Transcriptional Profiling Defines Unique Subtypes of Transit Amplifying Neural Progenitors Within the Neonatal Mouse Subventricular Zone
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. SVZ Cell Dissociation
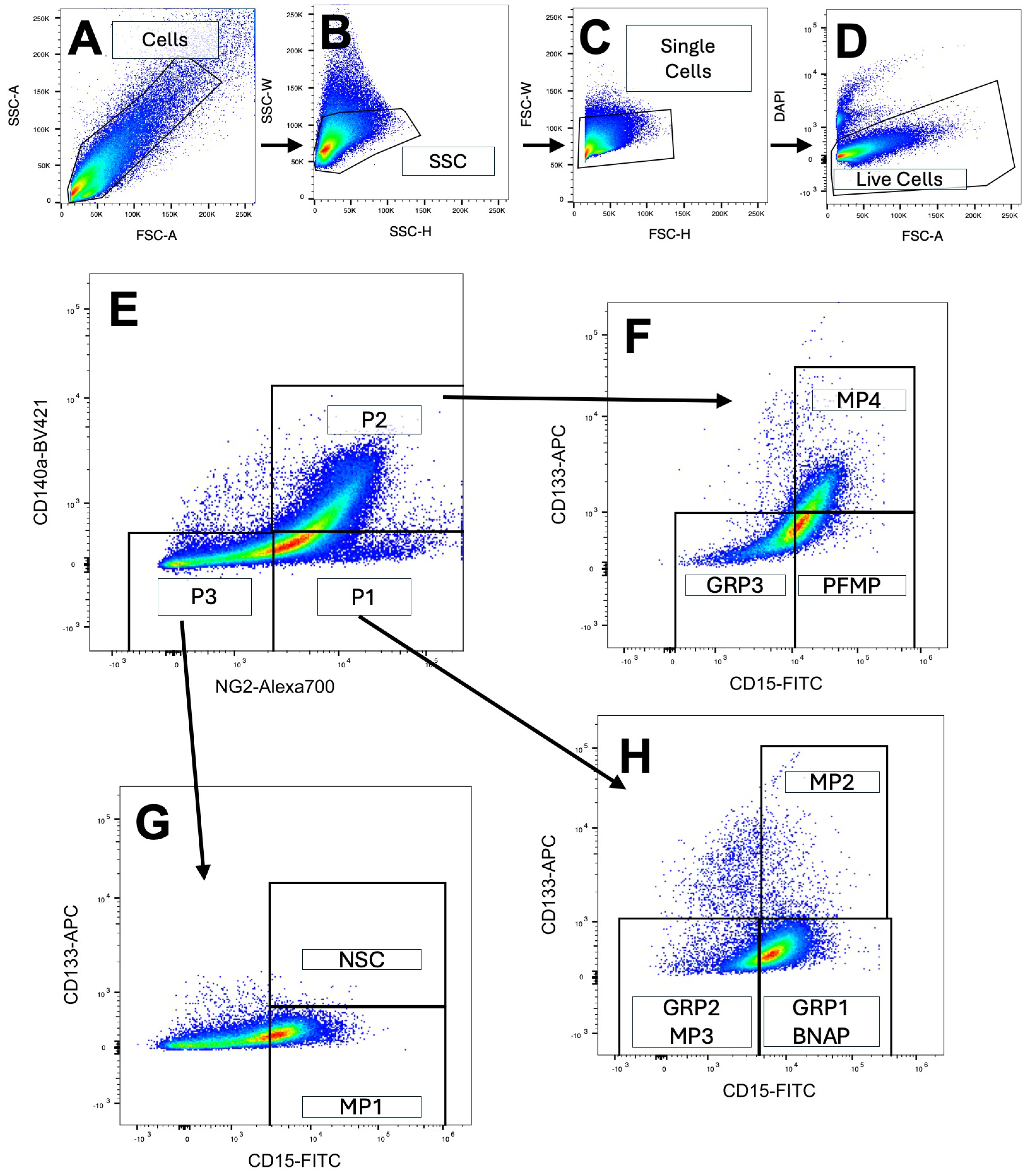
2.3. Flow Cytometry and Cell Sorting
2.4. RNA Sequencing
2.5. Bioinformatics Analysis
2.6. NSC Upset Plot Generation
3. Results
3.1. The Cells of the Subventricular Zone Differentially Express Surface Antigens That Identify Unique Subpopulations of Neural Progenitors
3.2. Neural Progenitor Populations Have Unique Gene Expression
3.3. Surface Markers That Define NP Populations Correlate with Gene Expression
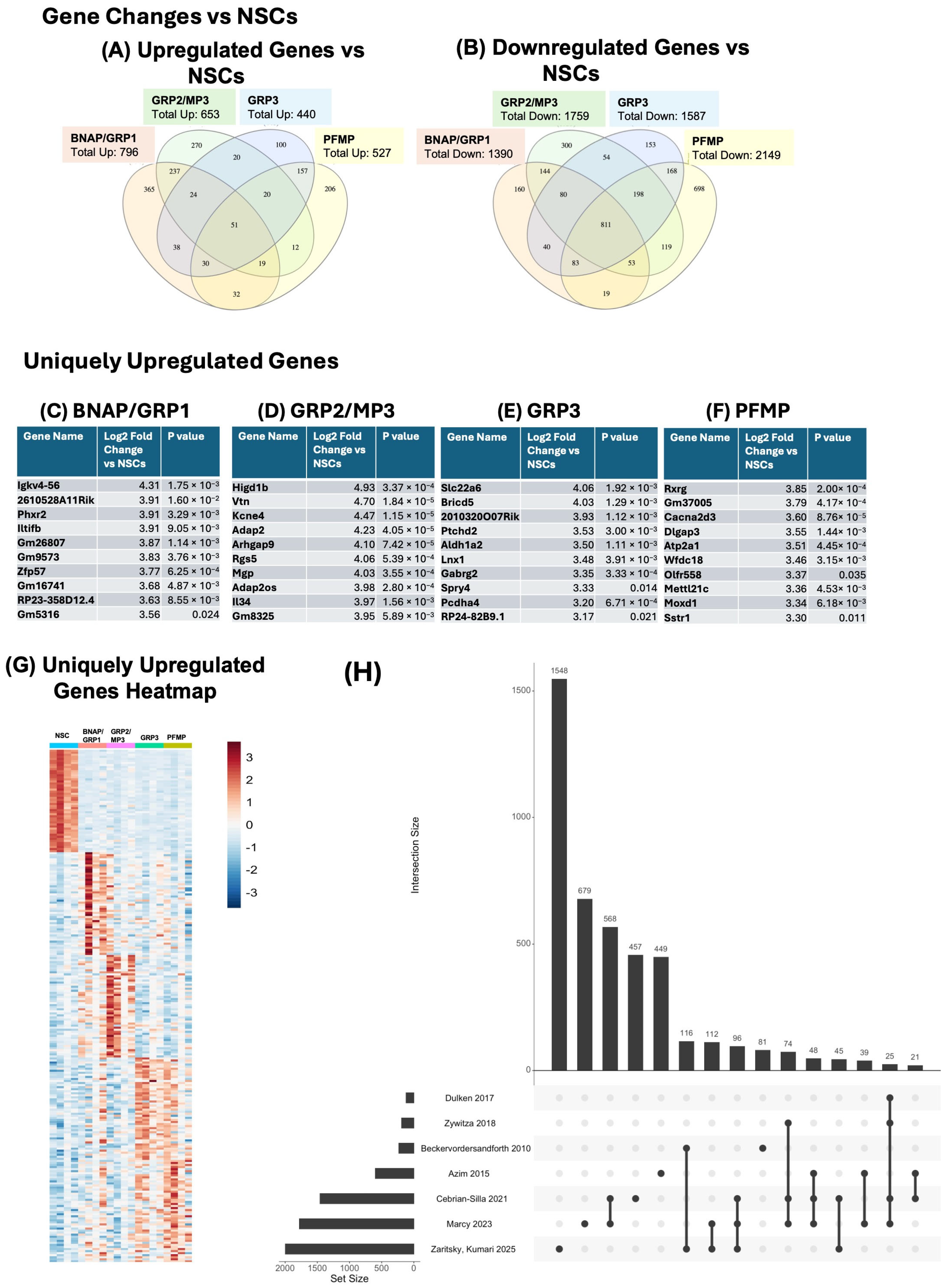
3.4. Each Neural Progenitor Population Upregulates Unique and Shared Genes Compared to Neural Stem Cells
3.5. BNAP/GRP1 Progenitors Share More Gene Expression Similarities with GRP2/MP3, While GRP3s Are More Similar to PFMPs
3.6. Each Neural Progenitor Population Expresses Unique Upregulated and Downregulated Genes
3.7. Comparison of NSC-Enriched Genes to Available Datasets
3.8. Each Neural Progenitor Has a Unique Signature of Up- and Down-Regulated Transcription Factors
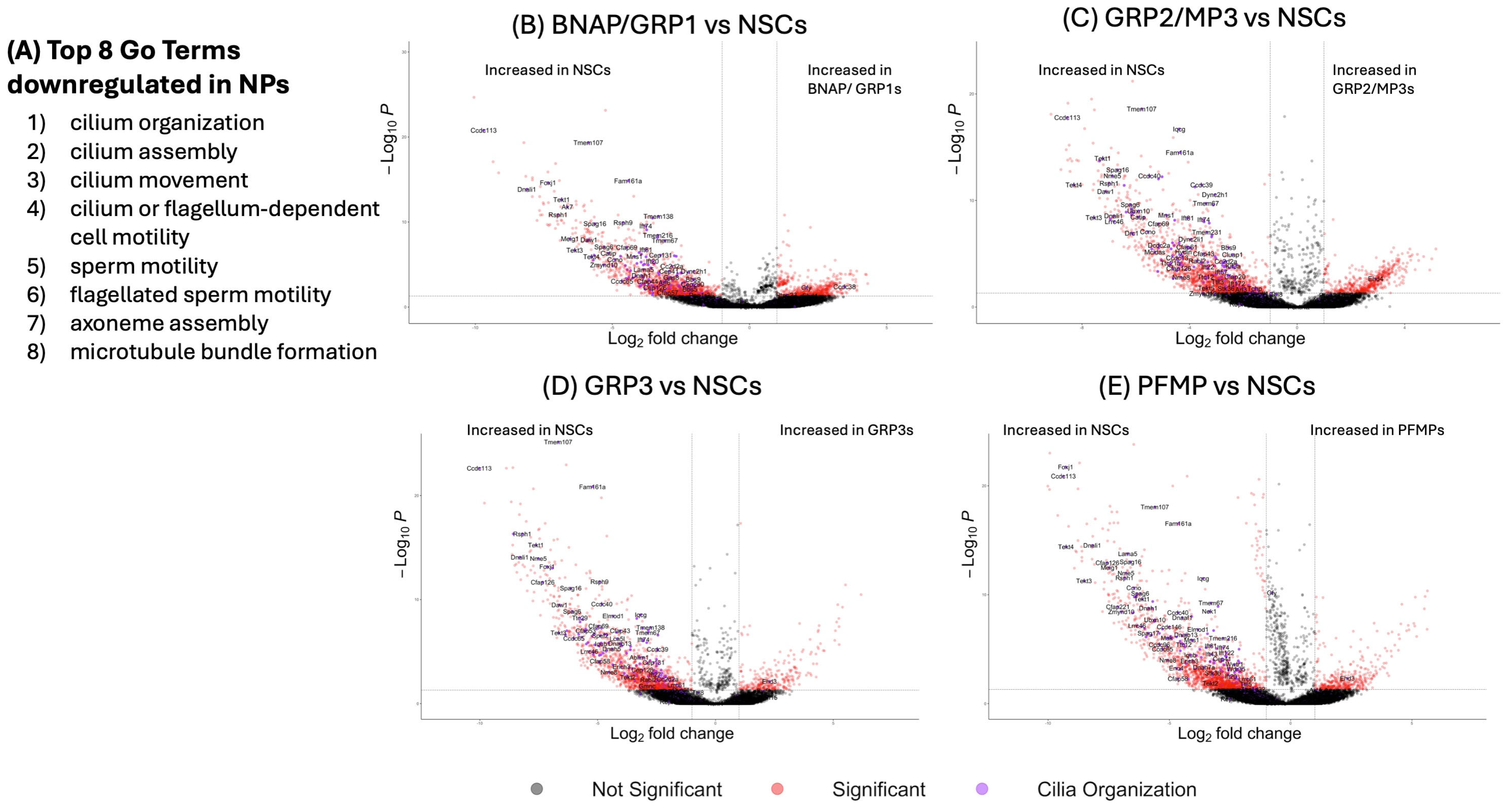
3.9. Neural Progenitor Gene Expression Profiles Indicate Unique Biological Functions in Neurodevelopment
3.9.1. Neural Stem Cells Downregulate Gene Expression in Cilia Organization as They Mature into Neural Progenitors
3.9.2. BNAP/GRP1 and GRP2/MP3 Express Many Immune-Related Genes and Pathways
3.9.3. GRP3 and PFMP Upregulate Genes Related to Neurogenesis and Gliogenesis
3.10. Transcription Factor Analysis Reveals Common and Unique Hubs Between Neural Progenitor Subtypes
4. Discussion
4.1. Shared TFs Are Expressed Across NP Subtypes
4.2. Neural Progenitors Express Unique Transcription Factors
4.3. Protein Interaction Networks and Gene Ontology
4.3.1. GRP1 and GRP2s Express Immune Signature Genes
4.3.2. GRP3 and PFMP Express Gene Networks Related to Neurogenesis
4.4. Transcription Factor Networks
4.5. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NSC | Neural Stem Cell |
| NP | Neural Progenitor |
| SVZ | Subventricular Zone |
| FACS | Fluorescence-Activated Cell Sorting |
| TF | Transcription Factor |
| MP | Multipotential Progenitor |
| GRP | Glial-Restricted Progenitor |
| qNSC | Quiescent Neural Stem Cell |
| aNSC | Activated Neural Stem Cell |
| BNAP/GRP1 | Bipotential Neuron Astrocyte Progenitor/Glial-Restricted Progenitor 1 |
| GRP2/MP3 | Glial-Restricted Progenitor 2/Multipotential Progenitor 3 |
| GRP3 | Glial Restricted Progenitor 3 |
| PFMP | PDGF-FGF-Responsive Multipotential Progenitor |
References
- Bayer, S.A.; Altman, J. Neocortical Development; Raven Press: New York, NY, USA, 1991. [Google Scholar]
- Sanai, N.; Nguyen, T.; Ihrie, R.A.; Mirzadeh, Z.; Tsai, H.H.; Wong, M.; Gupta, N.; Berger, M.S.; Huang, E.; Garcia-Verdugo, J.M.; et al. Corridors of migrating neurons in the human brain and their decline during infancy. Nature 2011, 478, 382–386. [Google Scholar] [CrossRef]
- Doetsch, F.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J. Neurosci. 1997, 17, 5046–5061. [Google Scholar] [CrossRef] [PubMed]
- Doetsch, F.; Caille, I.; Lim, D.A.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 1999, 97, 703–716. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Trupiano, M.X.; Simon, J.; Guo, J.; Anton, E.S. The essential role of primary cilia in cerebral cortical development and disorders. Curr. Top. Dev. Biol. 2021, 142, 99–146. [Google Scholar] [CrossRef] [PubMed]
- Fuentealba, L.C.; Rompani, S.B.; Parraguez, J.I.; Obernier, K.; Romero, R.; Cepko, C.L.; Alvarez-Buylla, A. Embryonic Origin of Postnatal Neural Stem Cells. Cell 2015, 161, 1644–1655. [Google Scholar] [CrossRef]
- Obernier, K.; Cebrian-Silla, A.; Thomson, M.; Parraguez, J.I.; Anderson, R.; Guinto, C.; Rodas Rodriguez, J.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Adult Neurogenesis Is Sustained by Symmetric Self-Renewal and Differentiation. Cell Stem Cell 2018, 22, 221–234.e228. [Google Scholar] [CrossRef]
- Ponti, G.; Obernier, K.; Guinto, C.; Jose, L.; Bonfanti, L.; Alvarez-Buylla, A. Cell cycle and lineage progression of neural progenitors in the ventricular-subventricular zones of adult mice. Proc. Natl. Acad. Sci. USA 2013, 110, E1045–E1054. [Google Scholar] [CrossRef]
- Young, K.M.; Fogarty, M.; Kessaris, N.; Richardson, W.D. Subventricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. J. Neurosci. 2007, 27, 8286–8296. [Google Scholar] [CrossRef]
- Merkle, F.T.; Tramontin, A.D.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc. Natl. Acad. Sci. USA 2004, 101, 17528–17532. [Google Scholar] [CrossRef]
- Menn, B.; Garcia-Verdugo, J.M.; Yaschine, C.; Gonzalez-Perez, O.; Rowitch, D.; Alvarez-Buylla, A. Origin of oligodendrocytes in the subventricular zone of the adult brain. J. Neurosci. 2006, 26, 7907–7918. [Google Scholar] [CrossRef]
- Bandeira, F.; Lent, R.; Herculano-Houzel, S. Changing numbers of neuronal and non-neuronal cells underlie postnatal brain growth in the rat. Proc. Natl. Acad. Sci. USA 2009, 106, 14108–14113. [Google Scholar] [CrossRef]
- Lyck, L.; Kroigard, T.; Finsen, B. Unbiased cell quantification reveals a continued increase in the number of neocortical neurones during early post-natal development in mice. Eur. J. Neurosci. 2007, 26, 1749–1764. [Google Scholar] [CrossRef]
- Ernst, C. Proliferation and Differentiation Deficits are a Major Convergence Point for Neurodevelopmental Disorders. Trends Neurosci. 2016, 39, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-J.; Dai, S.-K.; Duan, C.-H.; Zhang, Z.-H.; Liu, P.-P.; Liu, C.; Du, H.-Z.; Lu, X.-K.; Hu, S.; Li, L.; et al. ASH2L regulates postnatal neurogenesis through Onecut2-mediated inhibition of TGF-β signaling pathway. Cell Death Differ. 2023, 30, 1943–1956. [Google Scholar] [CrossRef] [PubMed]
- Sheu, J.R.; Hsieh, C.Y.; Jayakumar, T.; Tseng, M.F.; Lee, H.N.; Huang, S.W.; Manubolu, M.; Yang, C.H. A Critical Period for the Development of Schizophrenia-Like Pathology by Aberrant Postnatal Neurogenesis. Front. Neurosci. 2019, 13, 635. [Google Scholar] [CrossRef] [PubMed]
- Velloso, F.J.; Shankar, S.; Parpura, V.; Rakic, P.; Levison, S.W. Neural Stem Cells in Adult Mammals are not Astrocytes. ASN Neuro 2022, 14, 17590914221134739. [Google Scholar] [CrossRef]
- Martínez-Cerdeño, V.; Noctor, S.C. Neural Progenitor Cell Terminology. Front. Neuroanat. 2018, 12, 104. [Google Scholar] [CrossRef]
- Moore, L.; Bain, J.M.; Loh, J.M.; Levison, S.W. PDGF-responsive progenitors persist in the subventricular zone across the lifespan. ASN Neuro 2014, 6, AN20120041. [Google Scholar] [CrossRef]
- Zywitza, V.; Misios, A.; Bunatyan, L.; Willnow, T.E.; Rajewsky, N. Single-Cell Transcriptomics Characterizes Cell Types in the Subventricular Zone and Uncovers Molecular Defects Impairing Adult Neurogenesis. Cell Rep. 2018, 25, 2457–2469.E8. [Google Scholar] [CrossRef]
- Mizrak, D.; Levitin, H.M.; Delgado, A.C.; Crotet, V.; Yuan, J.; Chaker, Z.; Silva-Vargas, V.; Sims, P.A.; Doetsch, F. Single-Cell Analysis of Regional Differences in Adult V-SVZ Neural Stem Cell Lineages. Cell Rep. 2019, 26, 394–406.E5. [Google Scholar] [CrossRef]
- Mizrak, D.; Bayin, N.S.; Yuan, J.; Liu, Z.; Suciu, R.M.; Niphakis, M.J.; Ngo, N.; Lum, K.M.; Cravatt, B.F.; Joyner, A.L.; et al. Single-Cell Profiling and SCOPE-Seq Reveal Lineage Dynamics of Adult Ventricular-Subventricular Zone Neurogenesis and NOTUM as a Key Regulator. Cell Rep. 2020, 31, 107805. [Google Scholar] [CrossRef]
- Dulken, B.W.; Leeman, D.S.; Boutet, S.C.; Hebestreit, K.; Brunet, A. Single-Cell Transcriptomic Analysis Defines Heterogeneity and Transcriptional Dynamics in the Adult Neural Stem Cell Lineage. Cell Rep. 2017, 18, 777–790. [Google Scholar] [CrossRef] [PubMed]
- Beckervordersandforth, R.; Tripathi, P.; Ninkovic, J.; Bayam, E.; Lepier, A.; Stempfhuber, B.; Kirchhoff, F.; Hirrlinger, J.; Haslinger, A.; Lie, D.C.; et al. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell 2010, 7, 744–758. [Google Scholar] [CrossRef] [PubMed]
- Panchision, D.M.; Chen, H.L.; Pistollato, F.; Papini, D.; Ni, H.T.; Hawley, T.S. Optimized flow cytometric analysis of central nervous system tissue reveals novel functional relationships among cells expressing CD133, CD15, and CD24. Stem Cells 2007, 25, 1560–1570. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.; Giese, N.A.; Richardson, W.D. Spinal cord oligodendrocytes develop from ventrally derived progenitor cells that express PDGF alpha-receptors. Development 1996, 122, 4085–4094. [Google Scholar] [CrossRef]
- Stallcup, W.B.; Beasley, L. Bipotential glial precursor cells of the optic nerve express the NG2 proteoglycan. J. Neurosci. 1987, 7, 2737–2744. [Google Scholar] [CrossRef]
- Buono, K.D.; Vadlamuri, D.; Gan, Q.; Levison, S.W. Leukemia inhibitory factor is essential for subventricular zone neural stem cell and progenitor homeostasis as revealed by a novel flow cytometric analysis. Dev. Neurosci. 2012, 34, 449–462. [Google Scholar] [CrossRef]
- Buono, K.D.; Goodus, M.T.; Moore, L.; Ziegler, A.N.; levison, S. Multimarker Flow Cytometric Characterization, Isolation and Differentiation of Neural Stem Cells and Progenitors of the Normal and Injured Mouse Subventricular Zone. In Neural Surface Antigens: From Basic Biology Towards Biomedical Applications; Prusac, J., Ed.; Elsevier Press: Amsterdam, The Netherlands, 2015; Volume 1, pp. 175–185. [Google Scholar]
- Velloso, F.J.; Kumari, E.; Buono, K.D.; Frondelli, M.J.; Levison, S.W. Analyzing mouse neural stem cell and progenitor cell proliferation using EdU incorporation and multicolor flow cytometry. STAR Protoc. 2022, 3, 101065. [Google Scholar] [CrossRef]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef]
- Cebrian-Silla, A.; Nascimento, M.A.; Redmond, S.A.; Mansky, B.; Wu, D.; Obernier, K.; Romero Rodriguez, R.; Gonzalez-Granero, S.; Garcia-Verdugo, J.M.; Lim, D.A.; et al. Single-cell analysis of the ventricular-subventricular zone reveals signatures of dorsal and ventral adult neurogenesis. eLife 2021, 10, e67436. [Google Scholar] [CrossRef]
- Marcy, G.; Foucault, L.; Babina, E.; Capeliez, T.; Texeraud, E.; Zweifel, S.; Heinrich, C.; Hernandez-Vargas, H.; Parras, C.; Jabaudon, D.; et al. Single-cell analysis of the postnatal dorsal V-SVZ reveals a role for Bmpr1a signaling in silencing pallial germinal activity. Sci. Adv. 2023, 9, eabq7553. [Google Scholar] [CrossRef]
- Azim, K.; Hurtado-Chong, A.; Fischer, B.; Kumar, N.; Zweifel, S.; Taylor, V.; Raineteau, O. Transcriptional Hallmarks of Heterogeneous Neural Stem Cell Niches of the Subventricular Zone. Stem Cells 2015, 33, 2232–2242. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, K.; Ojima, M.; Otabe, K.; Horie, M.; Koga, H.; Sekiya, I.; Muneta, T. Effects of Different Cell-Detaching Methods on the Viability and Cell Surface Antigen Expression of Synovial Mesenchymal Stem Cells. Cell Transpl. 2017, 26, 1089–1102. [Google Scholar] [CrossRef] [PubMed]
- Samuels, T.J.; Järvelin, A.I.; Ish-Horowicz, D.; Davis, I. Imp/IGF2BP levels modulate individual neural stem cell growth and division through myc mRNA stability. eLife 2020, 9, e51529. [Google Scholar] [CrossRef] [PubMed]
- Willardsen, M.; Hutcheson, D.A.; Moore, K.B.; Vetter, M.L. The ETS transcription factor Etv1 mediates FGF signaling to initiate proneural gene expression during Xenopus laevis retinal development. Mech. Dev. 2014, 131, 57–67. [Google Scholar] [CrossRef]
- Hasegawa, H.; Ashigaki, S.; Takamatsu, M.; Suzuki-Migishima, R.; Ohbayashi, N.; Itoh, N.; Takada, S.; Tanabe, Y. Laminar patterning in the developing neocortex by temporally coordinated fibroblast growth factor signaling. J. Neurosci. 2004, 24, 8711–8719. [Google Scholar] [CrossRef]
- Ling, K.-H.; Hewitt, C.A.; Beissbarth, T.; Hyde, L.; Banerjee, K.; Cheah, P.-S.; Cannon, P.Z.; Hahn, C.N.; Thomas, P.Q.; Smyth, G.K. Molecular networks involved in mouse cerebral corticogenesis and spatio-temporal regulation of Sox4 and Sox11 novel antisense transcripts revealed by transcriptome profiling. Genome Biol. 2009, 10, R104. [Google Scholar] [CrossRef]
- Girard, F.; Eichenberger, S.; Celio, M. Thrombospondin 4 deficiency in mouse impairs neuronal migration in the early postnatal and adult brain. Mol. Cell. Neurosci. 2014, 61, 176–186. [Google Scholar] [CrossRef]
- Zeisel, A.; Hochgerner, H.; Lönnerberg, P.; Johnsson, A.; Memic, F.; Van Der Zwan, J.; Häring, M.; Braun, E.; Borm, L.E.; La Manno, G. Molecular architecture of the mouse nervous system. Cell 2018, 174, 999–1014.E22. [Google Scholar] [CrossRef]
- Jones, F.S.; Holst, B.D.; Minowa, O.; De Robertis, E.M.; Edelman, G.M. Binding and transcriptional activation of the promoter for the neural cell adhesion molecule by HoxC6 (Hox-3.3). Proc. Natl. Acad. Sci. USA 1993, 90, 6557–6561. [Google Scholar] [CrossRef]
- Yan, T.-f.; Wu, M.-j.; Xiao, B.; Hu, Q.; Fan, Y.-H.; Zhu, X.-G. Knockdown of HOXC6 inhibits glioma cell proliferation and induces cell cycle arrest by targeting WIF-1 in vitro and vivo. Pathol.-Res. Pract. 2018, 214, 1818–1824. [Google Scholar] [CrossRef]
- Adachi, K.; Mirzadeh, Z.; Sakaguchi, M.; Yamashita, T.; Nikolcheva, T.; Gotoh, Y.; Peltz, G.; Gong, L.; Kawase, T.; Alvarez-Buylla, A.; et al. Beta-catenin signaling promotes proliferation of progenitor cells in the adult mouse subventricular zone. Stem Cells 2007, 25, 2827–2836. [Google Scholar] [CrossRef]
- Ille, F.; Sommer, L. Wnt signaling: Multiple functions in neural development. Cell. Mol. Life Sci. 2005, 62, 1100–1108. [Google Scholar] [CrossRef]
- Yu, J.M.; Kim, J.H.; Song, G.S.; Jung, J.S. Increase in proliferation and differentiation of neural progenitor cells isolated from postnatal and adult mice brain by Wnt-3a and Wnt-5a. Mol. Cell. Biochem. 2006, 288, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Soon, H.R.; Gaunt, J.R.; Bansal, V.A.; Lenherr, C.; Sze, S.K.; Ch’ng, T.H. Seizure enhances SUMOylation and zinc-finger transcriptional repression in neuronal nuclei. iScience 2023, 26, 107707. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Feng, D.; Lu, Z.; Hu, Z.; Wu, H.; Lian, Y.; Li, D.; Yan, Z.; Li, Y.; Wang, X. Neurod1 mediates the reprogramming of NG2 glial into neurons in vitro. Gene Expr. Patterns 2023, 47, 119305. [Google Scholar] [CrossRef] [PubMed]
- Allen Institute for Brain Science. Allen Mouse Brain Atlas [Dataset]. 2004. Available online: https://mouse.brain-map.org/ (accessed on 6 August 2025).
- Manning, L.; Ohyama, K.; Saeger, B.; Hatano, O.; Wilson, S.A.; Logan, M.; Placzek, M. Regional morphogenesis in the hypothalamus: A BMP-Tbx2 pathway coordinates fate and proliferation through Shh downregulation. Dev. Cell 2006, 11, 873–885. [Google Scholar] [CrossRef]
- Decaesteker, B.; Denecker, G.; Van Neste, C.; Dolman, E.M.; Van Loocke, W.; Gartlgruber, M.; Nunes, C.; De Vloed, F.; Depuydt, P.; Verboom, K. TBX2 is a neuroblastoma core regulatory circuitry component enhancing MYCN/FOXM1 reactivation of DREAM targets. Nat. Commun. 2018, 9, 4866. [Google Scholar] [CrossRef]
- Liu, X.; Miao, Z.; Wang, Z.; Zhao, T.; Xu, Y.; Song, Y.; Huang, J.; Zhang, J.; Xu, H.; Wu, J.; et al. TBX2 overexpression promotes proliferation and invasion through epithelial-mesenchymal transition and ERK signaling pathway. Exp. Ther. Med. 2019, 17, 723–729. [Google Scholar] [CrossRef]
- McIntyre, A.J.; Angel, C.Z.; Smith, J.S.; Templeman, A.; Beattie, K.; Beattie, S.; Ormrod, A.; Devlin, E.; McGreevy, C.; Bothwell, C.; et al. TBX2 acts as a potent transcriptional silencer of tumour suppressor genes through interaction with the CoREST complex to sustain the proliferation of breast cancers. Nucleic Acids Res. 2022, 50, 6154–6173. [Google Scholar] [CrossRef]
- Souroullas, G.P.; Salmon, J.M.; Sablitzky, F.; Curtis, D.J.; Goodell, M.A. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell 2009, 4, 180–186. [Google Scholar] [CrossRef]
- Wang, S.; Ren, D.; Arkoun, B.; Kaushik, A.-L.; Matherat, G.; Lécluse, Y.; Filipp, D.; Vainchenker, W.; Raslova, H.; Plo, I. Lyl-1 regulates primitive macrophages and microglia development. Commun. Biol. 2021, 4, 1382. [Google Scholar] [CrossRef] [PubMed]
- Jalali, A.; Bassuk, A.G.; Kan, L.; Israsena, N.; Mukhopadhyay, A.; McGuire, T.; Kessler, J.A. HeyL promotes neuronal differentiation of neural progenitor cells. J. Neurosci. Res. 2011, 89, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Oliver, C.H.; Nichols, J.; Watson, C.J. The KRAB domain zinc finger protein, Zfp157, is expressed in multiple tissues during mouse embryogenesis and in specific cells in adult mammary gland and skin. genesis 2013, 51, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Hoser, M.; Potzner, M.R.; Koch, J.M.; Bösl, M.R.; Wegner, M.; Sock, E. Sox12 deletion in the mouse reveals nonreciprocal redundancy with the related Sox4 and Sox11 transcription factors. Mol. Cell. Biol. 2008, 28, 4675–4687. [Google Scholar] [CrossRef]
- Bergsland, M.; Werme, M.; Malewicz, M.; Perlmann, T.; Muhr, J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006, 20, 3475–3486. [Google Scholar] [CrossRef]
- Li, X.; Newbern, J.M.; Wu, Y.; Morgan-Smith, M.; Zhong, J.; Charron, J.; Snider, W.D. MEK Is a Key Regulator of Gliogenesis in the Developing Brain. Neuron 2012, 75, 1035–1050. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y. ETV5 is Essential for Neuronal Differentiation of Human Neural Progenitor Cells by Repressing NEUROG2 Expression. Stem Cell Rev. Rep. 2019, 15, 703–716. [Google Scholar] [CrossRef]
- Kastner, P.; Mark, M.; Ghyselinck, N.; Krezel, W.; Dupé, V.; Grondona, J.M.; Chambon, P. Genetic evidence that the retinoid signal is transduced by heterodimeric RXR/RAR functional units during mouse development. Development 1997, 124, 313–326. [Google Scholar] [CrossRef]
- Molotkova, N.; Molotkov, A.; Duester, G. Role of retinoic acid during forebrain development begins late when Raldh3 generates retinoic acid in the ventral subventricular zone. Dev. Biol. 2007, 303, 601–610. [Google Scholar] [CrossRef]
- Janesick, A.; Wu, S.C.; Blumberg, B. Retinoic acid signaling and neuronal differentiation. Cell. Mol. Life Sci. 2015, 72, 1559–1576. [Google Scholar] [CrossRef]
- Wang, T.W.; Zhang, H.; Parent, J.M. Retinoic acid regulates postnatal neurogenesis in the murine subventricular zone-olfactory bulb pathway. Development 2005, 132, 2721–2732. [Google Scholar] [CrossRef] [PubMed]
- Krezel, W.; Dupe, V.; Mark, M.; Dierich, A.; Kastner, P.; Chambon, P. RXR gamma null mice are apparently normal and compound RXR alpha +/–/RXR beta –/–/RXR gamma –/– mutant mice are viable. Proc. Natl. Acad. Sci. USA 1996, 93, 9010–9014. [Google Scholar] [CrossRef] [PubMed]
- Chiang, M.Y.; Misner, D.; Kempermann, G.; Schikorski, T.; Giguère, V.; Sucov, H.M.; Gage, F.H.; Stevens, C.F.; Evans, R.M. An essential role for retinoid receptors RARbeta and RXRgamma in long-term potentiation and depression. Neuron 1998, 21, 1353–1361. [Google Scholar] [CrossRef] [PubMed]
- Baldassarro, V.A.; Krężel, W.; Fernandez, M.; Schuhbaur, B.; Giardino, L.; Calza, L. The role of nuclear receptors in the differentiation of oligodendrocyte precursor cells derived from fetal and adult neural stem cells. Stem Cell Res. 2019, 37, 101443. [Google Scholar] [CrossRef]
- de la Fuente, A.G.; Errea, O.; van Wijngaarden, P.; Gonzalez, G.A.; Kerninon, C.; Jarjour, A.A.; Lewis, H.J.; Jones, C.A.; Nait-Oumesmar, B.; Zhao, C.; et al. Vitamin D receptor-retinoid X receptor heterodimer signaling regulates oligodendrocyte progenitor cell differentiation. J. Cell Biol. 2015, 211, 975–985. [Google Scholar] [CrossRef]
- Maekawa, M.; Yamaguchi, K.; Nakamura, T.; Shibukawa, R.; Kodanaka, I.; Ichisaka, T.; Kawamura, Y.; Mochizuki, H.; Goshima, N.; Yamanaka, S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature 2011, 474, 225–229. [Google Scholar] [CrossRef]
- Scoville, D.W.; Kang, H.S.; Jetten, A.M. GLIS1-3: Emerging roles in reprogramming, stem and progenitor cell differentiation and maintenance. Stem Cell Investig. 2017, 4, 80. [Google Scholar] [CrossRef]
- Liodis, P.; Denaxa, M.; Grigoriou, M.; Akufo-Addo, C.; Yanagawa, Y.; Pachnis, V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J. Neurosci. 2007, 27, 3078–3089. [Google Scholar] [CrossRef]
- Vogt, D.; Hunt, R.F.; Mandal, S.; Sandberg, M.; Silberberg, S.N.; Nagasawa, T.; Yang, Z.; Baraban, S.C.; Rubenstein, J.L. Lhx6 directly regulates Arx and CXCR7 to determine cortical interneuron fate and laminar position. Neuron 2014, 82, 350–364. [Google Scholar] [CrossRef]
- Garay, P.A.; McAllister, A.K. Novel roles for immune molecules in neural development: Implications for neurodevelopmental disorders. Front. Synaptic Neurosci. 2010, 2, 136. [Google Scholar] [CrossRef]
- Stolp, H. Neuropoietic cytokines in normal brain development and neurodevelopmental disorders. Mol. Cell. Neurosci. 2013, 53, 63–68. [Google Scholar] [CrossRef]
- Tran, P.B.; Banisadr, G.; Ren, D.; Chenn, A.; Miller, R.J. Chemokine receptor expression by neural progenitor cells in neurogenic regions of mouse brain. J. Comp. Neurol. 2007, 500, 1007–1033. [Google Scholar] [CrossRef] [PubMed]
- Veerasammy, S.; Kumari, E.; Goodus, M.T.; Neuberger, E.J.; Santhakumar, V.; Levison, S.W. Consequences of inflammation within neural stem cell niches on development and regeneration. In Frontiers in Stem Cell and Regenerative Medicine; Atta-Ur-Rahman, Anjum, S., Eds.; Bentham: Sharjah, United Arab Emirates, 2017; Volume 2, pp. 23–71. [Google Scholar]
- Tanabe, S.; Yamashita, T. The role of immune cells in brain development and neurodevelopmental diseases. Int. Immunol. 2018, 30, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Mócsai, A.; Ruland, J.; Tybulewicz, V.L. The SYK tyrosine kinase: A crucial player in diverse biological functions. Nat. Rev. Immunol. 2010, 10, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jain, S.; Li, P.; Lin, J.X.; Oh, J.; Qi, C.; Gao, Y.; Sun, J.; Sakai, T.; Naghashfar, Z.; et al. Transcription factors IRF8 and PU.1 are required for follicular B cell development and BCL6-driven germinal center responses. Proc. Natl. Acad. Sci. USA 2019, 116, 9511–9520. [Google Scholar] [CrossRef]
- Kierdorf, K.; Erny, D.; Goldmann, T.; Sander, V.; Schulz, C.; Perdiguero, E.G.; Wieghofer, P.; Heinrich, A.; Riemke, P.; Hölscher, C. Microglia emerge from erythromyeloid precursors via Pu. 1-and Irf8-dependent pathways. Nat. Neurosci. 2013, 16, 273–280. [Google Scholar] [CrossRef]
- Chan, S.F.; Huang, X.; McKercher, S.R.; Zaidi, R.; Okamoto, S.-i.; Nakanishi, N.; Lipton, S.A. Transcriptional profiling of MEF2-regulated genes in human neural progenitor cells derived from embryonic stem cells. Genom. Data 2015, 3, 24–27. [Google Scholar] [CrossRef]
- Covacu, R.; Arvidsson, L.; Andersson, A.s.; Khademi, M.; Erlandsson-Harris, H.; Harris, R.A.; Svensson, M.A.; Olsson, T.; Brundin, L. TLR activation induces TNF-α production from adult neural stem/progenitor cells. J. Immunol. 2009, 182, 6889–6895. [Google Scholar] [CrossRef]
- Lan, X.; Chen, Q.; Wang, Y.; Jia, B.; Sun, L.; Zheng, J.; Peng, H. TNF-α affects human cortical neural progenitor cell differentiation through the autocrine secretion of leukemia inhibitory factor. PLoS ONE 2012, 7, e50783. [Google Scholar] [CrossRef]
- Liu, Y.-P.; Lin, H.-I.; Tzeng, S.-F. Tumor necrosis factor-α and interleukin-18 modulate neuronal cell fate in embryonic neural progenitor culture. Brain Res. 2005, 1054, 152–158. [Google Scholar] [CrossRef]
- Wedel, M.; Fröb, F.; Elsesser, O.; Wittmann, M.-T.; Lie, D.C.; Reis, A.; Wegner, M. Transcription factor Tcf4 is the preferred heterodimerization partner for Olig2 in oligodendrocytes and required for differentiation. Nucleic Acids Res. 2020, 48, 4839–4857. [Google Scholar] [CrossRef] [PubMed]
- Uittenbogaard, M.; Chiaramello, A. Expression of the bHLH transcription factor Tcf12 (ME1) gene is linked to the expansion of precursor cell populations during neurogenesis. Gene Expr. Patterns 2002, 1, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pol, S.U.; Haberman, A.K.; Wang, C.; O’Bara, M.A.; Sim, F.J. Transcription factor induction of human oligodendrocyte progenitor fate and differentiation. Proc. Natl. Acad. Sci. USA 2014, 111, E2885–E2894. [Google Scholar] [CrossRef] [PubMed]
- Freudenstein, D.; Lippert, M.; Popp, J.S.; Aprato, J.; Wegner, M.; Sock, E.; Haase, S.; Linker, R.A.; González Alvarado, M.N. Endogenous Sox8 is a critical factor for timely remyelination and oligodendroglial cell repletion in the cuprizone model. Sci. Rep. 2023, 13, 22272. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, G.; Butt, S.J.; Takebayashi, H.; Fishell, G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J. Neurosci. 2007, 27, 7786–7798. [Google Scholar] [CrossRef]
- Merchan-Sala, P.; Nardini, D.; Waclaw, R.R.; Campbell, K. Selective neuronal expression of the SoxE factor, Sox8, in direct pathway striatal projection neurons of the developing mouse brain. J. Comp. Neurol. 2017, 525, 2805–2819. [Google Scholar] [CrossRef]
- Cai, J.; Chen, Y.; Cai, W.H.; Hurlock, E.C.; Wu, H.; Kernie, S.G.; Parada, L.F.; Lu, Q.R. A crucial role for Olig2 in white matter astrocyte development. Development 2007, 134, 1887–1899. [Google Scholar] [CrossRef]
- Tatsumi, K.; Isonishi, A.; Yamasaki, M.; Kawabe, Y.; Morita-Takemura, S.; Nakahara, K.; Terada, Y.; Shinjo, T.; Okuda, H.; Tanaka, T.; et al. Olig2-Lineage Astrocytes: A Distinct Subtype of Astrocytes That Differs from GFAP Astrocytes. Front. Neuroanat. 2018, 12, 8. [Google Scholar] [CrossRef]
- Takouda, J.; Katada, S.; Imamura, T.; Sanosaka, T.; Nakashima, K. SoxE group transcription factor Sox8 promotes astrocytic differentiation of neural stem/precursor cells downstream of Nfia. Pharmacol. Res. Perspect. 2021, 9, e00749. [Google Scholar] [CrossRef]
- Sun, Y.; Meijer, D.H.; Alberta, J.A.; Mehta, S.; Kane, M.F.; Tien, A.-C.; Fu, H.; Petryniak, M.A.; Potter, G.B.; Liu, Z. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron 2011, 69, 906–917. [Google Scholar] [CrossRef]
- Del Aguila, A.; Adam, M.; Ullom, K.; Shaw, N.; Qin, S.; Ehrman, J.; Nardini, D.; Salomone, J.; Gebelein, B.; Lu, Q.R.; et al. Olig2 defines a subset of neural stem cells that produce specific olfactory bulb interneuron subtypes in the subventricular zone of adult mice. Development 2022, 149, dev200028. [Google Scholar] [CrossRef]
- Ono, K.; Takebayashi, H.; Ikeda, K.; Furusho, M.; Nishizawa, T.; Watanabe, K.; Ikenaka, K. Regional- and temporal-dependent changes in the differentiation of Olig2 progenitors in the forebrain, and the impact on astrocyte development in the dorsal pallium. Dev. Biol. 2008, 320, 456–468. [Google Scholar] [CrossRef]
- Lange, C.; Huttner, W.B.; Calegari, F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell 2009, 5, 320–331. [Google Scholar] [CrossRef]
- Lukaszewicz, A.I.; Anderson, D.J. Cyclin D1 promotes neurogenesis in the developing spinal cord in a cell cycle-independent manner. Proc. Natl. Acad. Sci. USA 2011, 108, 11632–11637. [Google Scholar] [CrossRef]
- Fujikake, K.; Sawada, M.; Hikita, T.; Seto, Y.; Kaneko, N.; Herranz-Pérez, V.; Dohi, N.; Homma, N.; Osaga, S.; Yanagawa, Y. Detachment of chain-forming neuroblasts by fyn-mediated control of cell–cell adhesion in the postnatal brain. J. Neurosci. 2018, 38, 4598–4609. [Google Scholar] [CrossRef]
- Dahlin, A.; Qiu, W.; Litonjua, A.A.; Lima, J.J.; Tamari, M.; Kubo, M.; Irvin, C.G.; Peters, S.P.; Wu, A.C.; Weiss, S.T. The phosphatidylinositide 3-kinase (PI3K) signaling pathway is a determinant of zileuton response in adults with asthma. Pharmacogenom. J. 2018, 18, 665–677. [Google Scholar] [CrossRef]
- Xie, S.; Ni, J.; Guo, H.; Luu, V.; Wang, Y.; Zhao, J.J.; Roberts, T.M. The role of the PIK3CA gene in the development and aging of the brain. Sci. Rep. 2021, 11, 291. [Google Scholar] [CrossRef]
- Pauklin, S.; Madrigal, P.; Bertero, A.; Vallier, L. Initiation of stem cell differentiation involves cell cycle-dependent regulation of developmental genes by Cyclin D. Genes Dev. 2016, 30, 421–433. [Google Scholar] [CrossRef]
- Billon, N.; Carlisi, D.; Datto, M.B.; van Grunsven, L.A.; Watt, A.; Wang, X.F.; Rudkin, B.B. Cooperation of Sp1 and p300 in the induction of the CDK inhibitor p21WAF1/CIP1 during NGF-mediated neuronal differentiation. Oncogene 1999, 18, 2872–2882. [Google Scholar] [CrossRef]
- Mondanizadeh, M.; Arefian, E.; Mosayebi, G.; Saidijam, M.; Khansarinejad, B.; Hashemi, S.M. MicroRNA-124 regulates neuronal differentiation of mesenchymal stem cells by targeting Sp1 mRNA. J. Cell. Biochem. 2015, 116, 943–953. [Google Scholar] [CrossRef]
- Yeo, S.; Bandyopadhyay, S.; Messing, A.; Brenner, M. Transgenic analysis of GFAP promoter elements. Glia 2013, 61, 1488–1499. [Google Scholar] [CrossRef]
- Cao, J.; Li, Y.; Zeng, F.; Liu, X.; Tao, T.; Qin, Z. Propofol Exposure Disturbs the Differentiation of Rodent Neural Stem Cells via an miR-124-3p/Sp1/Cdkn1b Axis. Front. Cell Dev. Biol. 2020, 8, 838. [Google Scholar] [CrossRef]
- Yin, B.-K.; Lázaro, D.; Wang, Z.-Q. TRRAP-mediated acetylation on Sp1 regulates adult neurogenesis. Comput. Struct. Biotechnol. J. 2023, 21, 472–484. [Google Scholar] [CrossRef]
- Opitz, O.G.; Rustgi, A.K. Interaction between Sp1 and cell cycle regulatory proteins is important in transactivation of a differentiation-related gene. Cancer Res. 2000, 60, 2825–2830. [Google Scholar]
- Palazuelos, J.; Klingener, M.; Aguirre, A. TGFbeta signaling regulates the timing of CNS myelination by modulating oligodendrocyte progenitor cell cycle exit through SMAD3/4/FoxO1/Sp1. J. Neurosci. 2014, 34, 7917–7930. [Google Scholar] [CrossRef]
- Melnikova, I.N.; Lin, H.-R.; Blanchette, A.R.; Gardner, P.D. Synergistic transcriptional activation by Sox10 and Sp1 family members. Neuropharmacology 2000, 39, 2615–2623. [Google Scholar] [CrossRef]
- Ozerdem, U.; Grako, K.A.; Dahlin-Huppe, K.; Monosov, E.; Stallcup, W.B. NG2 proteoglycan is expressed exclusively by mural cells during vascular morphogenesis. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2001, 222, 218–227. [Google Scholar] [CrossRef]
- Cho, H.; Kozasa, T.; Bondjers, C.; Betsholtz, C.; Kehrl, J.H. Pericyte-specific expression of Rgs5: Implications for PDGF and EDG receptor signaling during vascular maturation. FASEB J. 2003, 17, 1–17. [Google Scholar] [CrossRef]


| BNAP/GRP1 | GRP2/MP3 | GRP3 | PFMP | |||||
|---|---|---|---|---|---|---|---|---|
| TF | Log2FC vs. NSCs | p Value | Log2FC vs. NSCs | p Value | Log2FC vs. NSCs | p Value | Log2FC vs. NSCs | p Value |
| Foxj1 | −7.35 | 2.4 × 10−15 | −7.34 | 6.8 × 10−14 | −7.14 | 6.8 × 10−14 | −9.28 | 1.9 × 10−22 |
| Zfp474 | −5.39 | 8.0 × 10−5 | −6.86 | 4.8 × 10−7 | −6.73 | 4.8 × 10−7 | −7.76 | 3.6 × 10−9 |
| Trp73 | −4.36 | 8.8 × 10−5 | −5.38 | 2.8 × 10−7 | −5.98 | 2.8 × 10−7 | −5.61 | 7.1 × 10−6 |
| Emx1 | −4.17 | 2.0 × 10−5 | −5.33 | 1.5 × 10−5 | −4.26 | 1.5 × 10−5 | −4.46 | 2.8 × 10−6 |
| Rfx2 | −3.09 | 1.1 × 10−3 | −4.11 | 1.8 × 10−7 | −4.46 | 1.8 × 10−7 | −6.24 | 1.0 × 10−12 |
| Myb | −4.01 | 2.0 × 10−9 | −4.77 | 2.6 × 10−10 | −4.19 | 2.6 × 10−10 | −4.49 | 5.2 × 10−12 |
| Foxa2 | −2.97 | 0.031 | −4.01 | 2.5 × 10−3 | −4.17 | 2.5 × 10−3 | −5.10 | 1.2 × 10−4 |
| Id4 | −3.53 | 9.7 × 10−5 | −3.90 | 4.6 × 10−8 | −4.52 | 4.6 × 10−8 | −3.53 | 6.7 × 10−5 |
| Hopx | −3.17 | 3.8 × 10−5 | −3.77 | 7.0 × 10−8 | −3.88 | 7.0 × 10−8 | −3.54 | 9.3 × 10−7 |
| Pbx4 | −3.34 | 2.9 × 10−3 | −5.09 | 6.0 × 10−8 | −3.34 | 1.2 × 10−3 | −2.42 | 0.042 |
| A. BNAP/GRP1 Downregulated | ||
| TF | Log2FC vs. NSCs | p Value |
| Sox21 | −2.23 | 0.041 |
| Tsc22d1 | −1.94 | 7.43 × 10−3 |
| B. GRP2/MP3 Downregulated | ||
| TF | Log2FC vs. NSCs | p Value |
| Barx2 | −3.48 | 6.41E × 10−3 |
| Vsx2 | −3.26 | 0.015 |
| Msx1 | −3.01 | 0.019 |
| Nr2e1 | −3.01 | 2.71 × 10−3 |
| Smad9 | −2.96 | 9.08 × 10−3 |
| C. GRP3 Downregulated | ||
| TF | Log2FC vs. NSCs | p Value |
| Tgif1 | −2.27 | 0.046 |
| Fosb | −1.98 | 0.047 |
| Shox2 | −1.45 | 0.043 |
| Atf5 | −1.24 | 0.033 |
| D. PFMP Downregulated | ||
| TF | Log2FC vs. NSCs | p Value |
| Lbx2 | −4.70 | 4.67 × 10−5 |
| Nkx2-4 | −4.57 | 2.72 × 10−4 |
| T | −3.91 | 8.99 × 10−4 |
| Hoxc11 | −3.40 | 9.23 × 10−3 |
| Tbx1 | −3.18 | 0.024 |
| A. BNAP/GRP1 Upregulated | ||
| TF | Log2FC vs. NSCs | p Value |
| Zfp57 | 3.77 | 6.25 × 10−4 |
| Hoxc6 | 3.21 | 0.015 |
| Zfp955a | 2.70 | 0.015 |
| Lhx1 | 2.63 | 0.050 |
| Gm13154 | 2.51 | 0.031 |
| B. GRP2/MP3 Upregulated | ||
| TF | Log2FC vs. NSCs | p Value |
| Tbx2 | 3.18 | 0.018 |
| Lyl1 | 3.10 | 6.1 × 10−3 |
| Heyl | 2.85 | 0.044 |
| Rel | 2.74 | 3.1 × 10−3 |
| Hmga1-rs1 | 2.51 | 0.017 |
| C. GRP3 Upregulated | ||
| TF | Log2FC vs. NSCs | p Value |
| Sox12 | 1.97 | 0.011 |
| Etv5 | 1.94 | 8.9 × 10−3 |
| Zfp157 | 1.79 | 4.5 × 10−3 |
| Rfx7 | 1.13 | 0.047 |
| D. PFMP Upregulated | ||
| TF | Log2FC vs. NSCs | p Value |
| Rxrg | 3.85 | 2.0 × 10−4 |
| Glis1 | 2.84 | 0.040 |
| Lhx6 | 2.46 | 0.040 |
| Zfp846 | 1.97 | 0.041 |
| Zfp882 | 1.92 | 0.042 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaritsky, R.; Kumari, E.; Velloso, F.J.; Lemenze, A.; Husain, S.; Levison, S.W. Transcriptional Profiling Defines Unique Subtypes of Transit Amplifying Neural Progenitors Within the Neonatal Mouse Subventricular Zone. Biomolecules 2025, 15, 1438. https://doi.org/10.3390/biom15101438
Zaritsky R, Kumari E, Velloso FJ, Lemenze A, Husain S, Levison SW. Transcriptional Profiling Defines Unique Subtypes of Transit Amplifying Neural Progenitors Within the Neonatal Mouse Subventricular Zone. Biomolecules. 2025; 15(10):1438. https://doi.org/10.3390/biom15101438
Chicago/Turabian StyleZaritsky, Rebecca, Ekta Kumari, Fernando Janczur Velloso, Alexander Lemenze, Seema Husain, and Steven W. Levison. 2025. "Transcriptional Profiling Defines Unique Subtypes of Transit Amplifying Neural Progenitors Within the Neonatal Mouse Subventricular Zone" Biomolecules 15, no. 10: 1438. https://doi.org/10.3390/biom15101438
APA StyleZaritsky, R., Kumari, E., Velloso, F. J., Lemenze, A., Husain, S., & Levison, S. W. (2025). Transcriptional Profiling Defines Unique Subtypes of Transit Amplifying Neural Progenitors Within the Neonatal Mouse Subventricular Zone. Biomolecules, 15(10), 1438. https://doi.org/10.3390/biom15101438





