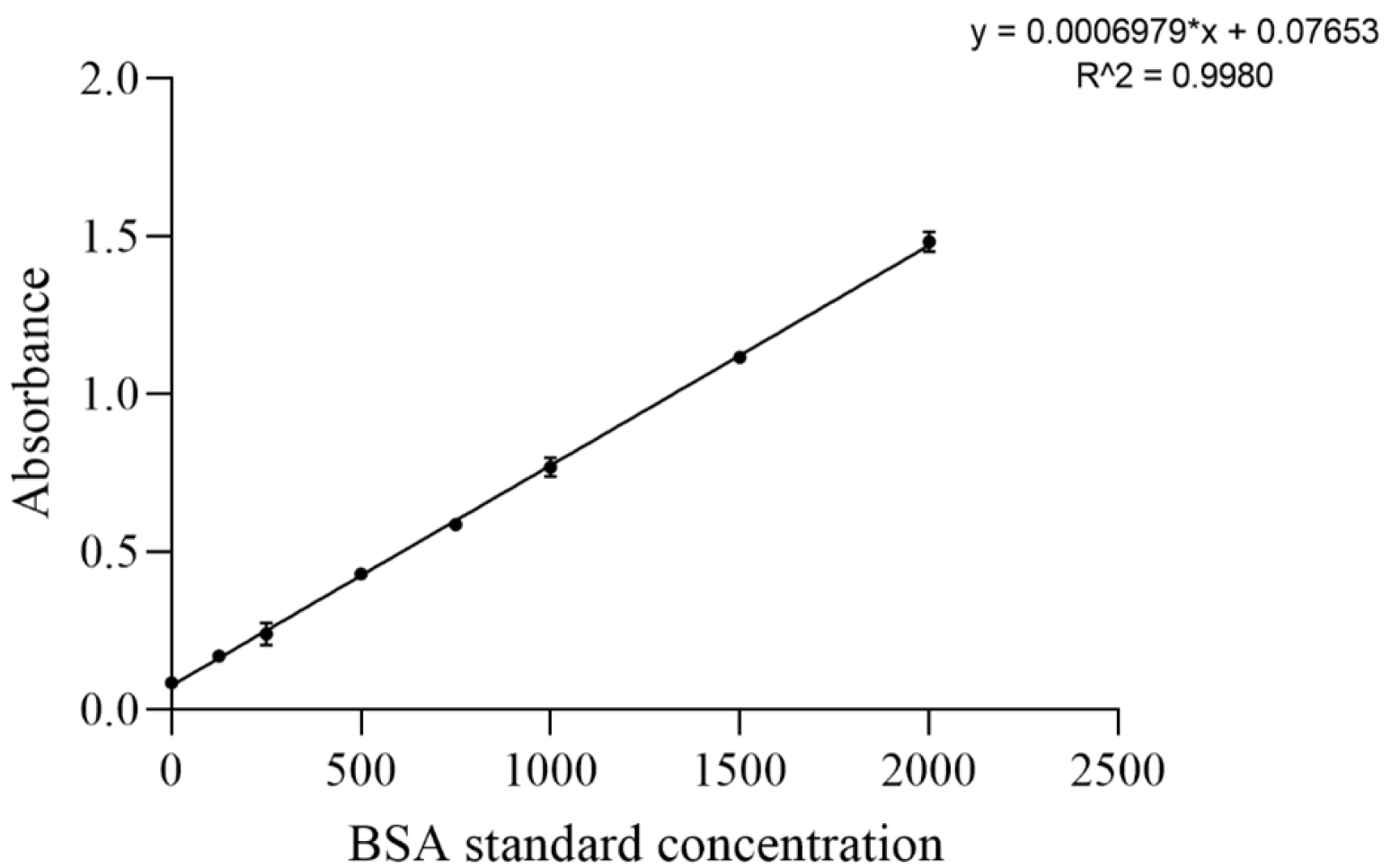
Appendix A.4. Standard Curves


References
- Brandelli, A. Bacterial keratinases: Useful enzymes for bioprocessing agroindustrial wastes and beyond. Food Bioprocess Technol. 2008, 1, 105–116. [Google Scholar] [CrossRef]
- Vidmar, B.; Vodovnik, M. Microbial keratinases: Enzymes with promising biotechnological applications. Food Technol. Biotechnol. 2018, 56, 312–328. [Google Scholar] [CrossRef]
- Lange, L.; Huang, Y.; Busk, P.K. Microbial decomposition of keratin in nature-a new hypothesis of industrial relevance. Appl. Microbiol. Biotechnol. 2016, 100, 2083–2096. [Google Scholar] [CrossRef]
- Bealer, E.J.; Onissema-Karimu, S.; Rivera-Galletti, A.; Francis, M.; Wilkowski, J.; Salas-de la Cruz, D.; Hu, X. Protein–Polysaccharide Composite Materials: Fabrication and Applications. Polymers 2020, 12, 464. [Google Scholar] [CrossRef]
- Shavandi, A.; Silva, T.; Bekhit, A.; Bekhit, A. Keratin: Dissolution, extraction and biomedical application. Biomater. Sci. 2017, 5, 1699–1735. [Google Scholar] [CrossRef] [PubMed]
- Rajabinejad, H.; Buciscanu, I.I.; Maier, S. Practical ways of extracting keratin from keratinous wastes and by-products: A review. Environ. Eng. Manag. J. 2016, 15, 1131–1147. [Google Scholar] [CrossRef]
- Qiu, J.; Wilkens, C.; Barrett, K.; Meyer, A.S. Microbial enzymes catalyzing keratin degradation: Classification, structure, function. Biotechnol. Adv. 2020, 44, 107607. [Google Scholar] [CrossRef] [PubMed]
- Gurav, R.G.; Jadhav, J.P. A novel source of biofertilizer from feather biomass for banana cultivation. Environ. Sci. Pollut. Res. 2013, 20, 4532–4539. [Google Scholar] [CrossRef]
- De Menezes, C.L.A.; Santos, R.D.C.; Santos, M.V.; Boscolo, M.; Da Silva, R.; Gomes, E.; Da Silva, R.R. Industrial sustainability of microbial keratinases: Production and potential applications. World J. Microbiol. Biotechnol. 2021, 37, 86. [Google Scholar] [CrossRef]
- Gupta, R.; Sharma, R.; Beg, Q.K. Revisiting microbial keratinases: Next generation proteases for sustainable biotechnology. Crit. Rev. Biotechnol. 2013, 33, 216–228. [Google Scholar] [CrossRef]
- Nnolim, N.E.; Nwodo, U.U. Microbial keratinase and the bio-economy: A three-decade meta-analysis of research exploit. AMB Express 2021, 11, 12. [Google Scholar] [CrossRef]
- Sanghvi, G.; Patel, H.; Vaishnav, D.; Oza, T.; Dave, G.; Kunjadia, P.; Sheth, N. A novel alkaline keratinase from Bacillus subtilis DP1 with potential utility in cosmetic formulation. Int. J. Biol. Macromol. 2016, 87, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Mohorcic, M.; Torkar, A.; Friedrich, J.; Kristl, J.; Murdan, S. An investigation into keratinolytic enzymes to enhance ungual drug delivery. Int. J. Pharm. 2007, 332, 196–201. [Google Scholar] [CrossRef]
- Cheng, Y.C.; Li, T.S.; Su, H.L.; Lee, P.C.; Wang, H.M.D. Transdermal Delivery Systems of Natural Products Applied to Skin Therapy and Care. Molecules 2020, 25, 5051. [Google Scholar] [CrossRef]
- Zhou, L.; Xie, X.; Wu, T.; Chen, M.; Yao, Q.; Zhu, H.; Zou, W. Compound enzymatic hydrolysis of feather waste to improve the nutritional value. Biomass Convers. Biorefinery 2022, 12, 287–298. [Google Scholar] [CrossRef]
- Shanmugasundaram, O.L.; Syed Zameer Ahmed, K.; Sujatha, K.; Ponnmurugan, P.; Srivastava, A.; Ramesh, R.; Sukumar, R.; Elanithi, K. Fabrication and characterization of chicken feather keratin/polysaccharides blended polymer coated nonwoven dressing materials for wound healing applications. Mater. Sci. Eng. Mater. Biol. Appl. 2018, 92, 26–33. [Google Scholar] [CrossRef]
- Bohacz, J. Changes in mineral forms of nitrogen and sulfur and enzymatic activities during composting of lignocellulosic waste and chicken feathers. Environ. Sci. Pollut. Res. Int. 2019, 26, 10333–10342. [Google Scholar] [CrossRef]
- Tamreihao, K.; Mukherjee, S.; Khunjamayum, R.; Devi, L.J.; Asem, R.S.; Ningthoujam, D.S. Feather degradation by keratinolytic bacteria and biofertilizing potential for sustainable agricultural production. J. Basic Microbiol. 2019, 59, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Latafat, N.; Siddiqui, M.H.; Vimal, A.; Bhargava, P. Biological Degradation of Keratin by Microbial Keratinase for Effective Waste Management and Potent Industrial Applications. Curr. Protein Pept. Sci. 2021, 22, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Nurdiawati, A.; Nakhshiniev, B.; Gonzales, H.B.; Yoshikawa, K. Nitrogen mineralization dynamics of liquid feather hydrolysates obtained by hydrothermal treatment. Appl. Soil Ecol. 2019, 134, 98–104. [Google Scholar] [CrossRef]
- Deniz, I.; Demir, T.; Oncel, S.S.; Hames, E.E.; Vardar-Sukan, F. Effect of Agitation and Aeration on Keratinase Production in Bioreactors Using Bioprocess Engineering Aspects. Protein J. 2021, 40, 388–395. [Google Scholar] [CrossRef]
- Gilbert, J. Metagenomics, Metadata, and Meta-analysis. In Encyclopedia of Metagenomics; Nelson, K.E., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–4. [Google Scholar] [CrossRef]
- Nazir, A. Review on metagenomics and its applications. Imp. J. Intersdiscip. Res. 2016, 2, 277–286. [Google Scholar]
- Ghosh, A.; Mehta, A.; Khan, A.M. Metagenomic Analysis and its Applications. In Encyclopedia of Bioinformatics and Computational Biology; Ranganathan, S., Gribskov, M., Nakai, K., Schönbach, C., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 184–193. [Google Scholar] [CrossRef]
- Streit, W.R.; Schmitz, R.A. Metagenomics–The key to the uncultured microbes. Curr. Opin. Microbiol. 2004, 7, 492–498. [Google Scholar] [CrossRef]
- Prayogo, F.A.; Budiharjo, A.; Kusumaningrum, H.P.; Wijanarka, W.; Suprihadi, A.; Nurhayati, N. Metagenomic applications in exploration and development of novel enzymes from nature: A review. J. Genet. Eng. Biotechnol. 2020, 18, 39. [Google Scholar] [CrossRef] [PubMed]
- Taewijit, S.; Waiyamai, K. CM-HMM: Inter-residue contact and HMM-profiles based enzyme subfamily prediction and structure analysis. In Proceedings of the 9th IEEE International Conference on Cognitive Informatics (ICCI’10), Beijing, China, 7–9 July 2010; pp. 863–868. [Google Scholar] [CrossRef]
- Li, S.C.; Bu, D.; Xu, J.; Li, M. Fragment-HMM: A new approach to protein structure prediction. Protein Sci. Publ. Protein Soc. 2008, 17, 1925–1934. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chaudhari, N. Protein secondary structure prediction with a hybrid RNN/HMM system. In Proceedings of the 2005 IEEE International Joint Conference on Neural Networks, Montreal, QC, Canada, 31 July–4 August 2005; Volume 1, pp. 538–541, ISSN 2161-4407. [Google Scholar] [CrossRef]
- Aydin, Z.; Altunbasak, Y.; Borodovsky, M. Protein secondary structure prediction with semi Markov HMMs. In Proceedings of the 2004 IEEE International Conference on Acoustics, Speech, and Signal Processing, Montreal, QC, Canada, 17–21 May 2004; pp. 2964–2967. [Google Scholar] [CrossRef]
- Asai, K.; Hayamizu, S.; Onizuka, K. HMM with protein structure grammar. In Proceedings of the Twenty-Sixth Hawaii International Conference on System Sciences, Wailea, HI, USA, 5–8 January 1993; IEEE: Piscataway, NJ, USA, 1993; pp. 783–791. [Google Scholar] [CrossRef]
- Asai, K.; Hayamizu, S.; Handa, K. Prediction of protein secondary structure by the hidden Markov model. Bioinformatics 1993, 9, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Lehti-Shiu, M.D.; Shiu, S.H. Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R. Soc. London Ser. Biol. Sci. 2012, 367, 2619–2639. [Google Scholar] [CrossRef]
- Bokros, N.; Popescu, S.C.; Popescu, G.V. Multispecies genome-wide analysis defines the MAP3K gene family in Gossypium hirsutum and reveals conserved family expansions. BMC Bioinform. 2019, 20, 99. [Google Scholar] [CrossRef]
- Bateman, A.; Birney, E.; Durbin, R.; Eddy, S.R.; Finn, R.D.; Sonnhammer, E.L. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 1999, 27, 260–262. [Google Scholar] [CrossRef]
- Hofmann, K. Sensitive protein comparisons with profiles and hidden Markov models. Briefings Bioinform. 2000, 1, 167–178. [Google Scholar] [CrossRef]
- Eddy, S.R. Multiple alignment using hidden Markov models. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995, 3, 114–120. [Google Scholar]
- Eddy, S.R. Hidden Markov models. Curr. Opin. Struct. Biol. 1996, 6, 361–365. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.Y.; Chen, Y.; Zhao, Y.H.; Wu, R. ORFeome-Based Search of Airway Epithelial Cell-Specific Novel Human β-Defensin Genes. Am. J. Respir. Cell Mol. Biol. 2003, 29, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Edlund, A.; Yang, Y.; McLean, J.S.; Yooseph, S. Metagenome and Metatranscriptome Analyses Using Protein Family Profiles. PLoS Comput. Biol. 2016, 12, e1004991. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Doerks, T.; Ponting, C.P.; Copley, R.R.; Bork, P. More than 1000 putative new human signalling proteins revealed by EST data mining. Nat. Genet. 2000, 25, 201–204. [Google Scholar] [CrossRef]
- Wittenberger, T.; Schaller, H.C.; Hellebrand, S. An expressed sequence tag (EST) data mining strategy succeeding in the discovery of new G-protein coupled receptors. J. Mol. Biol. 2001, 307, 799–813. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, Y.H.; Kalaslavadi, T.B.; Hamati, E.; Nehrke, K.; Le, A.D.; Ann, D.K.; Wu, R. Genome-wide search and identification of a novel gel-forming mucin MUC19/Muc19 in glandular tissues. Am. J. Respir. Cell Mol. Biol. 2004, 30, 155–165. [Google Scholar] [CrossRef]
- Robinson, S.L.; Piel, J.; Sunagawa, S. A roadmap for metagenomic enzyme discovery. Nat. Prod. Rep. 2021, 38, 1994–2023. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Kanokratana, P.; Eurwilaichitr, L.; Pootanakit, K.; Champreda, V. Identification of glycosyl hydrolases from a metagenomic library of microflora in sugarcane bagasse collection site and their cooperative action on cellulose degradation. J. Biosci. Bioeng. 2015, 119, 384–391. [Google Scholar] [CrossRef]
- Kanokratana, P.; Mhuantong, W.; Laothanachareon, T.; Tangphatsornruang, S.; Eurwilaichitr, L.; Pootanakit, K.; Champreda, V. Phylogenetic Analysis and Metabolic Potential of Microbial Communities in an Industrial Bagasse Collection Site. Microb. Ecol. 2013, 66, 322–334. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Mai, G.; Wang, Z.; Liu, Q.; Zhou, Y.; Ma, Y.; Liu, C. Metagenomic Insights Into a Cellulose-Rich Niche Reveal Microbial Cooperation in Cellulose Degradation. Front. Microbiol. 2019, 10, 618. [Google Scholar] [CrossRef] [PubMed]
- Biver, S.; Portetelle, D.; Vandenbol, M. Characterization of a new oxidant-stable serine protease isolated by functional metagenomics. SpringerPlus 2013, 2, 410. [Google Scholar] [CrossRef]
- Pessoa, T.B.A.; Rezende, R.P.; Marques, E.D.L.S.; Pirovani, C.P.; Dos Santos, T.F.; Dos Santos Gonçalves, A.C.; Romano, C.C.; Dotivo, N.C.; Freitas, A.C.O.; Salay, L.C.; et al. Metagenomic alkaline protease from mangrove sediment. J. Basic Microbiol. 2017, 57, 962–973. [Google Scholar] [CrossRef] [PubMed]
- Devi, S.G.; Fathima, A.A.; Sanitha, M.; Iyappan, S.; Curtis, W.R.; Ramya, M. Expression and characterization of alkaline protease from the metagenomic library of tannery activated sludge. J. Biosci. Bioeng. 2016, 122, 694–700. [Google Scholar] [CrossRef]
- López-López, O.; Cerdán, M.E.; González Siso, M.I. New Extremophilic Lipases and Esterases from Metagenomics. Curr. Protein Pept. Sci. 2014, 15, 445–455. [Google Scholar] [CrossRef]
- Hårdeman, F.; Sjöling, S. Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol. Ecol. 2007, 59, 524–534. [Google Scholar] [CrossRef]
- Schmeisser, C.; Steele, H.; Streit, W.R. Metagenomics, biotechnology with non-culturable microbes. Appl. Microbiol. Biotechnol. 2007, 75, 955–962. [Google Scholar] [CrossRef]
- Yeh, Y.F.; Chang, S.C.y.; Kuo, H.W.; Tong, C.G.; Yu, S.M.; Ho, T.H.D. A metagenomic approach for the identification and cloning of an endoglucanase from rice straw compost. Gene 2013, 519, 360–366. [Google Scholar] [CrossRef]
- Maruthamuthu, M.; Jiménez, D.J.; Stevens, P.; Van Elsas, J.D. A multi-substrate approach for functional metagenomics-based screening for (hemi)cellulases in two wheat straw-degrading microbial consortia unveils novel thermoalkaliphilic enzymes. BMC Genom. 2016, 17, 86. [Google Scholar] [CrossRef]
- Won, K.J.; Hamelryck, T.; Prügel-Bennett, A.; Krogh, A. An evolutionary method for learning HMM structure: Prediction of protein secondary structure. BMC Bioinform. 2007, 8, 357. [Google Scholar] [CrossRef]
- Söding, J. Protein homology detection by HMM–HMM comparison. Bioinformatics 2004, 21, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Jagadeesan, Y.; Meenakshisundaram, S.; Saravanan, V.; Balaiah, A. Sustainable production, biochemical and molecular characterization of thermo-and-solvent stable alkaline serine keratinase from novel Bacillus pumilus AR57 for promising poultry solid waste management. Int. J. Biol. Macromol. 2020, 163, 135–146. [Google Scholar] [CrossRef]
- Sharma, I.; Kango, N. Production and characterization of keratinase by Ochrobactrum intermedium for feather keratin utilization. Int. J. Biol. Macromol. 2021, 166, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Zaghloul, T.I.; Embaby, A.M.; Elmahdy, A.R. Biodegradation of chicken feathers waste directed by Bacillus subtilis recombinant cells: Scaling up in a laboratory scale fermentor. Bioresour. Technol. 2011, 102, 2387–2393. [Google Scholar] [CrossRef]
- Zhang, R.X.; Gong, J.S.; Su, C.; Qin, J.; Li, H.; Li, H.; Shi, J.S.; Xu, Z.H. Recombinant expression and molecular engineering of the keratinase from Brevibacillus parabrevis for dehairing performance. J. Biotechnol. 2020, 320, 57–65. [Google Scholar] [CrossRef]
- Rodrigues, C.J.C.; De Carvalho, C.C.C.R. Marine Bioprospecting, Biocatalysis and Process Development. Microorganisms 2022, 10, 1965. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Ramnani, P. Microbial keratinases and their prospective applications: An overview. Appl. Microbiol. Biotechnol. 2006, 70, 21–33. [Google Scholar] [CrossRef]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; Smetanin, N.; Verkuil, R.; Kabeli, O.; Shmueli, Y.; et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 2023, 379, 1123–1130. [Google Scholar] [CrossRef]
- Wu, W.L.; Chen, M.Y.; Tu, I.F.; Lin, Y.C.; EswarKumar, N.; Chen, M.Y.; Ho, M.C.; Wu, S.H. The discovery of novel heat-stable keratinases from Meiothermus taiwanensis WR-220 and other extremophiles. Sci. Rep. 2017, 7, 4658. [Google Scholar] [CrossRef] [PubMed]
- Jaouadi, B.; Abdelmalek, B.; Fodil, D.; Ferradji, F.Z.; Rekik, H.; Zaraî, N.; Bejar, S. Purification and characterization of a thermostable keratinolytic serine alkaline proteinase from Streptomyces sp. strain AB1 with high stability in organic solvents. Bioresour. Technol. 2010, 101, 8361–8369. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun. 2022, 13, 6968. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Schrödinger, L. The PyMOL Molecular Graphics System, Version 1.8; Schrodinger, LLC.: New York, NY, USA, 2015.
- Zhang, Y.; Skolnick, J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Yu, T.; Cui, H.; Li, J.C.; Luo, Y.; Jiang, G.; Zhao, H. Enzyme function prediction using contrastive learning. Science 2023, 379, 1358–1363. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Bjellqvist, B.; Basse, B.; Olsen, E.; Celis, J.E. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 1994, 15, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Bjellqvist, B.; Hughes, G.J.; Pasquali, C.; Paquet, N.; Ravier, F.; Sanchez, J.C.; Frutiger, S.; Hochstrasser, D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 1993, 14, 1023–1031. [Google Scholar] [CrossRef]
- Carter, P.; Wells, J.A. Dissecting the catalytic triad of a serine protease. Nature 1988, 332, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Coggill, P.; Bateman, A. The YARHG Domain: An Extracellular Domain in Search of a Function. PLoS ONE 2012, 7, e35575. [Google Scholar] [CrossRef][Green Version]
- Bryan, P.N. Protein engineering of subtilisin. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 2000, 1543, 203–222. [Google Scholar] [CrossRef]
- Robertus, J.D.; Kraut, J.; Alden, R.A.; Birktoft, J.J. Subtilisin. Stereochemical mechanism involving transition-state stabilization. Biochemistry 1972, 11, 4293–4303. [Google Scholar] [CrossRef]
- Bryan, P.; Pantoliano, M.W.; Quill, S.G.; Hsiao, H.Y.; Poulos, T. Site-directed mutagenesis and the role of the oxyanion hole in subtilisin. Proc. Natl. Acad. Sci. USA 1986, 83, 3743–3745. [Google Scholar] [CrossRef] [PubMed]
- Yaseen, A.; Nijim, M.; Williams, B.; Qian, L.; Li, M.; Wang, J.; Li, Y. FLEXc: Protein flexibility prediction using context-based statistics, predicted structural features, and sequence information. BMC Bioinform. 2016, 17, 281. [Google Scholar] [CrossRef]
- Berjanskii, M.V.; Wishart, D.S. Application of the random coil index to studying protein flexibility. J. Biomol. NMR 2008, 40, 31–48. [Google Scholar] [CrossRef]
- Deller, M.C.; Kong, L.; Rupp, B. Protein stability: A crystallographer’s perspective. Acta Crystallogr. Sect. F 2016, 72, 72–95. [Google Scholar] [CrossRef]
- Watson, M.D.; Monroe, J.; Raleigh, D.P. Size-Dependent Relationships between Protein Stability and Thermal Unfolding Temperature Have Important Implications for Analysis of Protein Energetics and High-Throughput Assays of Protein–Ligand Interactions. J. Phys. Chem. B 2018, 122, 5278–5285. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, T.; O’Donohoe, C.A.; Howe, N.; Malthouse, J.P.G. Importance of tetrahedral intermediate formation in the catalytic mechanism of the serine proteases chymotrypsin and subtilisin. Biochemistry 2012, 51, 6164–6170. [Google Scholar] [CrossRef]
- Brandelli, A.; Daroit, D.J.; Riffel, A. Biochemical features of microbial keratinases and their production and applications. Appl. Microbiol. Biotechnol. 2010, 85, 1735–1750. [Google Scholar] [CrossRef] [PubMed]
- Jana, A.; Kakkar, N.; Halder, S.K.; Das, A.J.; Bhaskar, T.; Ray, A.; Ghosh, D. Efficient valorization of feather waste by Bacillus cereus IIPK35 for concomitant production of antioxidant keratin hydrolysate and milk-clotting metallo-serine keratinase. J. Environ. Manag. 2022, 324, 116380. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.; Timorshina, S.; Osmolovskiy, A.; Misri, J.; Singh, R. Chicken Feather Waste Valorization Into Nutritive Protein Hydrolysate: Role of Novel Thermostable Keratinase From Bacillus pacificus RSA27. Front. Microbiol. 2022, 13, 882902. [Google Scholar] [CrossRef]
- Jaouadi, N.Z.; Rekik, H.; Badis, A.; Trabelsi, S.; Belhoul, M.; Yahiaoui, A.B.; Aicha, H.B.; Toumi, A.; Bejar, S.; Jaouadi, B. Biochemical and Molecular Characterization of a Serine Keratinase from Brevibacillus brevis US575 with Promising Keratin-Biodegradation and Hide-Dehairing Activities. PLoS ONE 2013, 8, e76722. [Google Scholar] [CrossRef]
- Pinto, É.S.M.; Dorn, M.; Feltes, B.C. The tale of a versatile enzyme: Alpha-amylase evolution, structure, and potential biotechnological applications for the bioremediation of n-alkanes. Chemosphere 2020, 250, 126202. [Google Scholar] [CrossRef]
- Friedrich, A.B.; Antranikian, G. Keratin Degradation by Fervidobacterium pennavorans, a Novel Thermophilic Anaerobic Species of the Order Thermotogales. Appl. Environ. Microbiol. 1996, 62, 2875–2882. [Google Scholar] [CrossRef]
- Jankiewicz, U.; Larkowska, E.; Swiontek Brzezinska, M. Production, characterization, gene cloning, and nematocidal activity of the extracellular protease from Stenotrophomonas maltophilia N4. J. Biosci. Bioeng. 2016, 121, 614–618. [Google Scholar] [CrossRef]
- Ebeling, W.; Hennrich, N.; Klockow, M.; Metz, H.; Orth, H.D.; Lang, H. Proteinase K from Tritirachium album Limber. Eur. J. Biochem. 1974, 47, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Busk, P.K.; Herbst, F.A.; Lange, L. Genome and secretome analyses provide insights into keratin decomposition by novel proteases from the non-pathogenic fungus Onygena corvina. Appl. Microbiol. Biotechnol. 2015, 99, 9635–9649. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, B.; Xu, M.; Jia, G.; Huang, Z.; Zhao, H.; Liu, G. Prokaryotic expression and characterization of a keratinolytic protease from Aspergillus niger. Biologia 2015, 70, 157–164. [Google Scholar] [CrossRef]
- Jarai, G.; Kirchherr, D.; Buxton, F.P. Cloning and characterization of the pepD gene of Aspergillus niger which codes for a subtilisin-like protease. Gene 1994, 139, 51–57. [Google Scholar] [CrossRef]
- Kang, E.; Jin, H.S.; La, J.W.; Sung, J.Y.; Park, S.Y.; Kim, W.C.; Lee, D.W. Identification of keratinases from Fervidobacterium islandicum AW-1 using dynamic gene expression profiling. Microb. Biotechnol. 2020, 13, 442–457. [Google Scholar] [CrossRef]
- Bohacz, J. Biodegradation of feather waste keratin by a keratinolytic soil fungus of the genus Chrysosporium and statistical optimization of feather mass loss. World J. Microbiol. Biotechnol. 2017, 33, 13. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, S.; Zhang, Q.; Gan, L.; Jiang, G.; Tian, Y.; Shi, B. Characterization and application of a novel halotolerant protease with no collagenase activity for cleaner dehairing of goatskin. Process. Biochem. 2022, 113, 203–215. [Google Scholar] [CrossRef]
- Dauparas, J.; Anishchenko, I.; Bennett, N.; Bai, H.; Ragotte, R.J.; Milles, L.F.; Wicky, B.I.M.; Courbet, A.; de Haas, R.J.; Bethel, N.; et al. Robust deep learning based protein sequence design using ProteinMPNN. bioRxiv 2022. [Google Scholar] [CrossRef] [PubMed]
- Hon, J.; Marusiak, M.; Martinek, T.; Kunka, A.; Zendulka, J.; Bednar, D.; Damborsky, J. SoluProt: Prediction of soluble protein expression in Escherichia coli. Bioinformatics 2021, 37, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Khurana, S.; Rawi, R.; Kunji, K.; Chuang, G.Y.; Bensmail, H.; Mall, R. DeepSol: A deep learning framework for sequence-based protein solubility prediction. Bioinformatics 2018, 34, 2605–2613. [Google Scholar] [CrossRef]
- Ghafoor, H.; Asim, M.N.; Ibrahim, M.A.; Dengel, A. ProSol-multi: Protein solubility prediction via amino acids multi-level correlation and discriminative distribution. Heliyon 2024, 10, e36041. [Google Scholar] [CrossRef]
| Gene | Gene Length (bp) | UniProt ID | Identity (%) | Similarity (%) | E-Value |
|---|---|---|---|---|---|
| ker820 | 445 | A0A2H4A2Y5 | 62 | 45.5 | |
| ker907 | 473 | A0A2H4A2Y5 | 62 | 43.2 |
| Keratinase | Isoelectric Point (pI) | Molecular Weight (kDa) | Predicted E.C. Number (s) | Confidence Level |
|---|---|---|---|---|
| ker820 | 8.92 | 45.0 | 3.4.21.62 | High |
| 3.4.24.12 | ||||
| ker907 | 7.86 | 50.1 | 3.4.21.62 | Medium |
| Keratinase | -Helix | Extended Strand | -Turn | Random Coil |
|---|---|---|---|---|
| ker820 | 26.01% | 24.94% | 5.62% | 43.43% |
| ker907 | 24.95% | 22.20% | 6.55% | 46.30% |
| Enzyme | Unpurified | Purified | Recovery |
|---|---|---|---|
| pET-28a(+)-ker820 | 1.384 U/mL | 15.868 U/mL | 91.64% |
| pET-28a(+)-ker907 | 1.365 U/mL | 12.737 U/mL | 81.32% |
| pGEX-4T-1-ker820 | 1.772 U/mL | 15.213 U/mL | 73.91% |
| pGEX-4T-1-ker907 | 1.068 U/mL | 11.684 U/mL | 89.31% |
| Enzyme | Substrate | (mg/mL) | (U/mg) |
|---|---|---|---|
| GST-ker820 | Casein | 9.81 | 120.99 |
| Feather | 40.94 | 44.40 | |
| GST-ker907 | Casein | 5.25 | 89.52 |
| Feather | 21.04 | 19.89 |
| Amino Acid | After Hydrolysis (mg/mL) | Before Hydrolysis (mg/mL) |
|---|---|---|
| Serine | 0.0251 | 0.0011 |
| -Aminoadipic acid | 0.0024 | - |
| Glycine | 0.0232 | - |
| L-Alanine | 0.0464 | 0.0031 |
| Cystine | 0.0374 | - |
| Methionine | 0.0087 | - |
| Valine | 0.4027 | 0.0012 |
| Isoleucine | 0.0913 | 0.0060 |
| Leucine | 0.0618 | 0.0009 |
| Tyrosine | 0.3940 | 0.0023 |
| Phenylalanine | 0.0362 | 0.0017 |
| -Alanine | 0.0567 | 0.0013 |
| -Aminobutyric acid | 0.0045 | - |
| Histidine | 0.0319 | - |
| Proline | 0.0529 | - |
| Tryptophan | 0.0032 | - |
| 3-Methyl-L-histidine | 0.0082 | 0.0006 |
| Ornithine | 0.0033 | - |
| Lysine | 0.0212 | 0.0003 |
| Arginine | 0.1825 | 0.0030 |
| Amino Acid | After Hydrolysis (mg/mL) | Before Hydrolysis (mg/mL) |
|---|---|---|
| Aspartic acid | 0.0013 | 0.0008 |
| Threonine | 0.0482 | - |
| Glutamic acid | 0.0465 | - |
| Serine | 0.0059 | - |
| Glycine | 0.0201 | 0.0012 |
| L-Alanine | 0.0629 | 0.0027 |
| Cystine | 0.0224 | - |
| Isoleucine | 0.8450 | 0.0018 |
| Leucine | 0.0723 | 0.0006 |
| Tyrosine | 0.3218 | 0.0012 |
| Phenylalanine | 0.0295 | 0.0013 |
| Lysine | 0.0110 | 0.0003 |
| -Alanine | 0.0473 | 0.0023 |
| 3-Methyl-L-histidine | 0.0017 | 0.0006 |
| Ornithine | 0.0021 | - |
| Proline | 0.0451 | - |
| Arginine | 0.1063 | 0.0022 |
| Proline | 0.0536 | 0.0028 |
| Ions | Relative Activity of GST-ker820 (%) | Relative Activity of GST-ker907 (%) |
|---|---|---|
| None | 100.00 | 100.00 |
| Mn2+ | 161.25 ± 1.57 | 123.89 ± 1.25 |
| Co2+ | 31.45 ± 3.14 | 49.24 ± 2.05 |
| Ag+ | 26.17 ± 1.52 | 63.52 ± 2.86 |
| Cu2+ | 126.87 ± 2.31 | 101.45 ± 2.76 |
| Cr2+ | 7.58 ± 2.25 | 14.12 ± 1.34 |
| K+ | 141.26 ± 2.06 | 98.17 ± 2.38 |
| Ca2+ | 87.72 ± 2.71 | 148.70 ± 2.56 |
| Mg2+ | 71.34 ± 1.54 | 64.27 ± 2.01 |
| Li+ | 7.12 ± 3.02 | 45.13 ± 1.34 |
| Zn2+ | 63.17 ± 0.91 | 121.81 ± 0.95 |
| Ni2+ | 97.24 ± 1.75 | 104.12 ± 1.57 |
| Fe3+ | 117.45 ± 1.92 | 130.68 ± 3.21 |
| EDTA | 71.49 ± 2.13 | 87.01 ± 1.38 |
| EGTA | 75.81 ± 1.92 | 79.52 ± 1.41 |
| Denaturants | Relative Activity of GST-ker820 (%) | Relative Activity of GST-ker907 (%) |
|---|---|---|
| DTT | 107.26 ± 1.94 | 102.87 ± 0.76 |
| GuHCl | 103.89 ± 2.48 | 110.75 ± 1.84 |
| Urea | 86.25 ± 2.05 | 77.87 ± 0.97 |
| SDS | 104.75 ± 2.16 | 102.28 ± 2.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Xu, G.; Yi, Z.; Tang, X. Efficient Mining and Characterization of Two Novel Keratinases from Metagenomic Database. Biomolecules 2025, 15, 1527. https://doi.org/10.3390/biom15111527
Zhang J, Xu G, Yi Z, Tang X. Efficient Mining and Characterization of Two Novel Keratinases from Metagenomic Database. Biomolecules. 2025; 15(11):1527. https://doi.org/10.3390/biom15111527
Chicago/Turabian StyleZhang, Jue, Guangxin Xu, Zhiwei Yi, and Xixiang Tang. 2025. "Efficient Mining and Characterization of Two Novel Keratinases from Metagenomic Database" Biomolecules 15, no. 11: 1527. https://doi.org/10.3390/biom15111527
APA StyleZhang, J., Xu, G., Yi, Z., & Tang, X. (2025). Efficient Mining and Characterization of Two Novel Keratinases from Metagenomic Database. Biomolecules, 15(11), 1527. https://doi.org/10.3390/biom15111527







