Long Non-Coding RNAs Contribute to Glucose Starvation-Induced Dedifferentiation in Lung Adenocarcinoma
Abstract
1. Introduction
2. Materials and Methods
- Cell lines.
- In Vitro Studies.
- Small interfering RNA transfection.
- Total protein extraction.
- Western blotting.
- RNA extraction.
- RT-qPCR.
- RNA-seq, data analysis, and material availability.
- ChIP-qPCR.
- meRIP (m6A-RIP).
- Quantification and statistical analysis.
3. Results
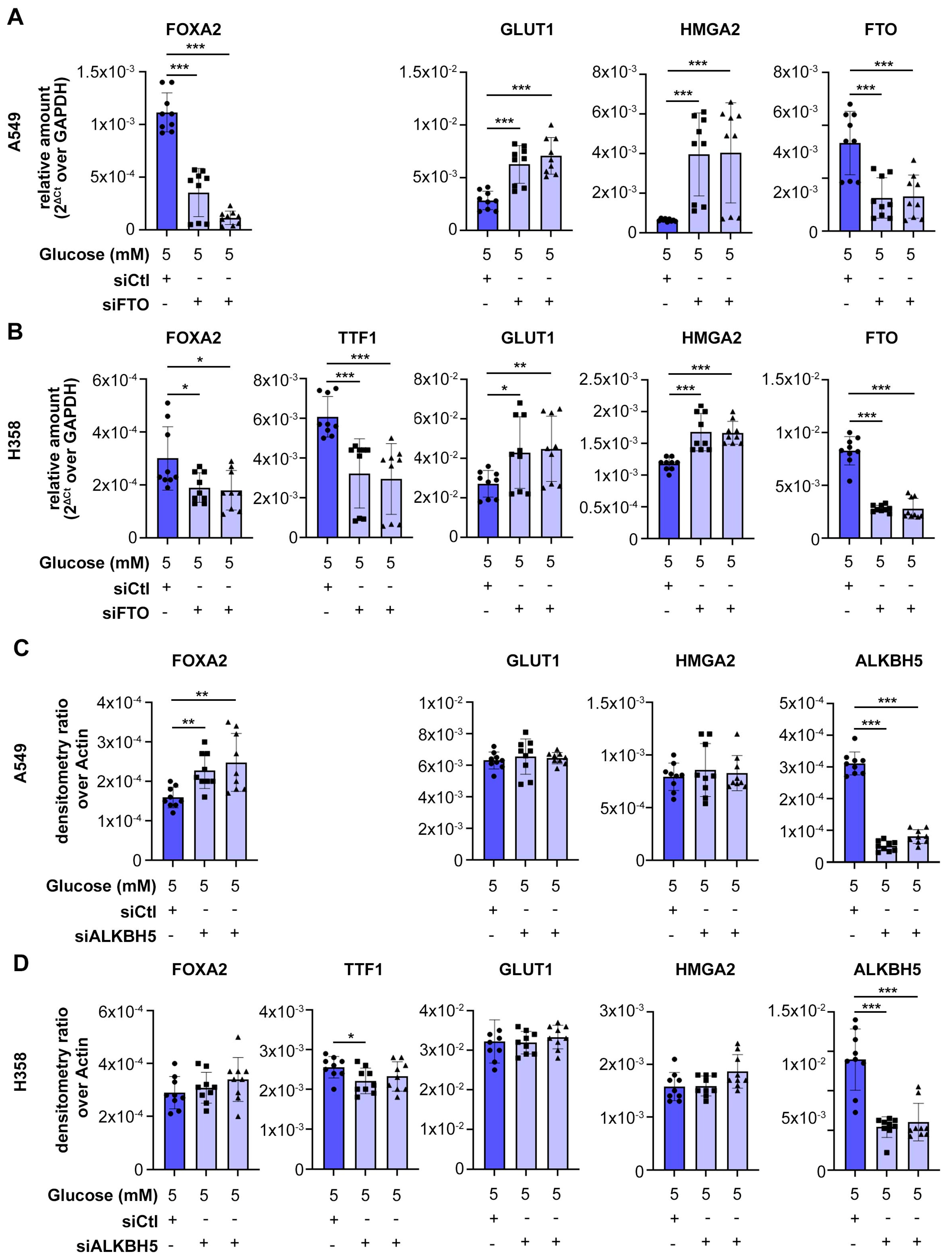
3.1. FTO Is Required to Maintain Cell Differentiation in Lung Cancer Cells
3.2. Long Non-Coding RNAs Are Upregulated by Glucose Deprivation
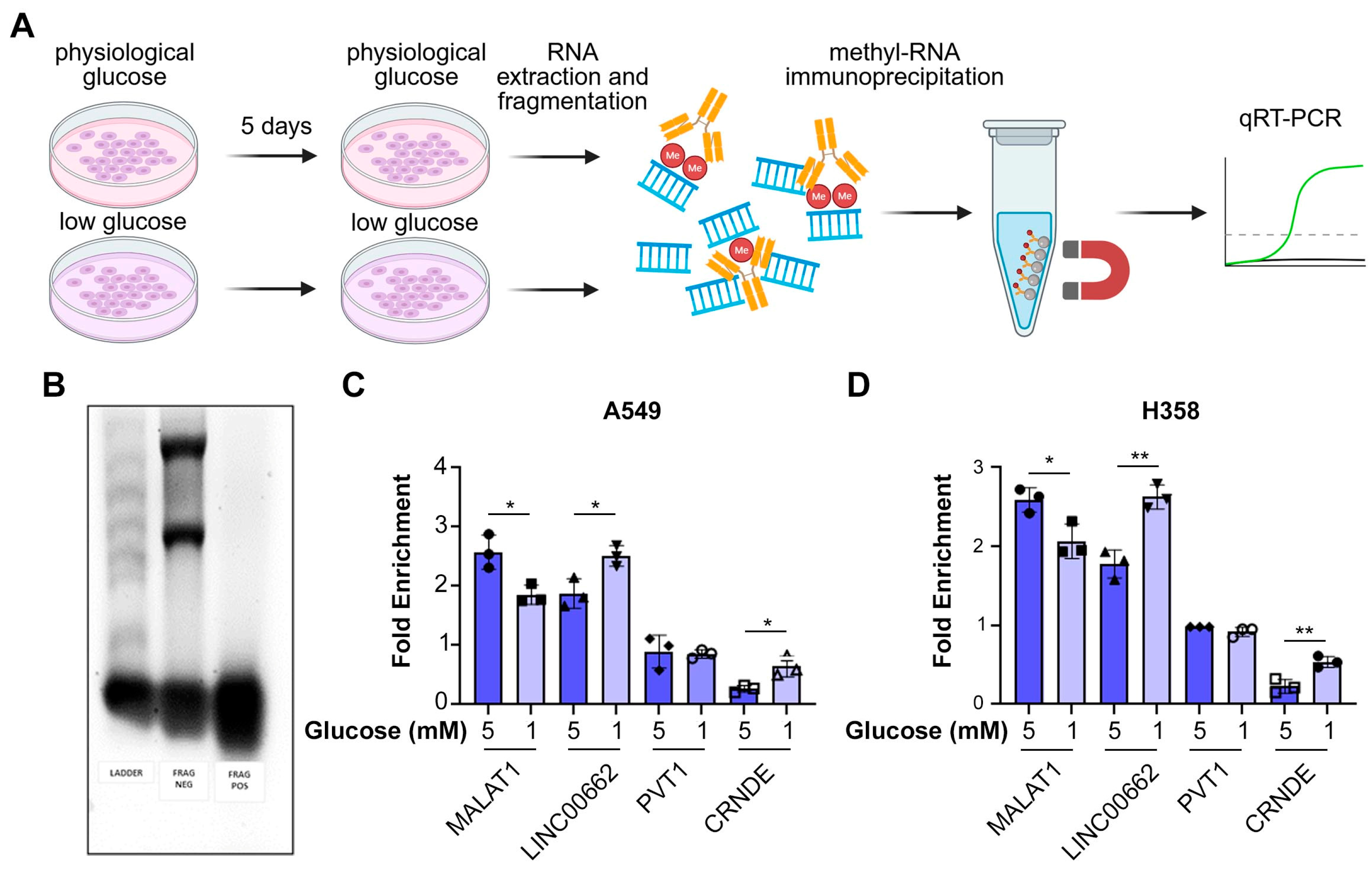
3.3. Certain lncRNAs Are Hypermethylated in Low Glucose
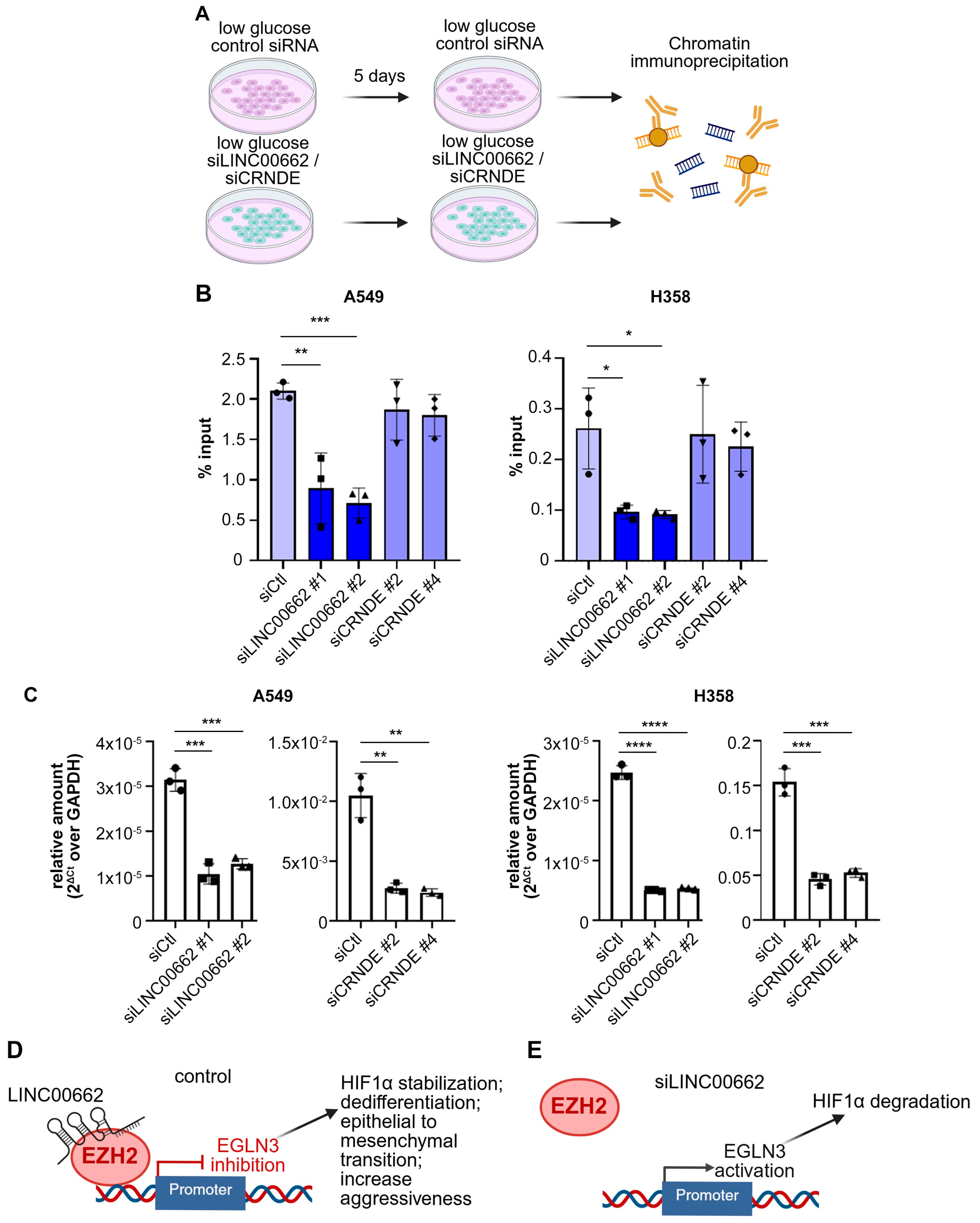
3.4. lncRNA LINC00662 Promotes EZH2 Recruitment on Target Gene Promoter in Low Glucose
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LUAD | Lung adenocarcinoma |
| α-KG | alpha-ketoglutarate |
| TCA | Tricarboxylic Acid Cycle |
| SDS-PAGE | Sodium dodecyl-sulfate Polyacrylamide Gel Electrophoresis |
| ChIP | Chromatin immunoprecipitation |
| MeRIP | Methyl-RNA immunoprecipitation |
| lncRNA | Long non-coding RNA |
References
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Noguchi, M.; Nicholson, A.G.; Geisinger, K.R.; Yatabe, Y.; Beer, D.G.; Powell, C.A.; Riely, G.J.; Van Schil, P.E.; et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 2011, 6, 244–285. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, B.; Min, Q.; Yang, X.; Yan, S.; Ma, Y.; Li, S.; Fan, J.; Wang, Y.; Dong, B.; et al. Spatial transcriptomics delineates molecular features and cellular plasticity in lung adenocarcinoma progression. Cell Discov. 2023, 9, 96. [Google Scholar] [CrossRef]
- Saggese, P.; Pandey, A.; Alcaraz, M.; Fung, E.; Hall, A.; Yanagawa, J.; Rodriguez, E.F.; Grogan, T.R.; Giurato, G.; Nassa, G.; et al. Glucose deprivation promotes pseudo-hypoxia and de-differentiation in lung adenocarcinoma. Cancer Res. 2024, 83, 305–327. [Google Scholar] [CrossRef]
- Pan, M.; Reid, M.A.; Lowman, X.H.; Kulkarni, R.P.; Tran, T.Q.; Liu, X.; Yang, Y.; Hernandez-Davies, J.E.; Rosales, K.K.; Li, H.; et al. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat. Cell Biol. 2016, 18, 1090–1101. [Google Scholar] [CrossRef]
- Tran, T.Q.; Hanse, E.A.; Habowski, A.N.; Li, H.; Gabra, M.B.I.; Yang, Y.; Lowman, X.H.; Ooi, A.M.; Liao, S.Y.; Edwards, R.A.; et al. alpha-Ketoglutarate attenuates Wnt signaling and drives differentiation in colorectal cancer. Nat. Cancer 2020, 1, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.P.; Yashinskie, J.J.; Koche, R.; Chandwani, R.; Tian, S.; Chen, C.C.; Baslan, T.; Marinkovic, Z.S.; Sanchez-Rivera, F.J.; Leach, S.D.; et al. alpha-Ketoglutarate links p53 to cell fate during tumour suppression. Nature 2019, 573, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.S.; Wang, H.; Li, X.; Chao, T.; Teav, T.; Christen, S.; Di Conza, G.; Cheng, W.C.; Chou, C.H.; Vavakova, M.; et al. alpha-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 2017, 18, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Carey, B.W.; Finley, L.W.; Cross, J.R.; Allis, C.D.; Thompson, C.B. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 2015, 518, 413–416. [Google Scholar] [CrossRef]
- Liu, Z.; Gao, L.; Cheng, L.; Lv, G.; Sun, B.; Wang, G.; Tang, Q. The roles of N6-methyladenosine and its target regulatory noncoding RNAs in tumors: Classification, mechanisms, and potential therapeutic implications. Exp. Mol. Med. 2023, 55, 487–501. [Google Scholar] [CrossRef]
- Huang, B.; Zhang, X.; Chen, J.; Wang, H. ALKBH5: A double-edged sword in cancer ferroptosis regulation: A review. Int. J. Biol. Macromol. 2025, 330, 147999. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.Y.; Du, Y.F.; Yan, H.C.; Fan, X.M.; Tian, F.Q.; Xu, P. The Clinical Relevance of FTO as a Demethylase Beyond Cancer: Molecular Mechanisms and Therapeutic Opportunities. Aging Dis. 2025. [Google Scholar] [CrossRef]
- Zhao, S.; Song, P.; Zhou, G.; Zhang, D.; Hu, Y. METTL3 promotes the malignancy of non-small cell lung cancer by N6-methyladenosine modifying SFRP2. Cancer Gene Ther. 2023, 30, 1094–1104. [Google Scholar] [CrossRef]
- Li, Y.; Su, R.; Deng, X.; Chen, Y.; Chen, J. FTO in cancer: Functions, molecular mechanisms, and therapeutic implications. Trends Cancer 2022, 8, 598–614. [Google Scholar] [CrossRef]
- Qu, J.; Yan, H.; Hou, Y.; Cao, W.; Liu, Y.; Zhang, E.; He, J.; Cai, Z. RNA demethylase ALKBH5 in cancer: From mechanisms to therapeutic potential. J. Hematol. Oncol. 2022, 15, 8. [Google Scholar] [CrossRef]
- Xiao, M.; Yang, H.; Xu, W.; Ma, S.; Lin, H.; Zhu, H.; Liu, L.; Liu, Y.; Yang, C.; Xu, Y.; et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012, 26, 1326–1338. [Google Scholar] [CrossRef]
- Loenarz, C.; Schofield, C.J. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 2008, 4, 152–156. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Xu, W.; Yang, H.; Liu, Y.; Yang, Y.; Wang, P.; Kim, S.H.; Ito, S.; Yang, C.; Xiao, M.T.; Liu, L.X.; et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 2011, 19, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Orstad, G.; Fort, G.; Parnell, T.J.; Jones, A.; Stubben, C.; Lohman, B.; Gillis, K.L.; Orellana, W.; Tariq, R.; Klingbeil, O.; et al. FoxA1 and FoxA2 control growth and cellular identity in NKX2-1-positive lung adenocarcinoma. Dev. Cell 2022, 57, 1866–1882.e10. [Google Scholar] [CrossRef]
- Li, C.M.; Gocheva, V.; Oudin, M.J.; Bhutkar, A.; Wang, S.Y.; Date, S.R.; Ng, S.R.; Whittaker, C.A.; Bronson, R.T.; Snyder, E.L.; et al. Foxa2 and Cdx2 cooperate with Nkx2-1 to inhibit lung adenocarcinoma metastasis. Genes Dev. 2015, 29, 1850–1862. [Google Scholar] [CrossRef]
- Perelman, M.G.; Brzezinski, R.Y.; Waissengrin, B.; Leshem, Y.; Bainhoren, O.; Rubinstein, T.A.; Perelman, M.; Rozenbaum, Z.; Havakuk, O.; Topilsky, Y.; et al. Sodium-glucose co-transporter-2 inhibitors in patients treated with immune checkpoint inhibitors. Cardiooncology 2024, 10, 2. [Google Scholar] [CrossRef]
- Gullino, P.M.; Clark, S.H.; Grantham, F.H. The Interstitial Fluid of Solid Tumors. Cancer Res. 1964, 24, 780–794. [Google Scholar] [PubMed]
- Ho, P.C.; Bihuniak, J.D.; Macintyre, A.N.; Staron, M.; Liu, X.; Amezquita, R.; Tsui, Y.C.; Cui, G.; Micevic, G.; Perales, J.C.; et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell 2015, 162, 1217–1228. [Google Scholar] [CrossRef]
- Mirzaei, S.; Gholami, M.H.; Hushmandi, K.; Hashemi, F.; Zabolian, A.; Canadas, I.; Zarrabi, A.; Nabavi, N.; Aref, A.R.; Crea, F.; et al. The long and short non-coding RNAs modulating EZH2 signaling in cancer. J. Hematol. Oncol. 2022, 15, 18. [Google Scholar] [CrossRef]
- Hao, A.; Wang, Y.; Stovall, D.B.; Wang, Y.; Sui, G. Emerging Roles of LncRNAs in the EZH2-regulated Oncogenic Network. Int. J. Biol. Sci. 2021, 17, 3268–3280. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Xiao, Y.; Tang, J.; Wu, J.; Ma, J.; Tian, B.; Zhou, Y.; Wang, H.; Yang, D.; Liao, Q.J.; et al. Role of lncRNA and EZH2 Interaction/Regulatory Network in Lung Cancer. J. Cancer 2018, 9, 4156–4165. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Shi, J.; Zhang, Y.; Xie, A.; Yu, L.; Zhang, C.; Lei, J.; Xu, H.; Leng, Z.; Li, T.; et al. LncTarD: A manually-curated database of experimentally-supported functional lncRNA-target regulations in human diseases. Nucleic Acids Res. 2020, 48, D118–D126. [Google Scholar] [CrossRef]
- Yang, J.H.; Li, J.H.; Shao, P.; Zhou, H.; Chen, Y.Q.; Qu, L.H. starBase: A database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011, 39, D202–209. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.Y.; Zhang, S.R.; Wang, L.H.; Wu, W.D.; Zhao, H. LINC00511 promotes the progression of non-small cell lung cancer through downregulating LATS2 and KLF2 by binding to EZH2 and LSD1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8377–8390. [Google Scholar] [CrossRef]
- Wang, Q.; Mao, X.; Luo, F.; Wang, J. LINC00511 promotes gastric cancer progression by regulating SOX4 and epigenetically repressing PTEN to activate PI3K/AKT pathway. J. Cell. Mol. Med. 2021, 25, 9112–9127. [Google Scholar] [CrossRef]
- Lu, Y.; Yu, Y.; Liu, F.; Han, Y.; Xue, H.; Sun, X.; Jiang, Y.; Tian, Z. LINC00511-dependent inhibition of IL-24 contributes to the oncogenic role of HNF4alpha in colorectal cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G338–G350. [Google Scholar] [CrossRef]
- Li, J.; Gao, W.; Zhao, Z.; Li, Y.; Yang, L.; Wei, W.; Ren, F.; Li, Y.; Yu, Y.; Duan, W.; et al. Ginsenoside Rg1 Reduced Microglial Activation and Mitochondrial Dysfunction to Alleviate Depression-Like Behaviour Via the GAS5/EZH2/SOCS3/NRF2 Axis. Mol. Neurobiol. 2022, 59, 2855–2873. [Google Scholar] [CrossRef]
- Zhao, D.; Li, Y.; Yu, M. LncRNA GAS5 facilitates nasopharyngeal carcinoma progression through epigenetically silencing PTEN via EZH2. RSC Adv. 2019, 9, 31691–31698. [Google Scholar] [CrossRef]
- Wang, Y.; Xie, Y.; Li, L.; He, Y.; Zheng, D.; Yu, P.; Yu, L.; Tang, L.; Wang, Y.; Wang, Z. EZH2 RIP-seq Identifies Tissue-specific Long Non-coding RNAs. Curr. Gene Ther. 2018, 18, 275–285. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Wang, Y.; He, X. The m(6)A demethylase FTO promotes renal epithelial-mesenchymal transition by reducing the m(6)A modification of lncRNA GAS5. Cytokine 2022, 159, 156000. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Liu, L.; Wu, H.; Zheng, Y.; Zhan, H.; Li, L. LncRNA GAS5 regulated by FTO-mediated m6A demethylation promotes autophagic cell death in NSCLC by targeting UPF1/BRD4 axis. Mol. Cell Biochem. 2024, 479, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gao, C.; Yang, Y.; Li, G.; Dong, J.; Ai, Y.; Chen, N.; Li, W. Long Noncoding RNA CRNDE/PRC2 Participated in the Radiotherapy Resistance of Human Lung Adenocarcinoma Through Targeting p21 Expression. Oncol. Res. 2018, 26, 1245–1255. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Ding, Y.; Zhuang, Y.; Zhang, C.; Han, L.; Li, W.; Guo, R.; Zhang, E. Copy number amplification-activated long non-coding RNA LINC00662 epigenetically inhibits BIK by interacting with EZH2 to regulate tumorigenesis in non-small cell lung cancer. J. Cancer 2022, 13, 1640–1651. [Google Scholar] [CrossRef]
- Wan, L.; Sun, M.; Liu, G.J.; Wei, C.C.; Zhang, E.B.; Kong, R.; Xu, T.P.; Huang, M.D.; Wang, Z.X. Long Noncoding RNA PVT1 Promotes Non-Small Cell Lung Cancer Cell Proliferation through Epigenetically Regulating LATS2 Expression. Mol. Cancer Ther. 2016, 15, 1082–1094. [Google Scholar] [CrossRef]
- Gou, X.; Zhao, X.; Wang, Z. Long noncoding RNA PVT1 promotes hepatocellular carcinoma progression through regulating miR-214. Cancer Biomarkers 2017, 20, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Zha, X.; Li, M.; Xia, X.; Wang, S. Insights into the m(6)A demethylases FTO and ALKBH5: Structural, biological function, and inhibitor development. Cell Biosci. 2024, 14, 108. [Google Scholar] [CrossRef]
- Xu, X.; Qiu, S.; Zeng, B.; Huang, Y.; Wang, X.; Li, F.; Yang, Y.; Cao, L.; Zhang, X.; Wang, J.; et al. N(6)-methyladenosine demethyltransferase FTO mediated m(6)A modification of estrogen receptor alpha in non-small cell lung cancer tumorigenesis. Oncogene 2024, 43, 1288–1302. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Shao, F.; Guo, D.; Wang, W.; Wang, J.; Zhu, R.; Gao, Y.; He, J.; Lu, Z. WNT/beta-catenin-suppressed FTO expression increases m(6)A of c-Myc mRNA to promote tumor cell glycolysis and tumorigenesis. Cell Death Dis. 2021, 12, 462. [Google Scholar] [CrossRef]
- He, P.C.; He, C. m(6) A RNA methylation: From mechanisms to therapeutic potential. EMBO J. 2021, 40, e105977. [Google Scholar] [CrossRef]
- Sendinc, E.; Shi, Y. RNA m6A methylation across the transcriptome. Mol. Cell 2023, 83, 428–441. [Google Scholar] [CrossRef]
- Lv, X.; Lian, Y.; Liu, Z.; Xiao, J.; Zhang, D.; Yin, X. Exosomal long non-coding RNA LINC00662 promotes non-small cell lung cancer progression by miR-320d/E2F1 axis. Aging 2021, 13, 6010–6024. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Su, Y.; Liu, Y.; Sun, P.; Wang, X. Long non-coding RNA Linc00662 promotes cell invasion and contributes to cancer stem cell-like phenotypes in lung cancer cells. J. Biochem. 2018, 164, 461–469. [Google Scholar] [CrossRef]
- Kim, H.J.; Cantor, H.; Cosmopoulos, K. Overcoming Immune Checkpoint Blockade Resistance via EZH2 Inhibition. Trends Immunol. 2020, 41, 948–963. [Google Scholar] [CrossRef]

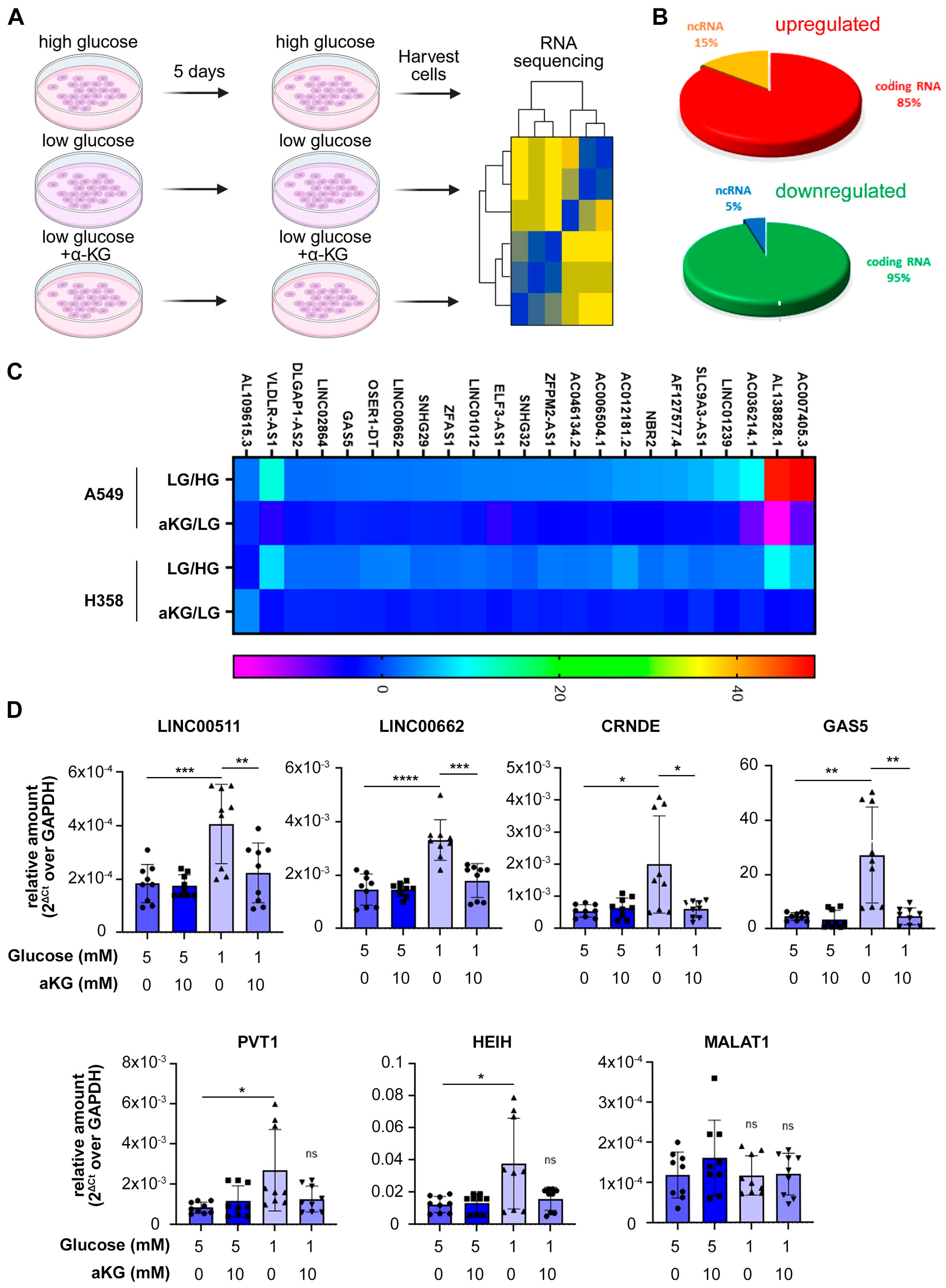
| GENE ID | A549 | H358 | ||||||
|---|---|---|---|---|---|---|---|---|
| LG/HG | α-KG/LG | LG/HG | α-KG/LG | |||||
| FC | padj | FC | padj | FC | padj | FC | padj | |
| AC007405.3 | 48.8 | 2.96 × 10−3 | −8.0 | 5.00 × 10−2 | 6.0 | 6.93 × 10−4 | −2.6 | 1.17 × 10−1 |
| AL138828.1 | 47.2 | 1.20 × 10−5 | −16.7 | 1.02 × 10−4 | 9.9 | 2.75 × 10−3 | −3.0 | 2.09 × 10−1 |
| AC036214.1 | 9.5 | 1.79 × 10−2 | −8.4 | 2.63 × 10−2 | 2.4 | 2.85 × 10−3 | −1.6 | 2.02 × 10−1 |
| LINC01239 | 7.2 | 2.32 × 10−3 | −2.2 | 1.48 × 10−1 | 2.3 | 2.65 × 10−2 | −2.6 | 1.28 × 10−2 |
| SLC9A3-AS1 | 5.6 | 6.21 × 10−10 | −2.4 | 1.88 × 10−3 | 1.9 | 4.35 × 10−2 | −1.2 | 7.94 × 10−1 |
| AF127577.4 | 4.8 | 4.15 × 10−6 | −2.6 | 6.43 × 10−3 | 2.7 | 6.39 × 10−3 | −2.5 | 2.04 × 10−2 |
| NBR2 | 4.5 | 1.20 × 10−16 | −3.5 | 3.82 × 10−12 | 2.0 | 3.47 × 10−4 | −1.8 | 4.75 × 10−3 |
| AC012181.2 | 4.4 | 1.27 × 10−2 | −3.8 | 2.68 × 10−2 | 4.5 | 1.84 × 10−2 | −1.9 | 4.52 × 10−1 |
| AC006504.1 | 3.6 | 5.58 × 10−4 | −2.5 | 1.96 × 10−2 | 2.7 | 1.77 × 10−2 | −2.3 | 8.47 × 10−2 |
| AC046134.2 | 3.3 | 2.34 × 10−4 | −3.0 | 1.06 × 10−3 | 2.4 | 3.18 × 10−2 | −1.7 | 2.67 × 10−1 |
| ZFPM2-AS1 | 3.2 | 5.31 × 10−5 | −3.3 | 6.44 × 10−5 | 2.6 | 1.27 × 10−3 | −2.1 | 3.26 × 10−2 |
| SNHG32 | 3.2 | 2.41 × 10−12 | −2.4 | 2.18 × 10−7 | 1.1 | 7.51 × 10−1 | −1.6 | 3.91 × 10−2 |
| ELF3-AS1 | 3.0 | 1.10 × 10−3 | −5.8 | 1.64 × 10−7 | 1.7 | 2.03 × 10−1 | −2.5 | 2.75 × 10−2 |
| LINC01012 | 3.0 | 1.29 × 10−4 | −2.4 | 2.69 × 10−3 | 2.2 | 1.03 × 10−2 | −1.8 | 1.00 × 10−1 |
| ZFAS1 | 2.6 | 1.39 × 10−12 | −1.7 | 1.15 × 10−4 | 1.9 | 2.41 × 10−5 | −2.0 | 7.39 × 10−6 |
| SNHG29 | 2.5 | 7.66 × 10−9 | −1.6 | 4.10 × 10−3 | 1.6 | 7.49 × 10−3 | −1.6 | 1.14 × 10−2 |
| LINC00662 | 2.3 | 2.97 × 10−4 | −2.0 | 5.01 × 10−3 | 3.0 | 9.27 × 10−6 | −2.2 | 2.82 × 10−3 |
| OSER1-DT | 2.3 | 2.49 × 10−4 | −1.8 | 9.82 × 10−3 | 2.9 | 2.91 × 10−5 | −1.9 | 2.43 × 10−2 |
| GAS5 | 2.2 | 7.72 × 10−7 | −1.5 | 1.46 × 10−2 | 1.8 | 1.27 × 10−3 | −1.8 | 2.32 × 10−3 |
| LINC02864 | 1.9 | 4.51 × 10−5 | −2.2 | 1.97 × 10−6 | 1.9 | 8.05 × 10−4 | −1.6 | 2.02 × 10−2 |
| DLGAP1-AS2 | 1.9 | 1.74 × 10−2 | −2.5 | 3.21 × 10−4 | 1.8 | 3.58 × 10−2 | −1.5 | 2.76 × 10−1 |
| VLDLR-AS1 | 11.1 | 1.15 × 10−7 | −5.5 | 1.58 × 10−4 | 7.7 | 2.60 × 10−5 | −2.6 | 8.65 × 10−2 |
| LncRNA | EZH2 Interaction (LncTarD) | FTO Interaction (StarBase) | References |
|---|---|---|---|
| LINC00511 | YES | YES | LINC00511 interacts with EZH2 [31,32,33] |
| GAS5 | YES | YES | GAS5 interacts with EZH2 [34,35,36]. GAS5 is de-methylated by FTO [37,38] |
| CRNDE | YES | YES | CRNDE interacts with EZH2 [39] |
| LINC00662 | NO | YES | LINC00662 interacts with EZH2 [40] |
| HEIH | YES | NO | |
| PVT1 | YES | YES | PVT1 interacts with EZH2 [41,42] |
| MALAT1 | NO | NO | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, A.; Saggese, P.; Soto, A.; Gomez, E.; Alcaraz, M., Jr.; Scafoglio, C. Long Non-Coding RNAs Contribute to Glucose Starvation-Induced Dedifferentiation in Lung Adenocarcinoma. Biomolecules 2025, 15, 1493. https://doi.org/10.3390/biom15111493
Pandey A, Saggese P, Soto A, Gomez E, Alcaraz M Jr., Scafoglio C. Long Non-Coding RNAs Contribute to Glucose Starvation-Induced Dedifferentiation in Lung Adenocarcinoma. Biomolecules. 2025; 15(11):1493. https://doi.org/10.3390/biom15111493
Chicago/Turabian StylePandey, Aparamita, Pasquale Saggese, Adriana Soto, Estefany Gomez, Martín Alcaraz, Jr., and Claudio Scafoglio. 2025. "Long Non-Coding RNAs Contribute to Glucose Starvation-Induced Dedifferentiation in Lung Adenocarcinoma" Biomolecules 15, no. 11: 1493. https://doi.org/10.3390/biom15111493
APA StylePandey, A., Saggese, P., Soto, A., Gomez, E., Alcaraz, M., Jr., & Scafoglio, C. (2025). Long Non-Coding RNAs Contribute to Glucose Starvation-Induced Dedifferentiation in Lung Adenocarcinoma. Biomolecules, 15(11), 1493. https://doi.org/10.3390/biom15111493







