Standard Sample Preparation for Serial Femtosecond Crystallography
Abstract
1. Introduction
1.1. Lysozyme
1.2. Thermolysin
1.3. Glucose Isomerase (Xylose Isomerase)
1.4. Proteinase K
1.5. Trypsin
1.6. Myoglobin
1.7. GFP and Its Derivatives
1.8. PYP
1.9. Thaumatin
1.10. Granulovirus
| Protein (Uniprot ID) | Size [kDa] | Major Use | References 1 | Reported Micro-Crystallization Conditions | Tertiary Structure |
|---|---|---|---|---|---|
| PYP (P16133) | 13.87 | Proof of principle | [64,65,67] | This work |  PDB ID 6P5G |
| Time-resolved study | [65,66,68] | ||||
| Sample delivery development | [72] | ||||
| Lysozyme (P00698) | 14.31 | Instrument commissioning | [33,86,87,88,89,90] | This work |  PDB ID 9I6N |
| Proof of principle | [90,91] | ||||
| Sample delivery development | [8,10,17,26,33,92,93,94] | ||||
| Myoglobin (P68082, P02185) | 16.95 | Instrument commissioning | [52,95,96] |  PDB ID 8BKH | |
| Time-resolved study | [48] | This work | |||
| Proof of principle | [50,52,97] [50,51] | ||||
| Sample delivery development | |||||
| Thaumatin (P02883) | 22.21 | Instrument commissioning | [39] | 100 mg/mL protein in ddH2O + 1.6 M sodium potassium tartrate [79] |  PDB ID 9FTS |
| Proof of principle | [75,77,78,79] | ||||
| Sample delivery development | [15,30,76,80,81] | ||||
| Trypsin (P00760) | 23.56 | Sample delivery development | [43,44,45,46] | 30 mg/mL protein in 25 mM HEPES (pH 7.0), 5 mM CaCl2 + 100 mM Tris (pH 8.5), 30%(w/v) PEG 3350, 200 mM Li2SO4 [43] (30 mg/mL protein + 10 mg/mL benzamidine in 20 mM HEPES (pH 7.0), 10 mM CaCl2) + (20% PEG 8000, 200 mM (NH4)2SO4, 100 mM Bis-Tris) [44] * (65 mg/mL protein + benzamidine in 3 mM CaCl2) + (11–14%(w/v) PEG 4000, 15% ethylene glycol, 200 mM SiSO4, 100 mM MES (pH 6.5)) [45] (15 mg/mL protein + 5 mg/mL benzamidine in 10 mM CaCl2, 20 mM HEPES (pH 7.0), 3.75% PEG 3350, 5% glycerol) + (15% PEG3350, 20% glycerol) (hanging drop) [46] |  PDB ID 7WA0 |
| iq-mEmerald | 27.1 | Sample delivery development | [63] | This work |  PDB ID 4KW4 |
| rsEGFP2 | 26.9 | Instrument commissioning | [98] | 20 mg/mL protein in 2M (NH4)2SO4, 20 mM NaCl, 120 mM HEPEs (pH 8.0) (seeding) [59] 20–24 mg/mL protein in 75 mM HEPES (pH 8.0), 20 mM NaCl, 1.1–1.3 M (NH4)2SO4 (seeding) [99] |  PDB ID 5O89 |
| Time-resolved study | [59,60,99] | ||||
| Proteinase K (P06873) | 29.1 | Instrument commissioning | [36,37,39,40] | 40 mg/mL protein in 20 mM MES (pH 6.5) + 100 mM MES (pH 6.5), 500 mM NaNO3, 100 mM CaCl2 [39] |  PDB ID 9FTX |
| Proof of principle | [38] | ||||
| Sample delivery development | [15,35,41,42] | ||||
| Thermolysin (P00800) | 34.86 | Proof of principle | [100,101] | 22.5 mg/mL protein in 100 mM MES (pH 6.5) + 10 mM CaCl2, 5% PEG 2000 [15] * 30 mg/mL protein in 50 mM NaOH + 15% (w/v) ammonium sulfate [16]. * 42.5 mg/mL protein + 40% PEG 2000 MME, 0.1 M MES (pH 6.5), 5 mM CaCl2 [18] 330 mg/mL protein + 45% DMSO in 50 mM Tris (7.5) + 1.45 M CaCl2 [44] |  PDB ID 5WR4 |
| Sample delivery development | [15,16,17,18,44,46] | ||||
| Glucose isomerase (P24300) | 43.33 (monomer) 173.32 (homo tetramer) | Instrument commissioning | [32] | 33 mg/mL protein in 6 mM Tris (pH 7.0), 0.91 M (NH4)2SO4, 1 mM MgSO4 [31] * 80 mg/mL protein + 35%(w/v) PEG3350, 0.2 M LiSO4, 10 mM HEPES (pH 7.5) [35] | 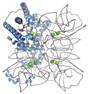 PDB ID 6KD2 |
| Sample delivery development | [26,27,28,29,30,31,33,34,35] | ||||
| Granulovirus (Granulin) (P87577) | 29.38 (monomer) 352.56 (homo 12-mer) | Proof of principle | [83] | Not applicable | 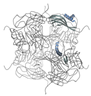 PDB ID 5G0Z |
| Sample delivery development | [84] |
2. Materials and Methods
2.1. Lysozyme
2.1.1. Materials
- Lysozyme: Carl Roth GmbH + Co. KG (Karlsruhe, Germany) Art.no. 8259;
- Sodium Acetate (NaOAc): Merck KGaA (Darmstadt, Germany) Art.no. 71183;
- Sodium chloride (NaCl): Carl Roth GmbH + Co. KG Art.no. P029;
- PEG 6000: Merck KGaA Art.no. 81260;
- Monoolein: Nu-Chek Prep (Elysian, MN, USA) Art.no. M-239;
- Gravity filters: CellTricsTM, Sysmex Deutschland GmbH (Norderstedt, Germany) (10 μm filter, Art. No. 04-0042-2314 and 20 μm filter, Art. No. 04-0042-2315);
- Gas-tight glass syringe: Hamilton Bonaduz AG (Bonaduz, Switzerland), Art.no. 202668;
- Syringe coupler:Rigaku Holdings Corporation (Tokyo, Japan), Art.no. EB-LCP-SUNION.
2.1.2. Prepared Solutions
- 0.5 M NaOAc, pH 3.5;
- 5 M NaCl;
- 50% (w/v) PEG 6000;
- Crystallization solution: 0.1 M NaOAc, pH 3.5, 5% PEG 6000 (w/v), 3.2 M NaCl, 0.2 µm filtered;
- Lysozyme solution: 100 mg/mL in 50 mM NaOAc, pH 3.5, 0.2 µm filtered;
- Storage buffer: 50 mM NaOAc, pH 3.5, 1.7 M NaCl, 0.2 µm filtered.
2.1.3. Crystallization
2.1.4. Storage Buffer Exchange
2.1.5. Crystal Filtration and Density Adjustment
2.1.6. Embedding in LCP
2.2. Myoglobin
2.2.1. Materials
- Myoglobin (equine skeletal muscle): Merck KGaA Art.no. M0630;
- Ammonium sulfate ((NH4)2SO4): Carl Roth GmbH Art.no. 9212.1;
- Tris (Tris-(hydroxymethyl)-amino methane): Carl Roth GmbH Art.no. 5429.3;
- Sodium dithionite: Merck KGaA Art.no. 71699;
- 40 μm frit filter: JR-1100-40P, Valco Instrument Co. Inc. (Houston, TX, USA);
- PreColumn: A-355, IDEX Health & Science LLC (Rohnert Park, CA, USA);
- Luer adapter: P-642, IDEX Health & Science LLC.
2.2.2. Prepared Solutions
- 4 M (NH4)2SO4, 0.2 µm filtered;
- 50 mM Tris buffer, pH 7.5, 0.2 µm filtered;
- 0.5 M sodium dithionite in degassed 3.3 M (NH4)2SO4, prepared in a glove box (GS MEGA 4).
2.2.3. Crystallization
2.2.4. Crystal Filtration
2.2.5. Deoxygenation
2.3. Iq-mEmerald
2.3.1. Materials
- Plasmid: iq-mEmerald expression vector pET17b-iq-mEmerald without using the fusion tag. The vector was purchased from Biocat GmbH (Heidelberg, Germany) using the amino acid sequence taken from the Fluorescent Protein Data Base (https://www.fpbase.org/protein/7G47U/ (accessed on 10 September 2025)) and pET17b as vector backbone.
- Recipient cell line: Escherichia coli BL21(DE3) (Thermo Fisher Scientific Inc. (Waltham, MA, USA), Art.no. EC0114).
- Glassware: 5 L flasks.
- Seed Beads: Jena Bioscience GmbH (Jena, Germany), Art.no. CO-501.
- Gravity filter: CellTrics™ 30 µm, Sysmex Deutschland GmbH, Art. Nr. 04-0042-2316.
- LB-medium: Carl Roth GmbH Art.no. 6673.4.
- Ampicillin: Carl Roth GmbH Art.no. K029.2.
- IPTG (isopropyl β-D-thiogalactopyranoside): Carl Roth GmbH Art.no. 2316.5.
- Tris (Tris-(hydroxymethyl)-amino methane): Carl Roth GmbH Art.no. 5429.3.
- Sodium chloride (NaCl): Carl Roth GmbH Art.no. P029.
- Ammonium sulfate ((NH4)2SO4): Carl Roth GmbH Art.no. 9212.1.
- Ethanol (99.9%): Merck KGaA Art.no. 1.00983.
- Hydrophobic interaction column (HIC), e.g., HiPrep Phenyl FF (High Sub) 16/10 Cytiva, Marlborough, MA, USA Art. Nr 28936545).
- 10 kDa cut-off concentrator: Merck KGaA Art.no. UFC9010.
2.3.2. Equipment
- Incubation shaker: Eppendorf SE (Hamburg, Germany) New BrunswickTM Innova®44/44R Shaker;
- FPLC: Cytiva ÄKTA pure™ chromatography system.
2.3.3. Prepared Solutions
- Antibiotic: 100 mg/mL Ampicillin stock solution in ethanol;
- 1 M IPTG (isopropyl β-D-thiogalactopyranoside); 0.2 µm filtered;
- Buffer A (Lysis Buffer): 20 mM Tris, pH 7.8, 150 mM NaCl; 0.2 µm filtered;
- Buffer B (HIC start buffer): 20 mM Tris, pH 7.8, 20% (NH4)2SO4 saturation, 0.2 µm filtered;
- Buffer C (HIC elution buffer): 20 mM Tris, pH 7.8, 0.2 µm filtered;
- 5 M NaCl, 0.2 µm filtered;
- 70% (NH4)2SO4 saturated solution;
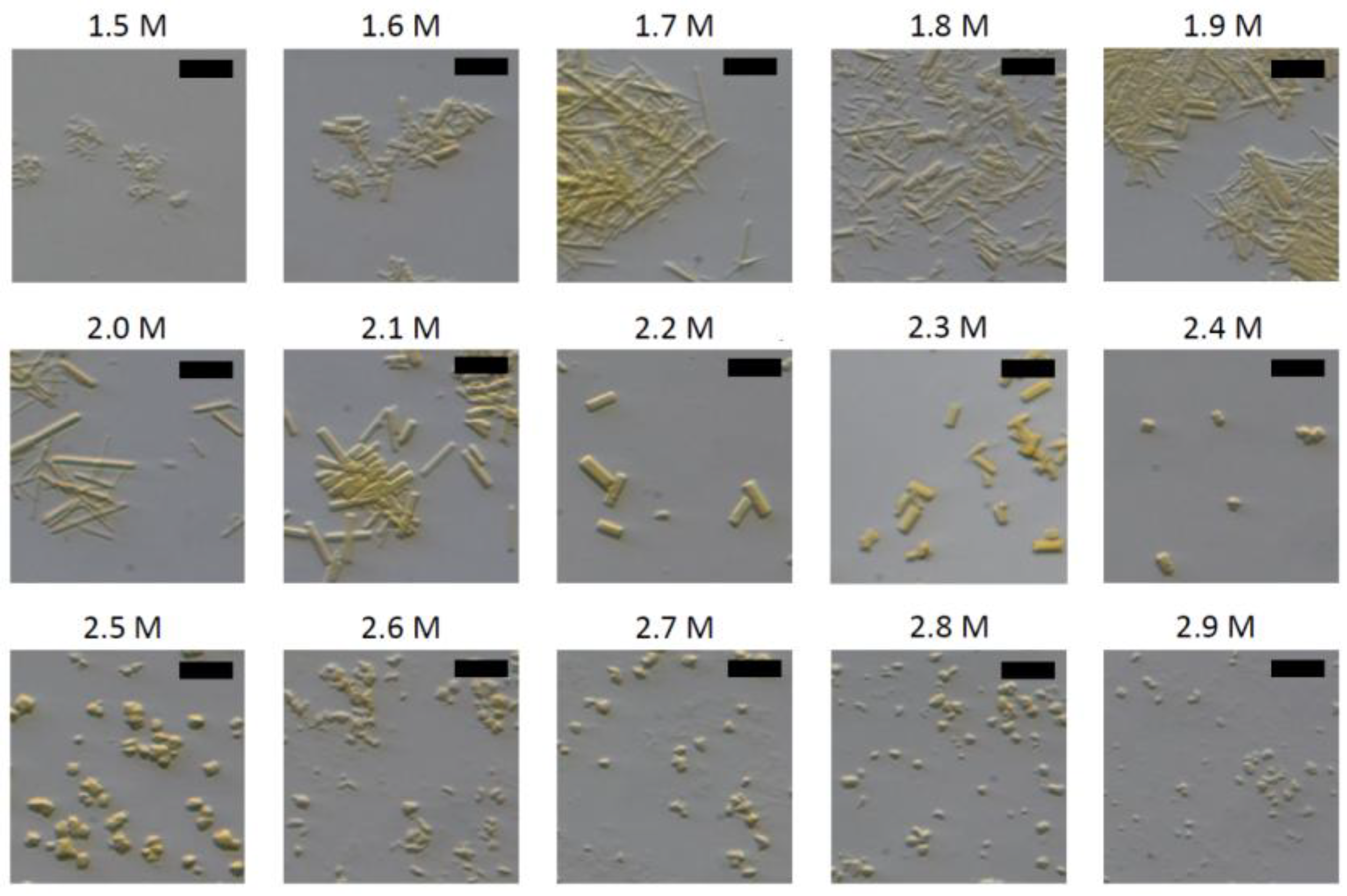
- Crystallization Buffers: 50 mM Tris, pH 8.0, 1.5–3 M (NH4)2SO4 concentrations in 0.1 M increments, 0.2 µm filtered.
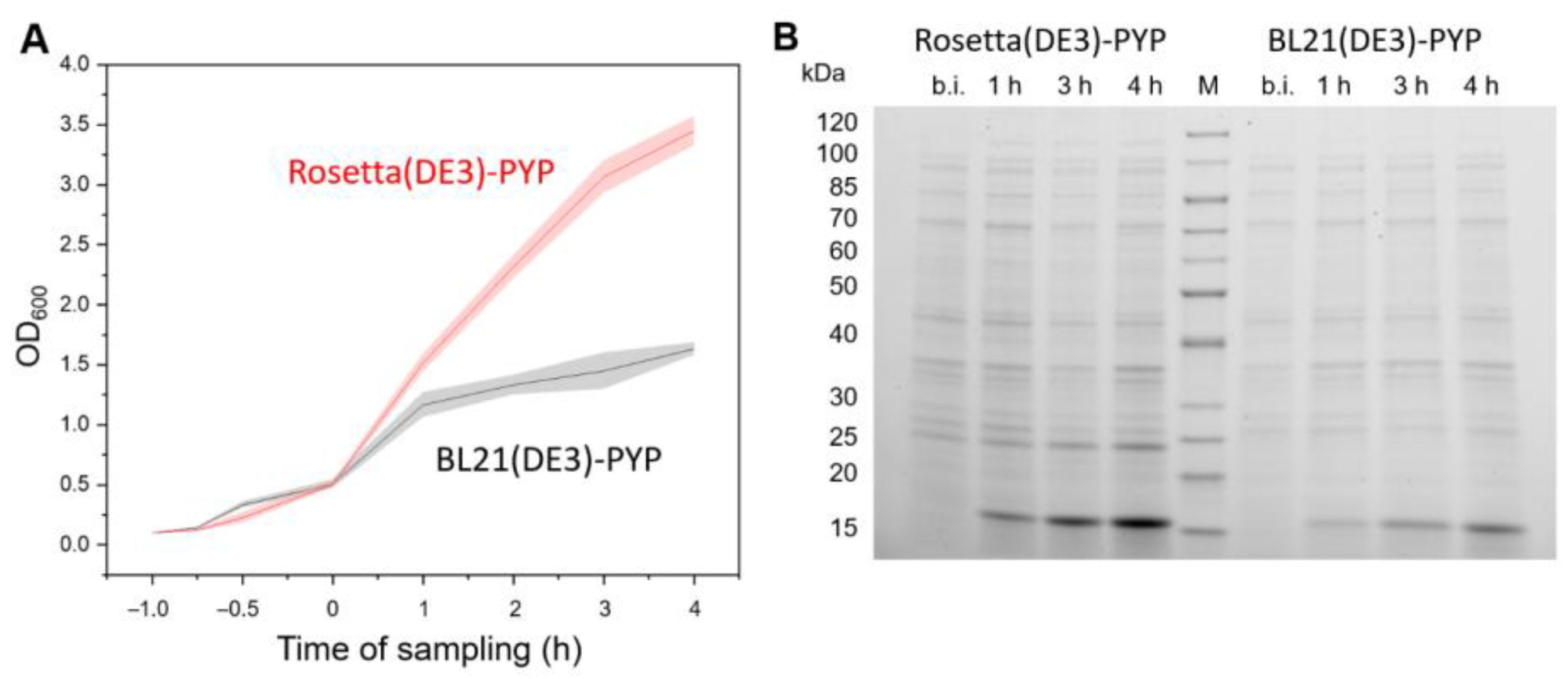
2.3.4. Expression
2.3.5. Purification
2.3.6. Crystallization
Seedstock
Needle Crystals
Cubic Crystals
2.3.7. Embedding in LCP
2.4. PYP (Photoactive Yellow Protein)
2.4.1. Materials
- Plasmid: pET-M11 [103]. The expression vector containing the codon-optimized gene sequence for PYP was purchased from Biocat GmbH (Heidelberg, Germany) using the full PYP sequence from UniprotKB: P16113.
- Recipient cell line: Escherichia coli Rosetta(DE3), Merck KGaA, Art.no. 70954-3.
- Glassware: 5 L flasks.
- Seed Beads: Jena Bioscience GmbH, Art.no. CO-501.
- PD-10 Buffer exchange columns: Cytiva, Marlborough, MA, USA, Art. No. 17085101.
- Gravity filter: CellTrics™ 30 µm, Sysmex Deutschland GmbH, Art. Nr. 04-0042-2316.
- NiNTA: Thermo Fisher Scientific Art.no. A50586.
- Gravity column: Carl Roth GmbH Art. No. 1518.1.
- LB-medium: Carl Roth GmbH Art.no. 6673.4.
- Kanamycin: Carl Roth GmbH Art.no. T832.4.
- Chloramphenicol: Thermo Scientific Chemicals Art.no. B20841.22.
- HEPES (N-2-Hydroxyethylpiperazine-N’-2-ethane sulphonic acid): Carl Roth GmbH Art.no. 9105.3.
- Sodium chloride (NaCl): Carl Roth GmbH Art.no. P029.
- Imidazole: Carl Roth GmbH Art.no. 3899.3.
- Tris (Tris-(hydroxymethyl)-amino methane): Carl Roth GmbH Art.no. 5429.3.
- Tri-sodium citrate dihydrate: Carl Roth GmbH Art.no. 3580.1.
- Citric acid: Carl Roth GmbH Art.no. 7624.1.
- p-Coumaric acid: Merck KGaA, Art.no. C9008.
- N,N’-Dicyclohexylcarbodiimide (DCC): Merck KGaA, Art.no. D80002.
- N,N-Dimethylformamide (DMF): Merck KGaA, Art.no. 227056.
- Sodium malonate (Na-malonate): Merck KGaA, Art.no. M4795.
- Beta-mercaptoethanol: Carl Roth GmbH Art. No. 4227.3.
- Glycerol: Carl Roth GmbH Art. No. 3783.2.
- 3 kDa cut-off concentrator: Merck KGaA Art.no. UFC9003.
2.4.2. Equipment
- Incubation shaker: New BrunswickTM Innova®44/44R Shaker;
- Anion exchange column: HiPrep Q HP 16/10 Cytiva Art.no. 29018182;
- FPLC: Cytiva ÄKTA pure™ chromatography system.
2.4.3. Prepared Solutions
- Kanamycin stock solution (100 mg/mL in ddH2O);
- Chloramphenicol stock solution (34 mg/mL in ethanol);
- Buffer A (Lysis Buffer): 20 mM HEPES, pH 7.4, 200 mM NaCl, 5 mM Imidazole, 0.2 µm filtered;
- Buffer B (Wash buffer): 20 mM HEPES, pH 7.4, 200 mM NaCl, 10 mM Imidazole, 0.2 µm filtered;
- Buffer C (Elution buffer): 20 mM HEPES, pH 7.4, 200 mM NaCl, 300 mM Imidazole, 0.2 µm filtered;
- Buffer D (TEV protease reaction buffer): 20 mM HEPES, pH 7.4, 200 mM NaCl, 0.2 µm filtered;
- Buffer E (Anion exchange start buffer): 25 mM Tris, 0.2 µm filtered;
- Buffer F (Anion exchange elution buffer): 25 mM Tris, 1 M NaCl, 0.2 µm filtered;
- Buffer G (Storage buffer): 50 mM Citrate Buffer, pH 6.0, 0.2 µm filtered;
- Buffer H (Crystallization buffer): 3.7 M sodium malonate, pH 7, 0.2 µm filtered;
- Tobacco Etch Virus (TEV) protease: prepared following the protocol from Berg et al. (2006) [104], in 50 mM Tris (pH 8.0), 200 mM NaCl, 5 mM beta-mercaptoethanol, and 10% (v/v) glycerol.
2.4.4. Expression
2.4.5. p-Coumaric Anhydride (pCA) Synthesis
2.4.6. Purification
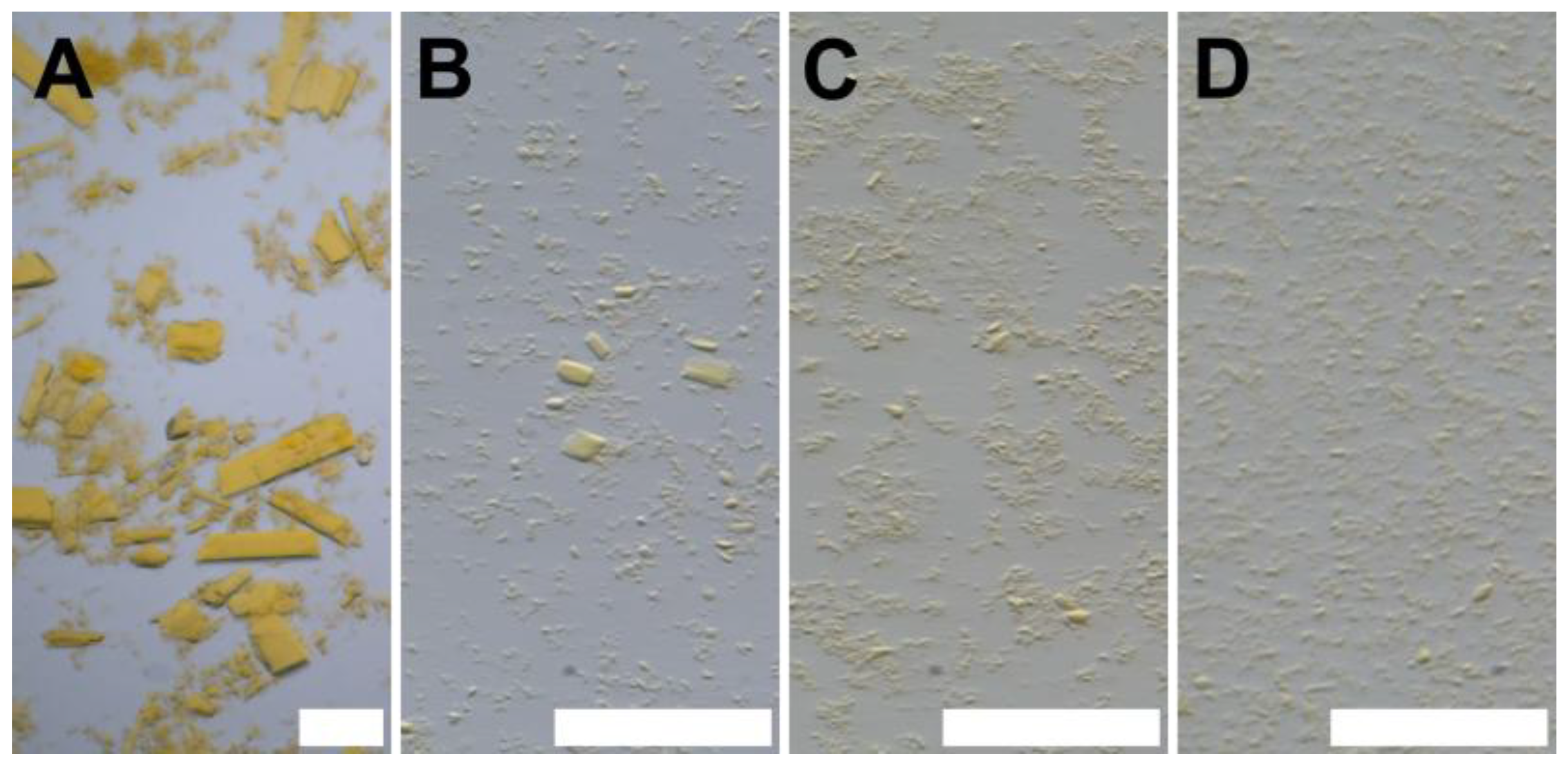
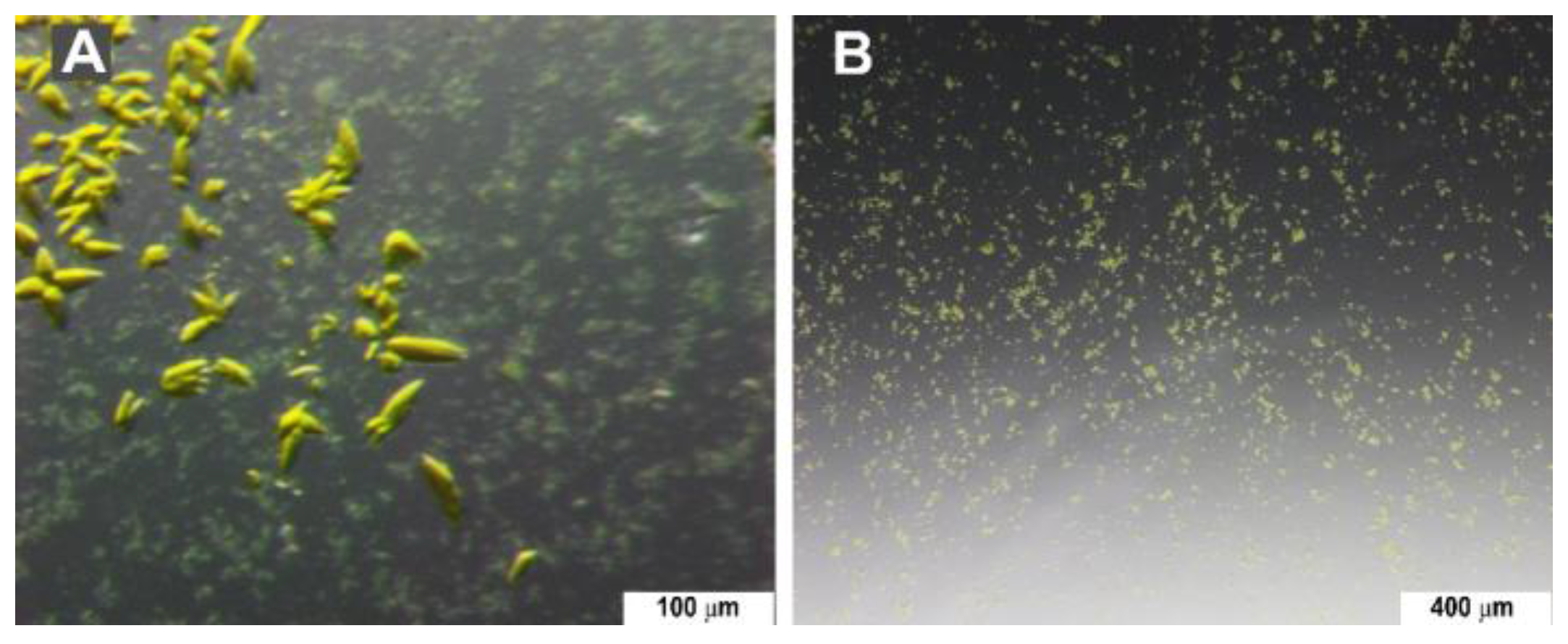
2.4.7. Crystallization
3. Results
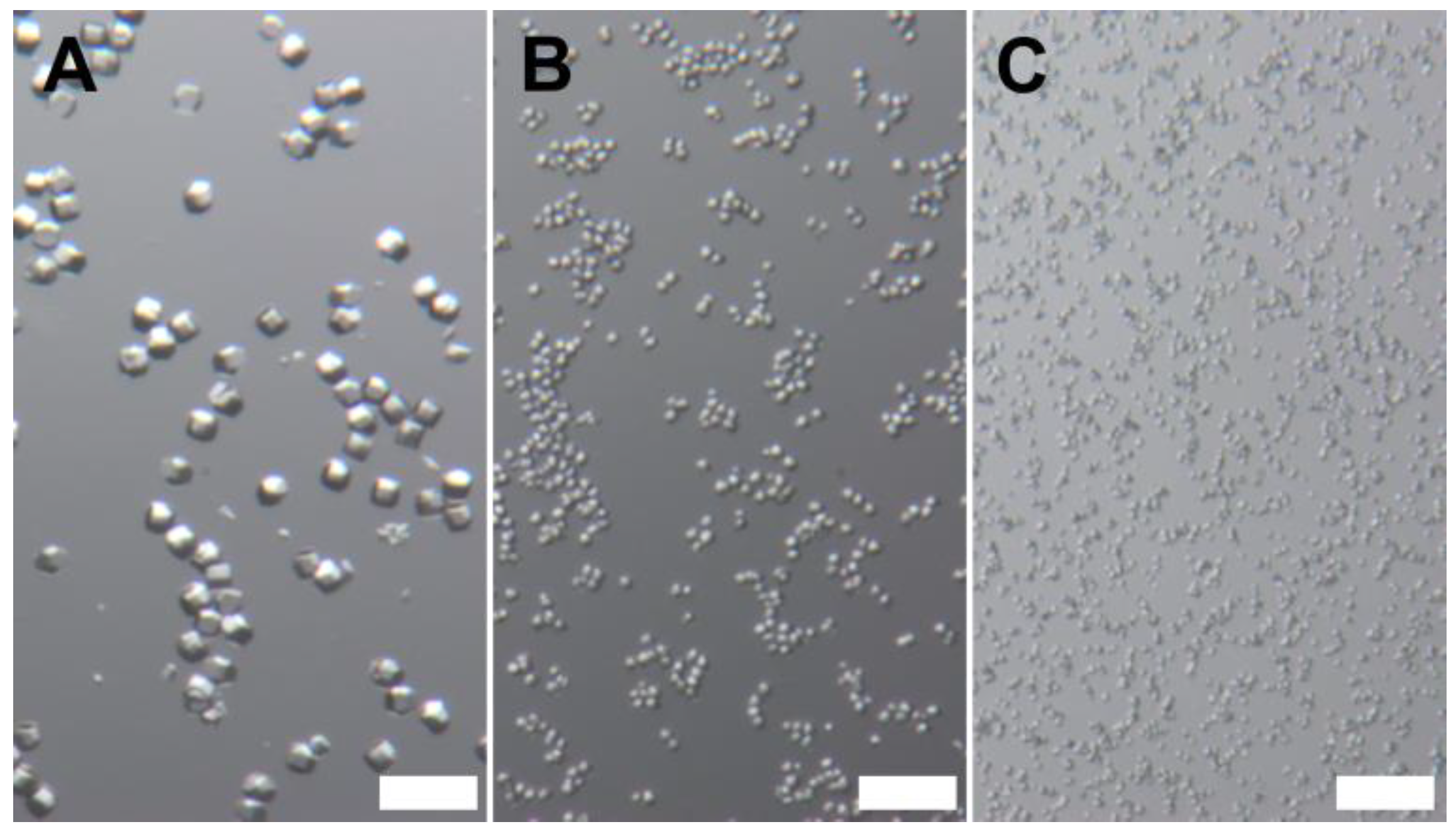
3.1. Lysozyme
3.2. Myoglobin
3.3. Iq-mEmerald
3.4. PYP
3.4.1. Upscaling of Protein Production
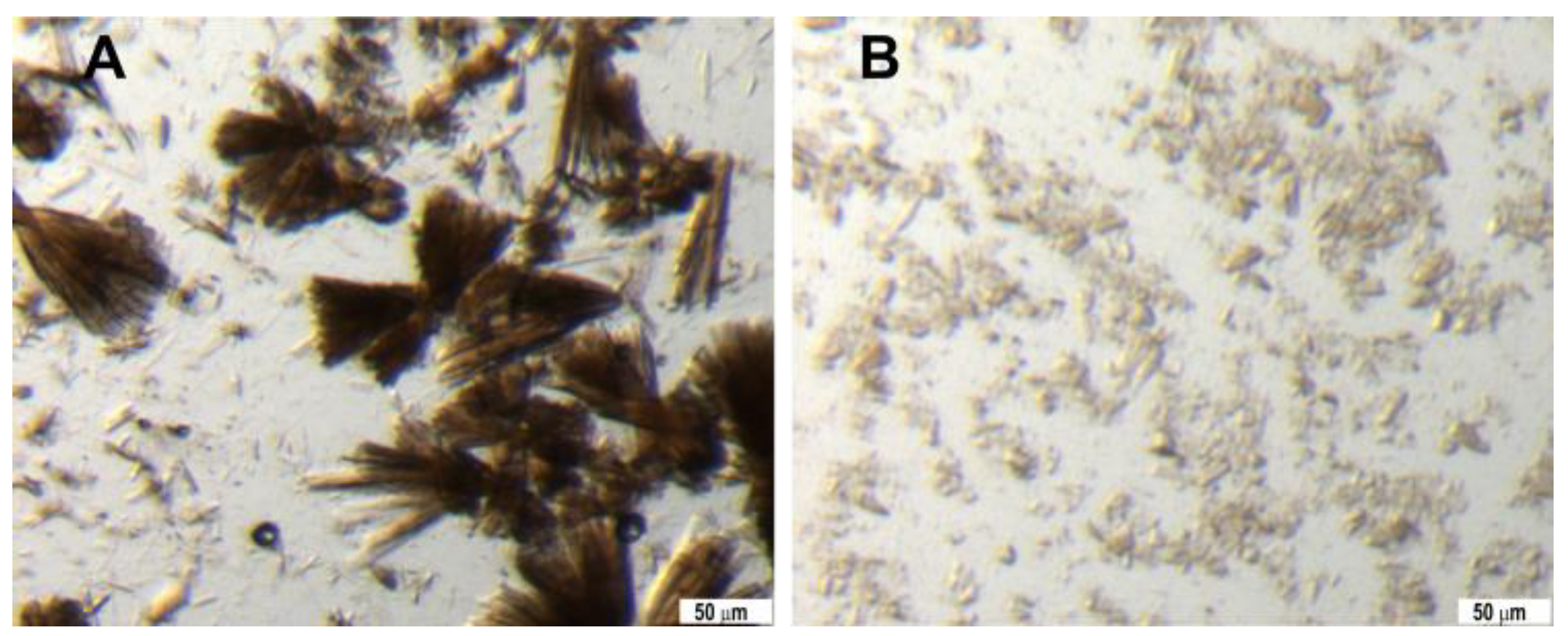
3.4.2. Seedstock Preparation and Crystallization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Chapman, H.N.; Fromme, P.; Barty, A.; White, T.A.; Kirian, R.A.; Aquila, A.; Hunter, M.S.; Schulz, J.; Deponte, D.P.; Weierstall, U.; et al. Femtosecond X-Ray Protein Nanocrystallography. Nature 2011, 470, 73–77. [Google Scholar] [CrossRef]
- Neutze, R.; Wouts, R.; Van Der Spoel, D.; Weckert, E.; Hajdu, J. Potential for Biomolecular Imaging with Femtosecond X-Ray Pulses. Nature 2000, 406, 752–757. [Google Scholar] [CrossRef]
- Barends, T.R.M.; Stauch, B.; Cherezov, V.; Schlichting, I. Serial Femtosecond Crystallography. Nat. Rev. Methods Prim. 2022, 2, 59. [Google Scholar] [CrossRef]
- Orville, A.M. Recent Results in Time Resolved Serial Femtosecond Crystallography at XFELs. Curr. Opin. Struct. Biol. 2020, 65, 193–208. [Google Scholar] [CrossRef]
- Kupitz, C.; Sierra, R.G. Preventing Bio-Bloopers and XFEL Follies: Best Practices from Your Friendly Instrument Staff. Crystals 2020, 10, 251. [Google Scholar] [CrossRef]
- Botha, S.; Fromme, P. Review of Serial Femtosecond Crystallography Including the COVID-19 Pandemic Impact and Future Outlook. Structure 2023, 31, 1306–1319. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.B.; Kim, J.M.; Seok, J.H.; Lee, J.H.; Jo, J.D.; Mun, J.Y.; Conrad, C.; Coe, J.; Nelson, G.; Hogue, B.; et al. Supersaturation-Controlled Microcrystallization and Visualization Analysis for Serial Femtosecond Crystallography. Sci. Rep. 2018, 8, 2541. [Google Scholar] [CrossRef] [PubMed]
- Fromme, R.; Ishchenko, A.; Metz, M.; Chowdhury, S.R.; Basu, S.; Boutet, S.; Fromme, P.; White, T.A.; Barty, A.; Spence, J.C.H.; et al. Serial Femtosecond Crystallography of Soluble Proteins in Lipidic Cubic Phase. IUCrJ 2015, 2, 545–551. [Google Scholar] [CrossRef]
- Stellato, F.; Oberthür, D.; Liang, M.; Bean, R.; Gati, C.; Yefanov, O.; Barty, A.; Burkhardt, A.; Fischer, P.; Galli, L.; et al. Room-Temperature Macromolecular Serial Crystallography Using Synchrotron Radiation. IUCrJ 2014, 1, 204–212. [Google Scholar] [CrossRef]
- Perrett, S.; Fadini, A.; Hutchison, C.D.M.; Bhattacharya, S.; Morrison, C.; Turkot, O.; Jakobsen, M.B.; Großler, M.; Licon-Salaiz, J.; Griese, F.; et al. Kilohertz Droplet-on-Demand Serial Femtosecond Crystallography at the European XFEL Station FX. Struct. Dyn. 2024, 11, 024310. [Google Scholar]
- DePonte, D.P.; Weierstall, U.; Schmidt, K.; Warner, J.; Starodub, D.; Spence, J.C.H.; Doak, R.B. Gas Dynamic Virtual Nozzle for Generation of Microscopic Droplet Streams. J. Phys. D Appl. Phys. 2008, 41, 195505. [Google Scholar] [CrossRef]
- Tolstikova, A.; Levantino, M.; Yefanov, O.; Hennicke, V.; Fischer, P.; Meyer, J.; Mozzanica, A.; Redford, S.; Crosas, E.; Opara, N.L.; et al. 1 KHz Fixed-Target Serial Crystallography Using a Multilayer Monochromator and an Integrating Pixel Detector. IUCrJ 2019, 6, 927–937. [Google Scholar] [CrossRef]
- Sierra, R.G.; Gati, C.; Laksmono, H.; Dao, E.H.; Gul, S.; Fuller, F.; Kern, J.; Chatterjee, R.; Ibrahim, M.; Brewster, A.S.; et al. Concentric-Flow Electrokinetic Injector Enables Serial Crystallography of Ribosome and Photosystem II. Nat. Methods 2015, 13, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Sierra, R.G.; Laksmono, H.; Kern, J.; Tran, R.; Hattne, J.; Alonso-Mori, R.; Lassalle-Kaiser, B.; Glöckner, C.; Hellmich, J.; Schafer, D.W.; et al. Nanoflow Electrospinning Serial Femtosecond Crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 1584–1587. [Google Scholar] [CrossRef] [PubMed]
- Lieske, J.; Cerv, M.; Kreida, S.; Komadina, D.; Fischer, J.; Barthelmess, M.; Fischer, P.; Pakendorf, T.; Yefanov, O.; Mariani, V.; et al. On-Chip Crystallization for Serial Crystallography Experiments and on-Chip Ligand-Binding Studies. IUCrJ 2019, 6, 714–728. [Google Scholar] [CrossRef] [PubMed]
- Roessler, C.G.; Agarwal, R.; Allaire, M.; Alonso-Mori, R.; Andi, B.; Bachega, J.F.R.; Bommer, M.; Brewster, A.S.; Browne, M.C.; Chatterjee, R.; et al. Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography. Structure 2016, 24, 631–640. [Google Scholar] [CrossRef]
- Park, J.; Park, S.; Kim, J.; Park, G.; Cho, Y.; Nam, K.H. Polyacrylamide Injection Matrix for Serial Femtosecond Crystallography. Sci. Rep. 2019, 9, 2525. [Google Scholar] [CrossRef]
- Naitow, H.; Matsuura, Y.; Tono, K.; Joti, Y.; Kameshima, T.; Hatsui, T.; Yabashi, M.; Tanaka, R.; Tanaka, T.; Sugahara, M.; et al. Protein-Ligand Complex Structure from Serial Femtosecond Crystallography Using Soaked Thermolysin Microcrystals and Comparison with Structures from Synchrotron Radiation. Acta Crystallogr. Sect. D Struct. Biol. 2017, 73, 702–709. [Google Scholar] [CrossRef]
- Matthews, B.W.; Weaver, L.H.; Kester, W.R. The Conformation of Thermolysin. J. Biol. Chem. 1974, 249, 8030–8044. [Google Scholar] [CrossRef]
- Dahlquist, F.W.; Long, J.W.; Bigbee, W.L. Role of Calcium in the Thermal Stability of Thermolysin. Biochemistry 1976, 15, 1103–1111. [Google Scholar] [CrossRef]
- Roche, R.S.; Voordouw, G.; Matthews, B.W. The Structural and Functional Roles of Metal Ions in Thermolysin. Crit. Rev. Biochem. Mol. Biol. 1978, 5, 1–23. [Google Scholar] [CrossRef]
- Leite, J.P.; Gales, L. Alzheimer’s Aβ 1-40 Peptide Degradation by Thermolysin: Evidence of Inhibition by a C-Terminal Aβ Product. FEBS Lett. 2019, 593, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Kovalevsky, A.Y.; Hanson, L.; Fisher, S.Z.; Mustyakimov, M.; Mason, S.A.; Trevor Forsyth, V.; Blakeley, M.P.; Keen, D.A.; Wagner, T.; Carrell, H.L.; et al. Metal Ion Roles and the Movement of Hydrogen during Reaction Catalyzed by D-Xylose Isomerase: A Joint X-Ray and Neutron Diffraction Study. Structure 2010, 18, 688–699. [Google Scholar] [CrossRef]
- Callens, M.; Kersters-Hilderson, H.; Van Opstal, O.; De Bruyne, C.K. Catalytic Properties of D-Xylose Isomerase from Streptomyces Violaceoruber. Enzym. Microb. Technol. 1986, 8, 696–700. [Google Scholar] [CrossRef]
- Danno, G. ichi Studies on D-Glucose-Isomerizing Enzyme of Bacillus Coagulans, Strain HN-68: Part III. Induced Formation of D-Glucose-Isomerizing Enzyme by D-Glucose-Grown Cells of Bacillus Coagulans, Strain HN-68. Agric. Biol. Chem. 1970, 34, 1658–1667. [Google Scholar] [CrossRef]
- Nam, K.H. Stable Sample Delivery in Viscous Media via a Capillary for Serial Crystallography. J. Appl. Crystallogr. 2020, 53, 45–50. [Google Scholar] [CrossRef]
- Nam, K.H. Shortening Injection Matrix for Serial Crystallography. Sci. Rep. 2020, 10, 107. [Google Scholar] [CrossRef]
- Nam, K.H. Polysaccharide-Based Injection Matrix for Serial Crystallography. Int. J. Mol. Sci. 2020, 21, 3332. [Google Scholar] [CrossRef]
- Nam, K.H. Beef Tallow Injection Matrix for Serial Crystallography. Sci. Rep. 2022, 12, 694. [Google Scholar] [CrossRef]
- Sugahara, M.; Mizohata, E.; Nango, E.; Suzuki, M.; Tanaka, T.; Masuda, T.; Tanaka, R.; Shimamura, T.; Tanaka, Y.; Suno, C.; et al. Grease Matrix as a Versatile Carrier of Proteins for Serial Crystallography. Nat. Methods 2015, 12, 61–63. [Google Scholar] [CrossRef]
- Lee, D.; Baek, S.; Park, J.; Lee, K.; Kim, J.; Lee, S.J.; Chung, W.K.; Lee, J.L.; Cho, Y.; Nam, K.H. Nylon Mesh-Based Sample Holder for Fixed-Target Serial Femtosecond Crystallography. Sci. Rep. 2019, 9, 6971. [Google Scholar] [CrossRef]
- Kim, Y.; Nam, K.H. Fixed-Target Pink-Beam Serial Synchrotron Crystallography at Pohang Light Source II. Crystals 2023, 13, 1544. [Google Scholar] [CrossRef]
- Mehrabi, P.; Schulz, E.C.; Agthe, M.; Horrell, S.; Bourenkov, G.; von Stetten, D.; Leimkohl, J.P.; Schikora, H.; Schneider, T.R.; Pearson, A.R.; et al. Liquid Application Method for Time-Resolved Analyses by Serial Synchrotron Crystallography. Nat. Methods 2019, 16, 979–982. [Google Scholar] [CrossRef]
- Mehrabi, P.; Sung, S.; von Stetten, D.; Prester, A.; Hatton, C.E.; Kleine-Döpke, S.; Berkes, A.; Gore, G.; Leimkohl, J.P.; Schikora, H.; et al. Millisecond Cryo-Trapping by the Spitrobot Crystal Plunger Simplifies Time-Resolved Crystallography. Nat. Commun. 2023, 14, 2365. [Google Scholar] [CrossRef]
- Norton-Baker, B.; Mehrabi, P.; Boger, J.; Schönherr, R.; Von Stetten, D.; Schikora, H.; Kwok, A.O.; Martin, R.W.; Miller, R.J.D.; Redecked, L.; et al. A Simple Vapor-Diffusion Method Enables Protein Crystallization inside the HARE Serial Crystallography Chip. Acta Crystallogr. Sect. D Struct. Biol. 2021, 77, 820–834. [Google Scholar] [CrossRef] [PubMed]
- Martin-Garcia, J.M.; Zhu, L.; Mendez, D.; Lee, M.Y.; Chun, E.; Li, C.; Hu, H.; Subramanian, G.; Kissick, D.; Ogata, C.; et al. High-Viscosity Injector-Based Pink-Beam Serial Crystallography of Microcrystals at a Synchrotron Radiation Source. IUCrJ 2019, 6, 412–425. [Google Scholar] [CrossRef]
- Meents, A.; Wiedorn, M.O.; Srajer, V.; Henning, R.; Sarrou, I.; Bergtholdt, J.; Barthelmess, M.; Reinke, P.Y.A.; Dierksmeyer, D.; Tolstikova, A.; et al. Pink-Beam Serial Crystallography. Nat. Commun. 2017, 8, 1281. [Google Scholar] [CrossRef]
- Botha, S.; Baitan, D.; Jungnickel, K.E.J.; Oberthür, D.; Schmidt, C.; Stern, S.; Wiedorn, M.O.; Perbandt, M.; Chapman, H.N.; Betzel, C. De Novo Protein Structure Determination by Heavy-Atom Soaking in Lipidic Cubic Phase and SIRAS Phasing Using Serial Synchrotron Crystallography. IUCrJ 2018, 5, 524–530. [Google Scholar] [CrossRef]
- Martin-Garcia, J.M.; Conrad, C.E.; Nelson, G.; Stander, N.; Zatsepin, N.A.; Zook, J.; Zhu, L.; Geiger, J.; Chun, E.; Kissick, D.; et al. Serial Millisecond Crystallography of Membrane and Soluble Protein Microcrystals Using Synchrotron Radiation. IUCrJ 2017, 4, 439–454. [Google Scholar] [CrossRef] [PubMed]
- Orlans, J.; Rose, S.L.; Ferguson, G.; Oscarsson, M.; Homs Puron, A.; Beteva, A.; Debionne, S.; Theveneau, P.; Coquelle, N.; Kieffer, J.; et al. Advancing Macromolecular Structure Determination with Microsecond X-Ray Pulses at a 4th Generation Synchrotron. Commun. Chem. 2025, 8, 6. [Google Scholar] [CrossRef]
- Zhao, F.Z.; Sun, B.; Yu, L.; Xiao, Q.J.; Wang, Z.J.; Chen, L.L.; Liang, H.; Wang, Q.S.; He, J.H.; Yin, D.C. A Novel Sample Delivery System Based on Circular Motion for: In Situ Serial Synchrotron Crystallography. Lab. Chip 2020, 20, 3888–3898. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Park, S.; Lee, K.; Kim, J.; Park, G.; Nam, K.H.; Baek, S.; Chung, W.K.; Lee, J.L.; Cho, Y.; et al. Application of a High-Throughput Microcrystal Delivery System to Serial Femtosecond Crystallography. J. Appl. Crystallogr. 2020, 53, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Okumura, H.; Sakai, N.; Murakami, H.; Mizuno, N.; Nakamura, Y.; Ueno, G.; Masunaga, T.; Kawamura, T.; Baba, S.; Hasegawa, K.; et al. In Situ Crystal Data-Collection and Ligand-Screening System at SPring-8. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2022, 78, 241–251. [Google Scholar] [CrossRef]
- Yin, X.; Scalia, A.; Leroy, L.; Cuttitta, C.M.; Polizzo, G.M.; Ericson, D.L.; Roessler, C.G.; Campos, O.; Ma, M.Y.; Agarwal, R.; et al. Hitting the Target: Fragment Screening with Acoustic in Situ Co-Crystallization of Proteins plus Fragment Libraries on Pin-Mounted Data-Collection Micromeshes. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 1177–1189. [Google Scholar] [CrossRef]
- Stubbs, J.; Hornsey, T.; Hanrahan, N.; Esteban, L.B.; Bolton, R.; Malý, M.; Basu, S.; Orlans, J.; de Sanctis, D.; Shim, J.U.; et al. Droplet Microfluidics for Time-Resolved Serial Crystallography. IUCrJ 2024, 11, 237–248. [Google Scholar] [CrossRef]
- Gao, Y.; Xu, W.; Shi, W.; Soares, A.; Jakoncic, J.; Myers, S.; Martins, B.; Skinner, J.; Liu, Q.; Bernstein, H.; et al. High-Speed Raster-Scanning Synchrotron Serial Microcrystallography with a High-Precision Piezo-Scanner. J. Synchrotron Radiat. 2018, 25, 1362–1370. [Google Scholar] [CrossRef]
- Kendrew, J.C.; Bodo, G.; Dintzis, H.M.; Parrish, R.G.; Wyckoff, H.; Phillips, D.C. A Three-Dimensional Model of the Myoglobin Molecule Obtained by x-Ray Analysis. Nature 1958, 181, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Barends, T.R.M.; Foucar, L.; Ardevol, A.; Nass, K.; Aquila, A.; Botha, S.; Doak, R.B.; Falahati, K.; Hartmann, E.; Hilpert, M.; et al. Direct Observation of Ultrafast Collective Motions in CO Myoglobin upon Ligand Dissociation. Science 2015, 350, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Levantino, M.; Schirò, G.; Lemke, H.T.; Cottone, G.; Glownia, J.M.; Zhu, D.; Chollet, M.; Ihee, H.; Cupane, A.; Cammarata, M. Ultrafast Myoglobin Structural Dynamics Observed with an X-Ray Free-Electron Laser. Nat. Commun. 2015, 6, 6772. [Google Scholar] [CrossRef]
- Owen, R.L.; Axford, D.; Sherrell, D.A.; Kuo, A.; Ernst, O.P.; Schulz, E.C.; Miller, R.J.D.; Mueller-Werkmeister, H.M. Low-Dose Fixed-Target Serial Synchrotron Crystallography. Acta Crystallogr. Sect. D Struct. Biol. 2017, 73, 373–378. [Google Scholar] [CrossRef]
- Oghbaey, S.; Sarracini, A.; Ginn, H.M.; Pare-Labrosse, O.; Kuo, A.; Marx, A.; Epp, S.W.; Sherrell, D.A.; Eger, B.T.; Zhong, Y.; et al. Fixed Target Combined with Spectral Mapping: Approaching 100% Hit Rates for Serial Crystallography. Acta Crystallogr. Sect. D Struct. Biol. 2016, 72, 944–955. [Google Scholar] [CrossRef]
- Mehrabi, P.; Bücker, R.; Bourenkov, G.; Ginn, H.M.; von Stetten, D.; Müller-Werkmeister, H.M.; Kuo, A.; Morizumi, T.; Eger, B.T.; Ou, W.L.; et al. Serial Femtosecond and Serial Synchrotron Crystallography Can Yield Data of Equivalent Quality: A Systematic Comparison. Sci. Adv. 2021, 7, eabf1380. [Google Scholar] [CrossRef]
- Ebrahim, A.; Moreno-Chicano, T.; Appleby, M.V.; Chaplin, A.K.; Beale, J.H.; Sherrell, D.A.; Duyvesteyn, H.M.E.; Owada, S.; Tono, K.; Sugimoto, H.; et al. Dose-Resolved Serial Synchrotron and XFEL Structures of Radiation-Sensitive Metalloproteins. IUCrJ 2019, 6, 543–551. [Google Scholar] [CrossRef]
- Remington, S.J. Green Fluorescent Protein: A Perspective. Protein Sci. 2011, 20, 1509–1519. [Google Scholar] [CrossRef]
- Lukyanov, K.A.; Chudakov, D.M.; Lukyanov, S.; Verkhusha, V.V. Photoactivatable Fluorescent Proteins. Nat. Rev. Mol. Cell Biol. 2005, 6, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Patterson, G.H.; Lippincott-Schwartz, J. A Photoactivatable GFP for Selective Photolabeling of Proteins and Cells. Science 2002, 297, 1873–1877. [Google Scholar] [CrossRef]
- Frommer, W.B.; Davidson, M.W.; Campbell, R.E. Genetically Encoded Biosensors Based on Engineered Fluorescent Proteins. Chem. Soc. Rev. 2009, 38, 2833–2841. [Google Scholar] [CrossRef] [PubMed]
- Coquelle, N.; Sliwa, M.; Woodhouse, J.; Schirò, G.; Adam, V.; Aquila, A.; Barends, T.R.M.; Boutet, S.; Byrdin, M.; Carbajo, S.; et al. Chromophore Twisting in the Excited State of a Photoswitchable Fluorescent Protein Captured by Time-Resolved Serial Femtosecond Crystallography. Nat. Chem. 2018, 10, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Woodhouse, J.; Nass Kovacs, G.; Coquelle, N.; Uriarte, L.M.; Adam, V.; Barends, T.R.M.; Byrdin, M.; de la Mora, E.; Bruce Doak, R.; Feliks, M.; et al. Photoswitching Mechanism of a Fluorescent Protein Revealed by Time-Resolved Crystallography and Transient Absorption Spectroscopy. Nat. Commun. 2020, 11, 741. [Google Scholar] [CrossRef]
- Adam, V.; Hadjidemetriou, K.; Jensen, N.; Shoeman, R.L.; Woodhouse, J.; Aquila, A.; Banneville, A.S.; Barends, T.R.M.; Bezchastnov, V.; Boutet, S.; et al. Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement**. ChemPhysChem 2022, 23, e202200192. [Google Scholar] [CrossRef]
- Beale, J.H.; Bolton, R.; Marshall, S.A.; Beale, E.V.; Carr, S.B.; Ebrahim, A.; Moreno-Chicano, T.; Hough, M.A.; Worrall, J.A.R.; Tews, I.; et al. Successful Sample Preparation for Serial Crystallography Experiments. J. Appl. Crystallogr. 2019, 52, 1385–1396. [Google Scholar] [CrossRef]
- Yu, X.; Strub, M.P.; Barnard, T.J.; Noinaj, N.; Piszczek, G.; Buchanan, S.K.; Taraska, J.W. An Engineered Palette of Metal Ion Quenchable Fluorescent Proteins. PLoS ONE 2014, 9, e95808. [Google Scholar] [CrossRef]
- Vakili, M.; Han, H.; Schmidt, C.; Wrona, A.; Kloos, M.; De Diego, I.; Dörner, K.; Geng, T.; Kim, C.; Koua, F.H.M.; et al. Mix-and-Extrude: High-Viscosity Sample Injection towards Time-Resolved Protein Crystallography. J. Appl. Crystallogr. 2023, 56, 1038–1045. [Google Scholar] [CrossRef]
- Hutchison, C.D.M.; van Thor, J.J. Populations and Coherence in Femtosecond Time Resolved X-Ray Crystallography of the Photoactive Yellow Protein. Int. Rev. Phys. Chem. 2017, 36, 117–143. [Google Scholar] [CrossRef]
- Konold, P.E.; Arik, E.; Weißenborn, J.; Arents, J.C.; Hellingwerf, K.J.; van Stokkum, I.H.M.; Kennis, J.T.M.; Groot, M.L. Confinement in Crystal Lattice Alters Entire Photocycle Pathway of the Photoactive Yellow Protein. Nat. Commun. 2020, 11, 4248. [Google Scholar] [CrossRef]
- Pandey, S.; Bean, R.; Sato, T.; Poudyal, I.; Bielecki, J.; Cruz Villarreal, J.; Yefanov, O.; Mariani, V.; White, T.A.; Kupitz, C.; et al. Time-Resolved Serial Femtosecond Crystallography at the European XFEL. Nat. Methods 2020, 17, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Pande, K.; Basu, S.; Tenboer, J. Room Temperature Structures beyond 1.5 Å by Serial Femtosecond Crystallography. Struct. Dyn. 2015, 2, 041708. [Google Scholar] [CrossRef]
- Schotte, F.; Cho, H.S.; Kaila, V.R.I.; Kamikubo, H.; Dashdorj, N.; Henry, E.R.; Graber, T.J.; Henning, R.; Wulff, M.; Hummer, G.; et al. Watching a Signaling Protein Function in Real Time via 100-Ps Time-Resolved Laue Crystallography. Proc. Natl. Acad. Sci. USA 2012, 109, 19256–19261. [Google Scholar] [CrossRef]
- Ihee, H.; Rajagopal, S.; Srajer, V.; Pahl, R.; Anderson, S.; Schmidt, M.; Schotte, F.; Anfinrud, P.A.; Wulff, M.; Moffat, K. Visualizing Reaction Pathways in Photoactive Yellow Protein from Nanoseconds to Seconds. Proc. Natl. Acad. Sci. USA 2005, 102, 7145–7150. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yabushita, A.; Taniguchi, S.; Chosrowjan, H.; Imamoto, Y.; Sueda, K.; Miyanaga, N.; Kobayashi, T. Ultrafast Time-Resolved Pump-Probe Spectroscopy of PYP by a Sub-8 Fs Pulse Laser at 400 Nm. J. Phys. Chem. B 2013, 117, 4818–4826. [Google Scholar] [CrossRef]
- Creelman, M.; Kumauchi, M.; Hoff, W.D.; Mathies, R.A. Chromophore Dynamics in the PYP Photocycle from Femtosecond Stimulated Raman Spectroscopy. J. Phys. Chem. B 2014, 118, 659–667. [Google Scholar] [CrossRef]
- Liu, Z.; Gu, K.; Shelby, M.; Roy, D.; Muniyappan, S.; Schmidt, M.; Narayanasamy, S.R.; Coleman, M.; Frank, M.; Kuhl, T.L. In Situ Counter-Diffusion Crystallization and Long-Term Crystal Preservation in Microfluidic Fixed Targets for Serial Crystallography. J. Appl. Crystallogr. 2024, 57, 1539–1550. [Google Scholar] [CrossRef]
- Kim, T.W.; Lee, J.H.; Choi, J.; Kim, K.H.; Van Wilderen, L.J.; Guerin, L.; Kim, Y.; Jung, Y.O.; Yang, C.; Kim, J.; et al. Protein Structural Dynamics of Photoactive Yellow Protein in Solution Revealed by Pump-Probe X-Ray Solution Scattering. J. Am. Chem. Soc. 2012, 134, 3145–3153. [Google Scholar] [CrossRef]
- Kim, J.G.; Kim, T.W.; Kim, J.; Ihee, H. Protein Structural Dynamics Revealed by Time-Resolved X-Ray Solution Scattering. Acc. Chem. Res. 2015, 48, 2200–2208. [Google Scholar] [CrossRef] [PubMed]
- Foos, N.; Seuring, C.; Schubert, R.; Burkhardt, A.; Svensson, O.; Meents, A.; Chapman, H.N.; Nanao, M.H. X-Ray and UV Radiation-Damage-Induced Phasing Using Synchrotron Serial Crystallography. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 366–378. [Google Scholar] [CrossRef]
- Gilbile, D.; Shelby, M.L.; Lyubimov, A.Y.; Wierman, J.L.; Monteiro, D.C.F.; Cohen, A.E.; Russi, S.; Coleman, M.A.; Frank, M.; Kuhl, T.L. Plug-and-Play Polymer Microfluidic Chips for Hydrated, Room-Temperature Fixed-Target Serial Crystallography. Lab. Chip 2021, 21, 4831–4845. [Google Scholar] [CrossRef]
- Nass, K.; Meinhart, A.; Barends, T.R.M.; Foucar, L.; Gorel, A.; Aquila, A.; Botha, S.; Doak, R.B.; Koglin, J.; Liang, M.; et al. Protein Structure Determination by Single-Wavelength Anomalous Diffraction Phasing of {X}-Ray Free-Electron Laser Data. IUCrJ 2016, 3, 180–191. [Google Scholar] [CrossRef]
- Nass, K.; Bacellar, C.; Cirelli, C.; Dworkowski, F.; Gevorkov, Y.; James, D.; Johnson, P.J.M.; Kekilli, D.; Knopp, G.; Martiel, I.; et al. Pink-Beam Serial Femtosecond Crystallography for Accurate Structure-Factor Determination at an {X}-Ray Free-Electron Laser. IUCrJ 2021, 8, 905–920. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.J.; Thompson, A.J.; Dijkstal, P.; Appleby, M.; Assmann, G.; Dworkowski, F.S.N.; Hiller, N.; Huang, C.-Y.; Mason, T.; Perrett, S.; et al. Damage before Destruction? {X}-Ray-Induced Changes in Single-Pulse Serial Femtosecond Crystallography. IUCrJ 2025, 12, 358–371. [Google Scholar] [CrossRef]
- Liu, Z.; Gu, K.K.; Shelby, M.L.; Gilbile, D.; Lyubimov, A.Y.; Russi, S.; Cohen, A.E.; Narayanasamy, S.R.; Botha, S.; Kupitz, C.; et al. A User-Friendly Plug-and-Play Cyclic Olefin Copolymer-Based Microfluidic Chip for Room-Temperature, Fixed-Target Serial Crystallography. Acta Crystallogr. D Struct. Biol. 2023, 79, 944–952. [Google Scholar] [CrossRef]
- Jaho, S.; Sallaz-Damaz, Y.; Budayova-Spano, M. Microdialysis On-Chip Crystallization of Soluble and Membrane Proteins with the MicroCrys Platform and in Situ X-Ray Diffraction Case Studies. CrystEngComm 2023, 25, 5513–5523. [Google Scholar] [CrossRef]
- Fan, J.; Jehle, J.A.; Wennmann, J.T. Population Structure of Cydia Pomonella Granulovirus Isolates Revealed by Quantitative Analysis of Genetic Variation. Virus Evol. 2021, 7, veaa073. [Google Scholar] [CrossRef]
- Gati, C.; Oberthuer, D.; Yefanov, O.; Bunker, R.D.; Stellato, F.; Chiu, E.; Yeh, S.M.; Aquila, A.; Basu, S.; Bean, R.; et al. Atomic Structure of Granulin Determined from Native Nanocrystalline Granulovirus Using an X-Ray Free-Electron Laser. Proc. Natl. Acad. Sci. USA 2017, 114, 2247–2252. [Google Scholar] [CrossRef]
- Awel, S.; Kirian, R.A.; Wiedorn, M.O.; Beyerlein, K.R.; Roth, N.; Horke, D.A.; Oberthür, D.; Knoska, J.; Mariani, V.; Morgan, A.; et al. Femtosecond X-Ray Diffraction from an Aerosolized Beam of Protein Nanocrystals. J. Appl. Crystallogr. 2018, 51, 133–139. [Google Scholar] [CrossRef]
- Bücker, R.; Hogan-Lamarre, P.; Mehrabi, P.; Schulz, E.C.; Bultema, L.A.; Gevorkov, Y.; Brehm, W.; Yefanov, O.; Oberthür, D.; Kassier, G.H.; et al. Serial Protein Crystallography in an Electron Microscope. Nat. Commun. 2020, 11, 996. [Google Scholar] [CrossRef] [PubMed]
- Boutet, S.; Lomb, L.; Williams, G.J.; Barends, T.R.M.; Aquila, A.; Doak, R.B.; Weierstall, U.; DePonte, D.P.; Steinbrener, J.; Shoeman, R.L.; et al. High-Resolution Protein Structure Determination by Serial Femtosecond Crystallography. Science 2012, 337, 362–364. [Google Scholar] [CrossRef] [PubMed]
- Wiedorn, M.O.; Oberthür, D.; Bean, R.; Schubert, R.; Werner, N.; Abbey, B.; Aepfelbacher, M.; Adriano, L.; Allahgholi, A.; Al-Qudami, N.; et al. Megahertz Serial Crystallography. Nat. Commun. 2018, 9, 4025. [Google Scholar] [CrossRef]
- Leonarski, F.; Nan, J.; Matej, Z.; Bertrand, Q.; Furrer, A.; Gorgisyan, I.; Bjelčić, M.; Kepa, M.; Glover, H.; Hinger, V.; et al. Kilohertz Serial Crystallography with the JUNGFRAU Detector at a Fourth-Generation Synchrotron Source. IUCrJ 2023, 10, 729–737. [Google Scholar] [CrossRef]
- Martin-Garcia, J.M.; Botha, S.; Hu, H.; Jernigan, R.; Castellví, A.; Lisova, S.; Gil, F.; Calisto, B.; Crespo, I.; Roy-Chowdhury, S.; et al. Serial Macromolecular Crystallography at ALBA Synchrotron Light Source. J. Synchrotron Radiat. 2022, 29, 896–907. [Google Scholar] [CrossRef]
- Monteiro, D.C.F.; Von Stetten, D.; Stohrer, C.; Sans, M.; Pearson, A.R.; Santoni, G.; Van Der Linden, P.; Trebbin, M. 3D-MiXD: 3D-Printed X-Ray-Compatible Microfluidic Devices for Rapid, Low-Consumption Serial Synchrotron Crystallography Data Collection in Flow. IUCrJ 2020, 7, 207–219. [Google Scholar] [CrossRef]
- Galli, L.; Son, S.K.; Barends, T.R.M.; White, T.A.; Barty, A.; Botha, S.; Boutet, S.; Caleman, C.; Doak, R.B.; Nanao, M.H.; et al. Towards Phasing Using High X-Ray Intensity. IUCrJ 2015, 2, 627–634. [Google Scholar] [CrossRef]
- Sugahara, M.; Nakane, T.; Masuda, T.; Suzuki, M.; Inoue, S.; Song, C.; Tanaka, R.; Nakatsu, T.; Mizohata, E.; Yumoto, F.; et al. Hydroxyethyl Cellulose Matrix Applied to Serial Crystallography. Sci. Rep. 2017, 7, 703. [Google Scholar] [CrossRef]
- Beyerlein, K.R.; Dierksmeyer, D.; Mariani, V.; Kuhn, M.; Sarrou, I.; Ottaviano, A.; Awel, S.; Knoska, J.; Fuglerud, S.; Jönsson, O.; et al. Mix-and-Diffuse Serial Synchrotron Crystallography. IUCrJ 2017, 4, 769–777. [Google Scholar] [CrossRef]
- Wranik, M.; Kepa, M.W.; Beale, E.V.; James, D.; Bertrand, Q.; Weinert, T.; Furrer, A.; Glover, H.; Gashi, D.; Carrillo, M.; et al. A Multi-Reservoir Extruder for Time-Resolved Serial Protein Crystallography and Compound Screening at X-Ray Free-Electron Lasers. Nat. Commun. 2023, 14, 7956. [Google Scholar] [CrossRef]
- Wierman, J.L.; Paré-Labrosse, O.; Sarracini, A.; Besaw, J.E.; Cook, M.J.; Oghbaey, S.; Daoud, H.; Mehrabi, P.; Kriksunov, I.; Kuo, A.; et al. Fixed-Target Serial Oscillation Crystallography at Room Temperature. IUCrJ 2019, 6, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.E.; Soltis, S.M.; González, A.; Aguila, L.; Alonso-Mori, R.; Barnes, C.O.; Baxter, E.L.; Brehmer, W.; Brewster, A.S.; Brunger, A.T.; et al. Goniometer-Based Femtosecond Crystallography with X-Ray Free Electron Lasers. Proc. Natl. Acad. Sci. USA 2014, 111, 17122–17127. [Google Scholar] [CrossRef] [PubMed]
- Barends, T.R.M.; Gorel, A.; Bhattacharyya, S.; Schirò, G.; Bacellar, C.; Cirelli, C.; Colletier, J.P.; Foucar, L.; Grünbein, M.L.; Hartmann, E.; et al. Influence of Pump Laser Fluence on Ultrafast Myoglobin Structural Dynamics. Nature 2024, 626, 905–911. [Google Scholar] [CrossRef] [PubMed]
- Schirò, G.; Woodhouse, J.; Weik, M.; Schlichting, I.; Shoeman, R.L. Simple and Efficient System for Photoconverting Light-Sensitive Proteins in Serial Crystallography Experiments. J. Appl. Crystallogr. 2017, 50, 932–939. [Google Scholar] [CrossRef]
- Fadini, A.; Hutchison, C.D.M.; Morozov, D.; Chang, J.; Maghlaoui, K.; Perrett, S.; Luo, F.; Kho, J.C.X.; Romei, M.G.; Morgan, R.M.L.; et al. Serial Femtosecond Crystallography Reveals That Photoactivation in a Fluorescent Protein Proceeds via the Hula Twist Mechanism. J. Am. Chem. Soc. 2023, 145, 15796–15808. [Google Scholar] [CrossRef]
- Kern, J.; Tran, R.; Alonso-Mori, R.; Koroidov, S.; Echols, N.; Hattne, J.; Ibrahim, M.; Gul, S.; Laksmono, H.; Sierra, R.G.; et al. Taking Snapshots of Photosynthetic Water Oxidation Using Femtosecond X-Ray Diffraction and Spectroscopy. Nat. Commun. 2014, 5, 4371. [Google Scholar] [CrossRef] [PubMed]
- Hattne, J.; Echols, N.; Tran, R.; Kern, J.; Gildea, R.J.; Brewster, A.S.; Alonso-Mori, R.; Glöckner, C.; Hellmich, J.; Laksmono, H.; et al. Accurate Macromolecular Structures Using Minimal Measurements from X-Ray Free-Electron Lasers. Nat. Methods 2014, 11, 545–548. [Google Scholar] [CrossRef]
- Samarkina, O.N.; Popova, A.G.; Gvozdik, E.Y.; Chkalina, A.V.; Zvyagin, I.V.; Rylova, Y.V.; Rudenko, N.V.; Lusta, K.A.; Kelmanson, I.V.; Gorokhovatsky, A.Y.; et al. Universal and Rapid Method for Purification of GFP-like Proteins by the Ethanol Extraction. Protein Expr. Purif. 2009, 65, 108–113. [Google Scholar] [CrossRef]
- Dümmler, A.; Lawrence, A.M.; de Marco, A. Simplified Screening for the Detection of Soluble Fusion Constructs Expressed in E. coli using a modular set of vectors. Microb. Cell Factories 2005, 4, 34. [Google Scholar] [CrossRef]
- Van Den Berg, S.; Löfdahl, P.Å.; Härd, T.; Berglund, H. Improved Solubility of TEV Protease by Directed Evolution. J. Biotechnol. 2006, 121, 291–298. [Google Scholar] [CrossRef]
- Schmidt, M.; Pandey, S.; Mancuso, A.; Beam, R. Time-Resolved Serial Femtosecond Crystallography at the European X-Ray Free Electron Laser. Protocol Exchange 2019, 1. [Google Scholar] [CrossRef]
- Kim, Y.; Ganesan, P.; Ihee, H. High-Throughput Instant Quantification of Protein Expression and Purity Based on Photoactive Yellow Protein Turn off/on Label. Protein Sci. 2013, 22, 1109–1117. [Google Scholar] [CrossRef]
- Sikorski, M.; Ramilli, M.; de Wijn, R.; Hinger, V.; Mozzanica, A.; Schmitt, B.; Han, H.; Bean, R.; Bielecki, J.; Bortel, G.; et al. First Operation of the JUNGFRAU Detector in 16-Memory Cell Mode at European XFEL. Front. Phys. 2023, 11, 1303247. [Google Scholar] [CrossRef]
- Kirkwood, H.J.; de Wijn, R.; Mills, G.; Letrun, R.; Kloos, M.; Vakili, M.; Karnevskiy, M.; Ahmed, K.; Bean, R.J.; Bielecki, J.; et al. A Multi-Million Image Serial Femtosecond Crystallography Dataset Collected at the European XFEL. Sci. Data 2022, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Kruglik, S.G.; Yoo, B.K.; Franzen, S.; Vos, M.H.; Martin, J.L.; Negrerie, M. Picosecond Primary Structural Transition of the Heme Is Retarded after Nitric Oxide Binding to Heme Proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 13678–13683. [Google Scholar] [CrossRef]
- Mizutani, Y.; Kitagawa, T. Ultrafast Dynamics of Myoglobin Probed by Time-Resolved Resonance Raman Spectroscopy. Chem. Rec. 2001, 1, 258–275. [Google Scholar] [CrossRef]
- Levantino, M.; Lemke, H.T.; Schirò, G.; Glownia, M.; Cupane, A.; Cammarata, M. Observing Heme Doming in Myoglobin with Femtosecond X-Ray Absorption Spectroscopy. Struct. Dyn. 2015, 2, 041713. [Google Scholar] [CrossRef]
- Larsen, D.S.; Vengris, M.; Van Stokkum, I.H.M.; Van Der Horst, M.A.; De Weerd, F.L.; Hellingwerf, K.J.; Van Grondelle, R. Photoisomerization and Photoionization of the Photoactive Yellow Protein Chromophore in Solution. Biophys. J. 2004, 86, 2538–2550. [Google Scholar] [CrossRef]
- Nakamura, R.; Hamada, N. Vibrational Energy Flow in Photoactive Yellow Protein Revealed by Infrared Pump-Visible Probe Spectroscopy. J. Phys. Chem. B 2015, 119, 5957–5961. [Google Scholar] [CrossRef] [PubMed]
- Baca, M.; Borgstahl, G.E.O.; Boissinot, M.; Burke, P.M.; Williams, D.W.R.; Slater, K.A.; Getzoff, E.D. Complete Chemical Structure of Photoactive Yellow Protein: Novel Thioester-Linked 4-Hydroxycinnamyl Chromophore and Photocycle Chemistry. Biochemistry 1994, 33, 14369–14377. [Google Scholar] [CrossRef]
- Genick, U.K.; Devanathan, S.; Meyer, T.E.; Canestrelli, I.L.; Williams, E.; Cusanovich, M.A.; Tollin, G.; Getzoff, E.D. Active Site Mutants Implicate Key Residues for Control of Color and Light Cycle Kinetics of Photoactive Yellow Protein. Biochemistry 1997, 36, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Thomson, B.; Both, J.; Wu, Y.; Parrish, R.M.; Martínez, T.J.; Boxer, S.G. Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation. J. Phys. Chem. B 2019, 123, 4844–4849. [Google Scholar] [CrossRef]
- Schulz, J.; Bielecki, J.; Doak, R.B.; Dörner, K.; Graceffa, R.; Shoeman, R.L.; Sikorski, M.; Thute, P.; Westphal, D.; Mancuso, A.P. A Versatile Liquid-Jet Setup for the European XFEL. J. Synchrotron Radiat. 2019, 26, 339–345. [Google Scholar] [CrossRef]
- Han, H.; Round, E.; Schubert, R.; Gül, Y.; Makroczyová, J.; Meza, D.; Heuser, P.; Aepfelbacher, M.; Barák, I.; Betzel, C.; et al. The XBI BioLab for Life Science Experiments at the European XFEL. J. Appl. Crystallogr. 2021, 54, 7–21. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmidt, C.; Lorenzen, K.; Schulz, J.; Han, H. Standard Sample Preparation for Serial Femtosecond Crystallography. Biomolecules 2025, 15, 1488. https://doi.org/10.3390/biom15111488
Schmidt C, Lorenzen K, Schulz J, Han H. Standard Sample Preparation for Serial Femtosecond Crystallography. Biomolecules. 2025; 15(11):1488. https://doi.org/10.3390/biom15111488
Chicago/Turabian StyleSchmidt, Christina, Kristina Lorenzen, Joachim Schulz, and Huijong Han. 2025. "Standard Sample Preparation for Serial Femtosecond Crystallography" Biomolecules 15, no. 11: 1488. https://doi.org/10.3390/biom15111488
APA StyleSchmidt, C., Lorenzen, K., Schulz, J., & Han, H. (2025). Standard Sample Preparation for Serial Femtosecond Crystallography. Biomolecules, 15(11), 1488. https://doi.org/10.3390/biom15111488







