Specific Phenylpropanoid Oligomerization in a Neutral Environment by the Recombinant Alkaline Laccase from Paramyrothecium roridum VKM F-3565
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Strain
2.2. Plasmid Construction and Laccase Gene Expression
2.2.1. Construction of an Expression Vector and Heterologous Expression of Recombinant RecLacF-3565 Laccase in E. coli Cells
2.2.2. Construction of an Expression Vector and Heterologous Expression of the Recombinant RecLacF-3565 Laccase in K. phaffii Cells
2.3. Purification of the Recombinant RecLacF-3565 Laccase
2.4. Laccase Activity Assay
2.5. Recombinant Laccase Characterization
2.5.1. Determination of Enzyme Molecular Mass
2.5.2. Enzyme Deglycosilation Assay and Molecular Modelling
2.5.3. Determination of pH and Temperature Optima of Enzyme
2.5.4. Enzyme Stability Determination
2.5.5. Determination of Enzyme Activity in the Presence of Inorganic Metal Salts, Ionic and Nonionic Detergents, Chelators and Organic Solvents
2.5.6. The Kinetic Data Analysis
2.6. Transformation of Phenylpropanoid and Lignin with the Recombinant Laccase
2.6.1. Analysis and Identification of Phenylpropanoid Transformation Products
2.6.2. Analysis of Lignin Transformation Products
3. Results and Discussion
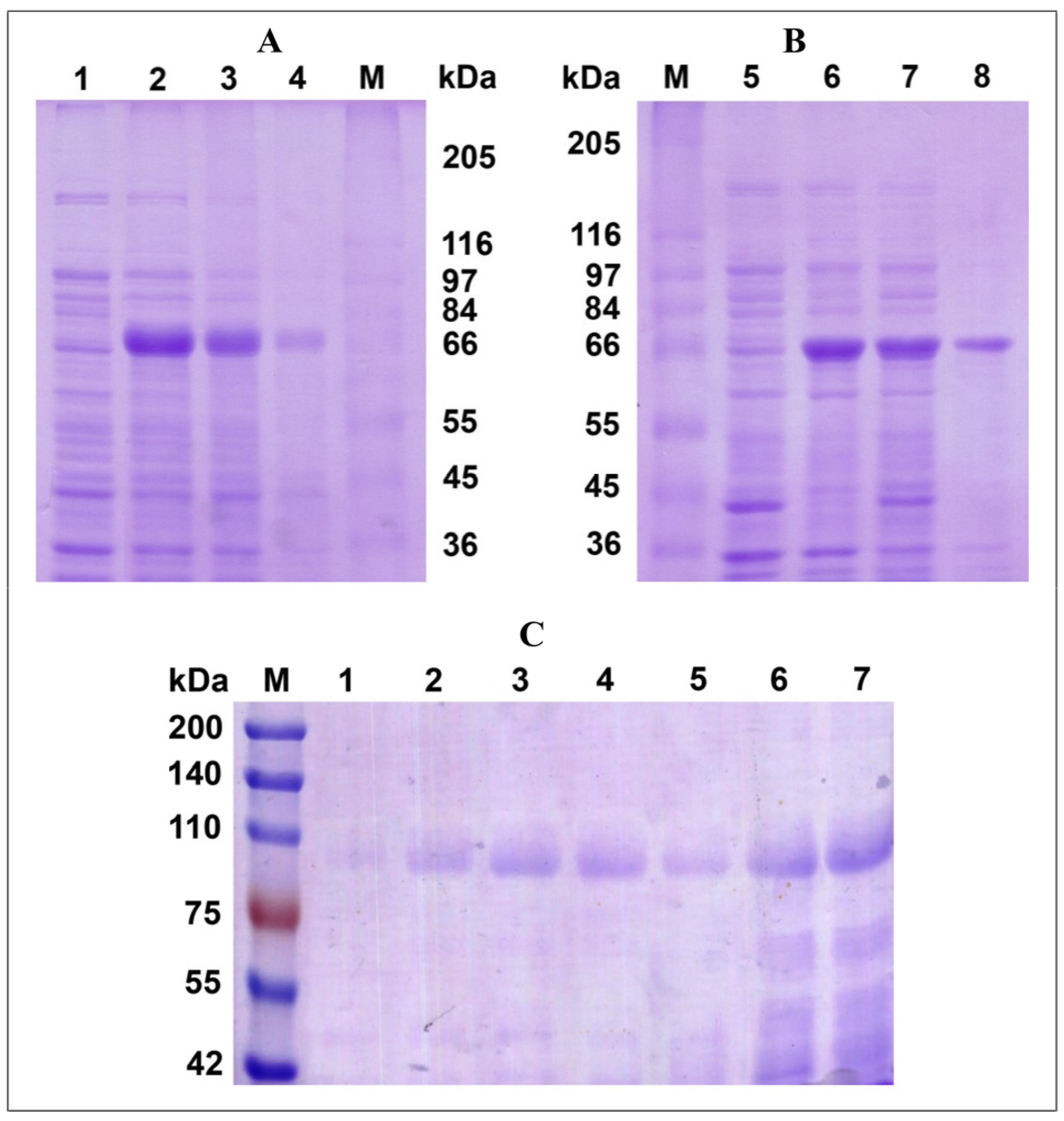
3.1. Production of the Recombinant Alkaliphilic Laccase from the Fungus of P. roridum VKM F-3565 in the E. coli Cells
3.2. Production of the Recombinant Alkaliphilic Laccase from the Fungus of P. roridum VKM F-3565 Strain in the Yeast Cells of K. phaffii
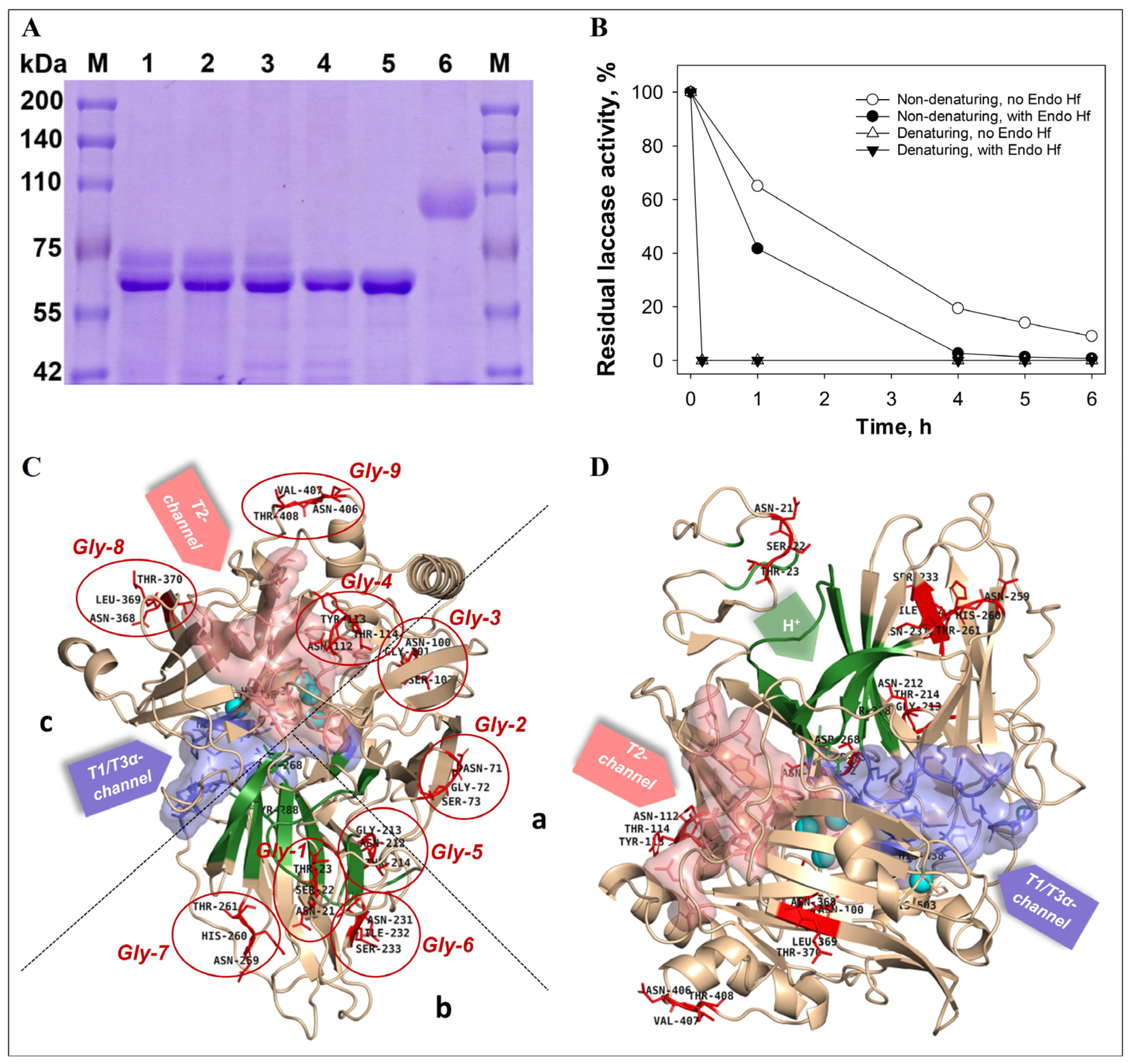
3.3. Role of Glycosylation in the Recombinant Laccase Functioning
3.4. Kinetic Properties of the Recombinant Laccase
3.5. Temperature and pH Optima, Stability of the Recombinant Laccase
3.6. The Influence of Metal Salts, Ionic and Nonionic Detergents, Cation Chelators and Organic Solvents on the RecLacF-3565 Laccase Activity
3.7. Transformation of Phenylpropanoids and Lignin by the Recombinant Laccase
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baldrian, P. Fungal laccases-occurrence and properties. FEMS Microbiol. Rev. 2006, 30, 215–242. [Google Scholar] [CrossRef]
- Maté, D.; Garcıá-Burgos, C.; Garcıá-Ruiz, E.; Ballesteros, A.O.; Camarero, S.; Alcalde, M. Laboratory evolution of high-redox potential laccases. Chem. Biol. 2013, 17, 1030–1041. [Google Scholar] [CrossRef]
- Kudanga, T.; Nemadziva, B.; Le Roes-Hill, M. Laccase catalysis for the synthesis of bioactive compounds. Appl. Microbiol. Biotechnol. 2017, 101, 13–33. [Google Scholar] [CrossRef]
- Cardullo, N.; Muccilli, V.; Tringali, C. Laccase-mediated synthesis of bioactive natural products and their analogues. RSC Chem. Biol. R. Soc. Chem. 2022, 3, 617–647. [Google Scholar] [CrossRef]
- Marienhagen, J.; Bott, M. Metabolic engineering of microorganisms for the synthesis of plant natural products. J. Biotechnol. 2013, 163, 166–178. [Google Scholar] [CrossRef]
- Scheibel, D.M.; Guo, D.; Luo, J.; Gitsov, I. A single enzyme mediates the “quasi-living” formation of multiblock copolymers with a broad biomedical potential. Biomacromolecules 2020, 21, 2132–2146. [Google Scholar] [CrossRef]
- Zhang, Y.; Lv, Z.; Zhou, J.; Xin, F.; Ma, J.; Wu, H.; Fang, Y.; Jiang, M.; Dong, W. Application of eukaryotic and prokaryotic laccases in biosensor and biofuel cells: Recent advances and electrochemical aspects. Appl. Microbiol. Biotechnol. 2018, 102, 10409–10423. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Pose-Boirazian, T.; Eibes, G.; McCoubrey, L.E.; Martínez-Costas, J.; Gaisford, S.; Goyanes, A.; Basit, A.W. A customizable 3D printed device for enzymatic removal of drugs in water. Water Res. 2022, 208, 117861. [Google Scholar] [CrossRef] [PubMed]
- Vicente, A.I.; Viña-Gonzalez, J.; Santos-Moriano, P.; Marquez-Alvarez, C.; Ballesteros, A.O.; Alcalde, M. Evolved alkaline fungal laccase secreted by Saccharomyces cerevisiae as useful tool for the synthesis of C–N heteropolymeric dye. J. Mol. Catal. B Enzym. 2016, 134, 323–330. [Google Scholar] [CrossRef]
- Witayakran, S.; Ragauskas, A.J. Synthetic applications of laccase in green chemistry. Adv. Synth. Catal. 2009, 351, 1187–1209. [Google Scholar] [CrossRef]
- Kolomytseva, M.P.; Myasoedova, N.M.; Chernykh, A.M.; Gaidina (“Samoilova”), A.S.; Shebanova, A.D.; Baskunov, B.P.; Aschenbrenner, J.; Rosengarten, J.F.; Renfeld, Z.V.; Gasanov, N.B.; et al. Laccase isoform diversity in basidiomycete Lentinus strigosus 1566: Potential for phenylpropanoid polymerization. Int. J. Biol. Macromol. 2019, 137, 1199–1210. [Google Scholar] [CrossRef] [PubMed]
- Janusz, G.; Pawlik, A.; Świderska-Burek, U.; Polak, J.; Sulej, J.; Jarosz-Wilkołazka, A.; Paszczyński, A. Laccase properties, physiological functions, and evolution. Int. J. Mol. Sci. 2020, 21, 966. [Google Scholar] [CrossRef]
- Scheibel, D.M.; Gitsov, I.P.I.; Gitsov, I. Enzymes in “Green” Synthetic Chemistry: Laccase and Lipase. Molecules 2024, 29, 989. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, H.; Yao, D.; Zhang, X.; Liu, J.; Fang, Z. Method for Recombinant Expression of Basidiomycete Laccase by Aspergillus niger. CN117187084A, 14 July 2023. [Google Scholar]
- Yaver, D.S.; Overjero, M.D.C.; Xu, F.; Nelson, B.A.; Brown, K.M.; Halkier, T.; Bernauer, S.; Brown, S.H.; Kauppinen, S. Molecular characterization of laccase genes from the basidiomycete Coprinus cinereus and heterologous expression of the laccase Lcc1. Appl. Environ. Microbiol. 1999, 65, 4943–4948. [Google Scholar] [CrossRef]
- Xu, G.; Wang, J.; Yin, Q.; Fang, W.; Xiao, Y.; Fang, Z. Expression of a thermo- and alkali-philic fungal laccase in Pichia pastoris and its application. Protein Expr. Purif. 2019, 154, 16–24. [Google Scholar] [CrossRef]
- Kilaru, S.; Hoegger, P.J.; Kües, U. The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr. Genet. 2006, 50, 45–60. [Google Scholar] [CrossRef]
- Yin, Q.; Zhou, G.; Peng, C.; Zhang, Y.; Kües, U.; Liu, J.; Xiao, Y.; Fang, Z. The first fungal laccase with an alkaline pH optimum obtained by directed evolution and its application in indigo dye decolorization. AMB Express 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Xiao, Y.; Yin, Q.; Fang, Z.; Zhang, X.; Fang, W. Fungal Laccase Mutant PIE5, and Expression Strain and Application Thereof. CN109295017B, 24 July 2018. [Google Scholar]
- Wahleithner, J.A.; Xu, F.; Brown, K.M.; Brown, S.H.; Golightly, E.J.; Halkier, T.; Kauppinen, S.; Pederson, A.; Schneider, P. The identification and characterization of four laccases from the plant pathogenic fungus Rhizoctonia solani. Curr. Genet. 1996, 29, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Rogalski, J.; Janusz, G.; Legieć, D.; Cho, N.-S.; Shin, S.-J.; Ohga, S. Purification of extracellular laccase from Rhizoctonia praticola. J. Fac. Agric. Kyushu Univ. 2011, 56, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, E.; Ruiz-Dueñas, F.J.; Kooistra, R.; Ram, A.; Martínez, A.T.; Martínez, M.J. Isolation of two laccase genes from the white-rot fungus Pleurotus eryngii and heterologous expression of the pel3 encoded protein. J. Biotechnol. 2008, 134, 9–19. [Google Scholar] [CrossRef]
- Soden, D.M.; O’callaghan, J.; Dobson, A.D.W. Molecular cloning of a laccase isozyme gene from Pleurotus sajor-caju and expression in the heterologous Pichia pastoris host. Microbiology 2002, 148, 4003–4014. [Google Scholar] [CrossRef]
- Rodríguez, E.D.; Pliego, M.R.; de Salas, C.F.; Aza, T.P.; Molpeceres, G.G.; Camarero, F.S. Tailor-Made Extremophilic Fungal Laccases. WO2023073180A1 (EP4174174A1), 3 May 2023. [Google Scholar]
- Hirai, H.; Kashima, Y.; Hayashi, K.; Sugiura, T.; Yamagishi, K.; Kawagishi, H.; Nishida, T. Efficient expression of laccase gene from white-rot fungus Schizophyllum commune in a transgenic tobacco plant. FEMS Microbiol. Lett. 2008, 286, 130–135. [Google Scholar] [CrossRef][Green Version]
- Yao, B.; Su, X.; Wang, X.; Luo, H.; Bai, Y.; Huang, H.; Wang, Y.; Wang, Y.; Tu, T. Laccase TvLac and Its Encoding Gene and Application. CN110272878A, 13 June 2019. [Google Scholar][Green Version]
- Theerachat, M.; Emond, S.; Cambon, E.; Bordes, F.; Marty, A.; Nicaud, J.-M.; Chulalaksananukul, W.; Guieysse, D.; Remaud-Siméon, M.; Morel, S. Engineering and production of laccase from Trametes versicolor in the yeast Yarrowia lipolytica. Biores. Technol. 2012, 125, 267–274. [Google Scholar] [CrossRef]
- Gouka, R.J.; van der Heiden, M.; Swarthoff, T.; Verrips, C.T. Cloning of a phenol oxidase gene from Acremonium murorum and its expression in Aspergillus awamori. Appl. Environ. Microbiol. 2001, 67, 2610–2616. [Google Scholar] [CrossRef]
- Yachi, H.; Sakai, K. Laccase. WO2022270590A1, 23 June 2022. [Google Scholar]
- Sylke, H.D.; Kerovuo, J.S.; Koch, O.; Habicher, T.; Robertson, D.; Desantis, G.; Mccann, R.; Luginbuhl, P. Laccases for Pulp Bio-Bleaching. PT2059587E, 19 January 2012. [Google Scholar]
- Renfeld, Z.V.; Chernykh, A.M.; Baskunov, B.P.; Gaidina, A.S.; Myasoedova, N.M.; Egorova, A.D.; Moiseeva, O.V.; Gorina, S.Y.; Kolomytseva, M.P. Unusual oligomeric laccase-like oxidases from ascomycete Curvularia geniculata VKM F-3561 polymerizing phenylpropanoids and phenolic compounds under neutral environmental conditions. Microorganisms 2023, 11, 2698. [Google Scholar] [CrossRef] [PubMed]
- Kruus, K.; Kiiskinen, L.-L.; Rättö, M.; Viikari, L.; Saloheimo, M. Novel Laccase Enzyme and the Gene Encoding the Enzyme. EP1294859A1, 26 March 2003. [Google Scholar]
- Kruus, K.; Kiiskinen, L.-L.; Rättö, M.; Viikari, L.; Saloheimo, M. Novel Laccase Enzyme and the Gene Encoding the Enzyme. US20050089980A1, 28 April 2005. [Google Scholar]
- Kruus, K.; Kiiskinen, L.-L.; Rättö, M.; Viikari, L.; Saloheimo, M. Laccase Enzyme and the Gene Encoding the Enzyme. US7183090B2, 27 February 2007. [Google Scholar]
- Andberg, M.; Hakulinen, N.; Auer, S.; Saloheimo, M.; Koivula, A.; Rouvinen, J.; Kruus, K. Essential role of the C-terminus in Melanocarpus albomyces laccase for enzyme production, catalytic properties and structure. FEBS J. 2009, 276, 6285–6300. [Google Scholar] [CrossRef]
- Kiiskinen, L.L.; Viikari, L.; Kruus, K. Purification and characterisation of a novel laccase from the ascomycete Melanocarpus albomyces. Appl. Microbiol. Biotechnol. 2002, 59, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Kiiskinen, L.L. Characterization and heterologous production of a novel laccase from Melanocarpus albomyces. VTT Publ. 2004, 556, 1–94. [Google Scholar]
- Liu, H.; Tong, C.; Du, B.; Liang, S.; Lin, Y. Expression and characterization of LacMP, a novel fungal laccase of Moniliophthora perniciosa FA553. Biotechnol. Lett. 2015, 37, 1829–1835. [Google Scholar] [CrossRef]
- Berka, R.M.; Schneider, P.; Golightly, E.J.; Brown, S.H.; Madden, M.; Brown, K.M.; Halkier, T.; Mondorf, K.; Xu, F. Characterization of the gene encoding an extracellular laccase of Myceliophthora thermophila and analysis of the recombinant enzyme expressed in Aspergillus oryzae. Appl. Environ. Microbiol. 1997, 63, 3151–3157. [Google Scholar] [CrossRef] [PubMed]
- Sulistyaningdyah, W.T.; Ogawa, J.; Tanaka, H.; Maeda, C.; Shimizu, S. Characterization of alkaliphilic laccase activity in the culture supernatant of Myrothecium verrucaria 24G-4 in comparison with bilirubin oxidase. FEMS Microbiol. Lett. 2004, 230, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Renfeld, Z.V.; Chernykh, A.M.; Egorova (Shebanova), A.D.; Baskunov, B.P.; Gaidina, A.S.; Myasoedova, N.M.; Moiseeva, O.V.; Kolomytseva, M.P. The laccase of Myrothecium roridum VKM F-3565: A new look at the fungal laccase tolerance to neutral alkaline conditions. ChemBioChem. 2023, 24, e202200600. [Google Scholar] [CrossRef]
- Yachi, H.; Sakai, K. Laccase. WO2022270589A1, 23 June 2022. [Google Scholar]
- Yang, X.; Gu, C.; Lin, Y. A novel fungal laccase from Sordaria macrospora k-hell: Expression, characterization, and application for lignin degradation. Bioprocess Biosyst. Eng. 2020, 43, 1133–1139. [Google Scholar] [CrossRef] [PubMed]
- del Río, J.C.; Rencoret, J.; Gutiérrez, A.; Elder, T.; Kim, H.; Ralph, J. Lignin monomers from beyond the canonical monolignol biosynthetic pathway: Another brick in the wall. ACS Sustainable Chem. Eng. 2020, 8, 4997–5012. [Google Scholar] [CrossRef]
- Karimi, R.; Rashidinejad, A. Lignans: Properties, health effects, and applications. In Handbook of Food Bioactive Ingredients; Jafari, S.M., Rashidinejad, A., Simal-Gandara, J., Eds.; Springer Nature: Cham, Switzerland, 2023; pp. 545–570. [Google Scholar] [CrossRef]
- Cunha, W.R.; Andrade e Silva, M.L.; Sola Veneziani, R.C.; Ambrosio, S.R.; Bastos, J.K. Lignans: Chemical and biological properties. In Phytochemicals—A Global Perspective of Their Role in Nutrition and Health; Rao, V., Ed.; IntechOpen: London, UK, 2012; pp. 213–234. [Google Scholar] [CrossRef]
- Madshus, I.H. Regulation of intracellular pH in eukaryotic cells. Biochem. J. 1988, 250, 1–8. [Google Scholar] [CrossRef]
- Poolman, B. Physicochemical homeostasis in bacteria. FEMS Microbiol. Rev. 2023, 47, 1–7. [Google Scholar] [CrossRef]
- Podieiablonskaia, E.V.; Kolomytseva, M.P.; Myasoedova, N.M.; Baskunov, B.P.; Chernykh, A.M.; Classen, T.; Pietruszka, J.; Golovleva, L.A. Myrothecium verrucaria F-3851, a producer of laccases transforming phenolic compounds at neutral and alkaline conditions. Microbiology 2017, 86, 370–376. [Google Scholar] [CrossRef]
- Xia, Y.; Li, K.; Li, J.; Wang, T.; Gu, L.; Xun, L. T5 exonuclease-dependent assembly offers a low-cost method for efficient cloning and site-directed mutagenesis. NAR 2019, 47, e15. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Perez, J.; Jeffries, T.W. Mineralization of C-ring-labeled synthetic lignin correlates with the production of lignin peroxidase, not of manganese peroxidase or laccase. Appl. Environ. Microbiol. 1990, 56, 1806–1812. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nat. Publ. Gr. 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Diezel, W.; Kopperschläger, G.; Hofmann, E. An improved procedure for protein staining in polyacrylamide gels with a new type of Coomassie Brilliant Blue. Anal. Biochem. 1972, 48, 617–620. [Google Scholar] [CrossRef]
- Xu, F. Effects of redox potential and hydroxide inhibition on the pH activity profile of fungal laccases. J. Biol. Chem. 1997, 272, 924–928. [Google Scholar] [CrossRef]
- Lineweaver, H.; Burk, D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]
- Yang, J.; Li, W.; Ng, T.B.; Deng, X.; Lin, J.; Ye, X. Laccases: Production, expression regulation, and applications in pharmaceutical. Biodegrad. Front. Microbiol. 2017, 8, 832. [Google Scholar] [CrossRef] [PubMed]
- Arregui, L.; Ayala, M.; Gómez-Gil, X.; Gutiérrez-Soto, G.; Hernández-Luna, C.E.; de los Santos, M.H.; Levin, L.; Rojo-Domínguez, A.; Romero-Martínez, D.; Saparrat, M.C.N.; et al. Laccases: Structure, function, and potential application in water bioremediation. Microb. Cell Fact. 2019, 18, 200. [Google Scholar] [CrossRef]
- Aza, P.; de Salas, F.; Molpeceres, G.; Rodríguez-Escribano, D.; de la Fuente, I.; Camarero, S. Protein engineering approaches to enhance fungal laccase production in S. cerevisiae. Int. J. Mol. Sci. 2021, 22, 1157. [Google Scholar] [CrossRef]
- Flourat, A.; Peru, A.M.; Haudrechy, A.; Renault, J.-H.; Allais, F. First total synthesis of (β-5)-(β-O-4) dihydroxytrimer and dihydrotrimer of coniferyl alcohol (G): Advanced lignin model compounds. Front. Chem. 2019, 7, 842. [Google Scholar] [CrossRef] [PubMed]
- Morreel, K.; Dima, O.; Kim, H.; Lu, F.; Niculaes, C.; Vanholme, R.; Dauwe, R.; Goeminne, G.; Inzé, D.; Messens, E.; et al. Mass spectrometry-based sequencing of lignin oligomers. Plant Physiol. 2010, 153, 1464–1478. [Google Scholar] [CrossRef]
- Adelakun, O.E.; Kudanga, T.; Parker, A.; Green, I.R.; Roes-Hilla, M.; Burton, S.G. Laccase-catalyzed dimerization of ferulic acid amplifies antioxidant activity. J. Mol. Catal. B Enzym. 2012, 74, 29–35. [Google Scholar] [CrossRef]
- Aljawish, A.; Chevalot, I.; Paris, C.; Muniglia, L. Enzymatic oxidation of ferulic acid as a way of preparing new derivatives. BioTech 2022, 11, 55. [Google Scholar] [CrossRef] [PubMed]
- Zerva, A.; Pentari, C.; Termentzi, A.; America, A.H.P.; Zouraris, D.; Bhattacharya, S.K.; Karantonis, A.; Zervakis, G.I.; Topakas, E. Discovery of two novel laccase-like multicopper oxidases from Pleurotus citrinopileatus and their application in phenolic oligomer synthesis. Biotechnol. Biofuels. 2021, 14, 83. [Google Scholar] [CrossRef]
- Carunchio, F.; Crescenzi, C.; Girelli, A.M.; Messina, A.; Tarola, A.M. Oxidation of ferulic acid by laccase: Identification of the products and inhibitory effects of some dipeptides. Talanta 2001, 55, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Bunzel, M.; Ralph, J.; Funk, C.; Steinhart, H. Isolation and identification of a ferulic acid dehydrotrimer from saponified maize bran insoluble fiber. Eur. Food Res. Technol. 2003, 217, 128–133. [Google Scholar] [CrossRef]
- Bunzel, M.; Ralph, J.; Funka, C.; Steinhart, H. Structural elucidation of new ferulic acid-containing phenolic dimers and trimers isolated from maize bran. Tetrahedron Lett. 2005, 46, 5845–5850. [Google Scholar] [CrossRef]
- Funk, C.; Ralph, J.; Steinhart, H.; Bunzel, M. Isolation and structural characterisation of 8–O–4/8–O–4- and 8–8/8–O–4-coupled dehydrotriferulic acids from maize bran. Phytochemistry 2005, 66, 363–371. [Google Scholar] [CrossRef]
- Kishimoto, T.; Hiyama, A.; Toda, H.; Urabe, D. Effect of pH on the dehydrogenative polymerization of monolignols by laccases from Trametes versicolor and Rhus vernicifera. ACS Omega 2022, 7, 9846–9852. [Google Scholar] [CrossRef]
- Okusa, K.; Miyakoshi, T.; Chen, C.-L. Comparative studies on dehydrogenative polymerization of coniferyl alcohol by laccases and peroxidases. Part 1. Preliminary Results. Holzforschung 1996, 50, 15–23. [Google Scholar] [CrossRef]
- Hilgers, R.; Vincken, J.P.; Gruppen, H.; Kabel, M.A. Laccase/mediator systems: Their reactivity toward phenolic lignin structures. ACS Sustain. Chem. Eng. 2018, 6, 2037–2046. [Google Scholar] [CrossRef]
- Ambika; Kumar, V.; Chandra, D.; Thakur, V.; Sharma, U.; Singh, D. Depolymerization of lignin using laccase from Bacillus sp. PCH94 for production of valuable chemicals: A sustainable approach for lignin valorization. Int. J. Biol. Macromol. 2023, 234, 123601. [Google Scholar] [CrossRef]
- Zhou, M.; Fakayode, O.A.; Ren, M.; Li, H.; Liang, J.; Yagoub, A.E.A.; Fan, Z.; Zhou, C. Laccase-catalyzed lignin depolymerization in deep eutectic solvents: Challenges and prospects. Biores. Bioproc. 2023, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Li, J.; Li, P.; Cai, C.; Chen, S.; Ding, B.; Liu, S.; Ge, M.; Jin, M. Efficient lignin biodegradation triggered by alkali-tolerant ligninolytic bacteria through improving lignin solubility in alkaline solution. J. Biores. Bioprod. 2023, 8, 461–477. [Google Scholar] [CrossRef]



| Laccase Source | Production Host | GenBank Acc. No. | MW (SW), kDa | T-Optimum, °C | pH- Optimum | Tested Substrates | Enzyme Yield or Specific Activity | Reference |
|---|---|---|---|---|---|---|---|---|
| Basidiomycetes | ||||||||
| Coprinus cinereus laccase Lcc9 | Aspergillus niger MA70.15 | BK004119.1 | No info | No info | 6.0 7.5 | SGZ dyes (M2GE, KD8B, KM8B, K7R, 6B, IC) | 150 U/L 3138 ± 62 U/L | [14] |
| Coprinus cinereus (Lcc1) | Aspergillus oryzae | AF118267.1 | (66) | 6.0–7.0 | SGZ | 135 mg/L | [15] | |
| Coprinopsis cinerea strain Okayama 7, Lcc9 | Pichia pastoris GS115/pPIC9K | BK004119.1 | 60.2 | 60–70 | 6.0 (with SGZ, 70–75% activity under pH 7.0–8.0) | SGZ, dyes | 1.750–3.138 U/L (315.3 U/mg) | [16,17] |
| Coprinopsis cinerea (Coprinus cinereus) PIE5—mutant Lcc9 | Pichia pastoris GS115/pPIC9K | no | (61) | 60 | 8.5 8.0 7.0 | guaiacol 2,6-DMP indigo blue | 318.4 U/mg (towards ABTS) | [18,19] |
| Rhizoctonia solani wt-lcc4 | Aspergillus oryzae | Z54277.1 | 130 (66) | No info | 6.0–7.0 | SGZ | No info | [20] |
| Rhizoctonia praticola Lac I, Lac IIa, Lac IIb | Not recombinant | no | 215 175 (78) 68 | 60 | 7.4 | SGZ | 2075.9 nkat/mg 1022.0 nkat/mg 692.3 nkat/mg | [21] |
| Pleurotus eryngii (PEL3) | Aspergillus niger MGG029strain/pPEL3G expression vectors | AY686700.1 | (58) | No info | 6.0 | 2,6-DMP | 1.3 U/mg (for pPEL3G), 2.4 U/mg | [22] |
| Pleurotus sajor-caju strain P32-1, Psc lac4 gene | Pichia pastoris GS115 | AF297528.1 | (59) | No info | 6.0 6.5 7.0 | 2,6-DMP SGZ guaiacol | 2100 U/mg (towards ABTS), 4.85 mg/L | [23] |
| Laccases from Pycnoporus cinnabarinus and basidiomycete PM1 L3, L4, L5 | Pichia pastoris, Saccharomyces cerevisiae | No info | No info | No info | 7.0–8.0 8.0–9.0 | 2,6-DMP guaiacol | No info | [24] |
| Schizophyllum commune scL12, scL genes | Nicotiana tabacum | AB015758.1 | No info | No info | 6.0 | 2,6-DMP, trichlorophenol | 13.1 nkat/g 6.7 nkat/g | [25] |
| Trametes versicolor Lcc4, LCC5 | Pichia pastoris GS115, expression vector pPIC9 | U44431.1 Q12717.1 | No info | No info | 7.0 | aflatoxin B1, zearalenone | No info | [26] |
| Trametes versicolor DSM 11,269 (Lcc1) | Y. lipolytica MY1212 | X84683.1 | 75 | 50 | 4.0–7.0 | dyes (Amaranth, Direct Blue 71, Reactive Black 5, RBBR, Acid Violet 17) | 0.25 U/mL | [27] |
| Ascomycetes | ||||||||
| Acremonium murorum war. murorum CBS 157.72 | Aspergillus awamori AWC4.20, expression vector pAWGLA2, pUR7893 Saccharomyces cerevisiae strain VW-K1 | AJ271104.1 | (67) | 60 | 8.5–9.0 | SGZ | 600 mg/L | [28] |
| Chrysocorona lucknowensis strain 5537 | Escherichia coli | PA565680.1 | No info | No info | 7.0 | Protein–protein cross-linking (pea, soybean, and wheat proteins) using DL-catechin as mediator | No info | [29] |
| Cochliobolus heterostrophus (BD22449) Fusarium verticillioides (BD22865) | Aspergillus | No info | No info | No info | 8.0 | SGZ | No info | [30] |
| Curvularia geniculata VKM F-3561 oxidase I oxidase II | Not recombinant | OR250480.1 | 1035 (>200) 870 (>200) | 70 | 7.0–7.5, 7.5–8.5 6.8–7.0 7.0–8.0 | SGZ 2,6-DMP ferulic acid coniferyl alcohol | 15.4 U/mg 22.9 U/mg | [31] |
| Melanocarpus albomyces IMI 255,989 strain | Saccharomyces cerevisiae | AJ571698.1 | (80) | 70 | 7.5 | guaiacol | No info | [32,33,34] |
| Melanocarpus albomyces, lac1 gene | Saccharomyces cerevisiae, expression vector pYES2 | AJ571698.1 | (100) | No info | 4.0–5.0 (70% activity under pH 7.0) | 2,6-DMP | 4.5 nkat/mL (towards ABTS), 7.0 mg/L (102 nkat/mg) | [35] |
| Melanocarpus albomyces VTT D-96490 | Not recombinant | AJ571698.1 | (80) | 60–70 | 6.0–7.0 5.0–7.5 | SGZ guaiacol | 836 nkat/mg (towards ABTS) | [36] |
| Melanocarpus albomyces VTT D-96490 | Trichoderma reesei RutC-30, pLLK13 | AJ571698.1 | (71.3) | No info | 6–7.5 5.0–7.5 | SGZ, guaiacol | 193 nkat/mL, 230 mg/L | [37] |
| Moniliophthora perniciosa FA553, LacMP gene | Pichia pastoris X33, expression plasmid pPICZaA-6AA-LacMP | ABRE01017453.1 | (57) | 45–60 | 7.5 6.5 6.5 | SGZ 2,6-DMP guaiacol | 232 U/L | [38] |
| Myceliophthora thermophila CBS 117.65, lcc1 gene | Aspergillus oryzae strain HowB104, expression vector pMWR3, pUC18, pRaMB5 | XM_003663693.1 | 100–140 (85) | No info | 6.5 | SGZ | 11–19 mg/L | [39] |
| Myrothecium verrucaria 24G-4 | Not recombinant | No info | (62) | 70 | 9.0 | 4-aminoantipyrine and phenol polymerization | 1.23 U/mg (towards 4-aminoantipyrine and phenol) | [40] |
| Myrothecium roridum VKM F-3565 | Not recombinant | OP168887 | 80 (80) | 65 | 7.8 7.4 6.5 7.0 | SGZ 2,6-DMP ferulic acid coniferyl alcohol | 93.9 U/mg | [41] |
| Paramyrothecium sp. strain MM13-F2103 | Escherichia coli | PA564876.1 | No info | No info | 8.5–9.0 | protein–protein cross-linking (pea, soybean, and wheat proteins) using mediators (L-DOPA and DL-catechin) | No info | [42] |
| Sordaria macrospora k-hell, lacSM gene | Escherichia coli Top10 BL21(DE3) pET-30a | NC_089376.1 (locus_tag = “SMAC4_06098”) corresponding to XP_024511624.1 | (67) | 55–60 | 6.0 7.0 5.0 | guaiacol SGZ 2,6-DMP | 239 U/L (towards guaiacol) | [43] |
| Substrates | RecLacF-3565 | WtLacF-3565* | ||||||
|---|---|---|---|---|---|---|---|---|
| pHopt | Km, µM | kcat, min−1 | kcat/Km, min−1·µM−1 | pHopt | Km, µM | kcat, min−1 | kcat/Km, min−1·µM−1 | |
| ABTS | 2.5 | 3703.0 | 13,851.0 | 3.7 | 2.8 | 1135.0 | 2291.6 | 2.0 |
| Syringaldazine | 7.0 | 6.0 | 162.0 | 27.0 | 7.8 | 7.5 | 240.0 | 32.0 |
| 2,6-dimethoxyphenol | 6.0 | 203 | 187.2 | 0.9 | 7.4 | 3500.0 | 851.7 | 0.3 |
| Ferulic acid | 7.5 | 16.7 | 36.0 | 2.2 | 6.5 | 6.2 | 462.4 | 74.6 |
| Coniferyl alcohol | 7.5 | 109.0 | 91.0 | 0.8 | 7.0 | 29.4 | 375.9 | 12.8 |
| Salts | Laccase Activity, % | |||||
|---|---|---|---|---|---|---|
| pH 5.0 | pH 7.2 | |||||
| 5 mM | 10 mM | 100 mM | 5 mM | 10 mM | 100 mM | |
| NaCl | 105.2 ± 1.3 | 99.4 ± 4.8 | 69.7 ± 1.3 | 100.0 ± 0.9 | 96.8 ± 2.0 | 92.8 ± 2.8 |
| KCl | 104.3 ± 7.3 | 101.0 ± 2.8 | 74.3 ± 0.5 | 98.5 ± 3.3 | 94.4 ± 0.5 | 89.8 ± 3.8 |
| NH4Cl | 103.0 ± 2.2 | 101.2 ± 2.5 | 76.0 ± 1.2 | 91.5 ± 1.8 | 90.9 ± 0.1 | 87.4 ± 2.3 |
| CuCl2·2H2O | 203.6 ± 2.3 | 114.1 ± 1.8 | 67.5 ± 1.9 | - | - | - |
| MgCl2·6H2O | 130.0 ± 0.3 | 130.3 ± 2.7 | 66.2 ± 3.0 | 96.0 ± 1.2 | 91.0 ± 0.5 | 77.6 ± 2.4 |
| MnCl2·4H2O | 153.6 ± 0.5 | 152.7 ± 2.2 | 76.9 ± 1.7 | 82.8 ± 1.6 | 78.3 ± 0.5 | 52.6 ± 2.9 |
| CaCl2·H2O | 138.1 ± 2.8 | 143.5 ± 1.0 | 84.6 ± 0.1 | 97.6 ± 8.3 | 97.4 ± 0.2 | 80.8 ± 0.8 |
| CoCl2·6H2O | 149.3 ± 4.3 | 149.5 ± 1.7 | 70.7 ± 1.7 | 59.8 ± 3.2 | 61.1 ± 1.4 | 47.9 ± 3.3 |
| Na2SO4 | 115.0 ± 4.0 | 116.2 ± 5.5 | 167.0 ± 6.0 | 97.4 ± 6.1 | 93.6 ± 0.3 | 111.4 ± 2.0 |
| K2SO4 | 109.1 ± 2.8 | 125.1 ± 3.2 | 164.2 ± 3.6 | 96.1 ± 0.6 | 96.5 ± 0.2 | 98.8 ± 1.5 |
| (NH4)2SO4 | 112.7 ± 2.2 | 125.6 ± 2.2 | 172.4 ± 3.3 | 97.6 ± 0.3 | 97.0 ± 1.8 | 96.0 ± 0.3 |
| CuSO4·5H2O | 201.1 ± 3.7 | 239.7 ± 8.1 | 288.4 ± 8.3 | - | - | - |
| MgSO4·7H2O | 144.3 ± 4.0 | 151.2 ± 2.3 | 195.0 ± 1.4 | 95.2 ± 2.4 | 95.9 ± 5.9 | 94.2 ± 4.2 |
| MnSO4·5H2O | 168.5 ± 7.3 | 188.8 ± 0.6 | 245.3 ± 3.3 | 84.2 ± 1.2 | 80.6 ± 1.2 | 78.5 ± 0.9 |
| ZnSO4·7H2O | 182.7 ± 4.8 | 200.3 ± 5.8 | 266.7 ± 8.4 | 28.2 ± 1.8 | 30.0 ± 0.1 | 29.0 ± 1.4 |
| NaH2PO4·2H2O | 111.0 ± 3.0 | 115.4 ± 0.9 | 162.9 ± 8.1 | 141.7 ± 1.4 | 155.6 ± 0.8 | 101.2 ± 1.8 |
| KH2PO4 | 108.1 ± 0.6 | 113.8 ± 5.7 | 165.0 ± 5.5 | 116.1 ± 2.4 | 141.2 ± 1.8 | 113.0 ± 0.6 |
| Compound | Laccase Activity, % | |||||
|---|---|---|---|---|---|---|
| 0.25% | 0.5% | 1% | 2% | 10% | 20% | |
| pH 5.0 | ||||||
| Tween 20 | n.d. | 59.4 ± 1.1 | 38.0 ± 2.1 | n.d. | n.d. | n.d. |
| Tween 40 | n.d. | 64.3 ± 2.6 | 50.8 ± 1.5 | n.d. | n.d. | n.d. |
| Tween 60 | n.d. | 65.7 ± 1.1 | 52.0 ± 1.8 | n.d. | n.d. | n.d. |
| Tween 80 | n.d. | 59.8 ± 1.1 | 41.8 ± 0.4 | n.d. | n.d. | n.d. |
| Triton X-100 | n.d. | 37.0 ± 3.1 | 20.0 ± 0.5 | n.d. | n.d. | n.d. |
| SDS | n.d. | n.d. | 105.7 ± 0.7 | 115.5 ± 2.5 | n.d. | n.d. |
| EDTA | 28.4 ± 1.7 | 0.0 | 0.0 | 0.0 | n.d. | n.d. |
| CTAB | 0.0 | 0.0 | 0.0 | 0.0 | n.d. | n.d. |
| Ethanol | n.d. | n.d. | n.d. | n.d. | 51.6 ± 3.1 | 22.0 ± 1.9 |
| Acetonitrile | n.d. | n.d. | n.d. | n.d. | 18.1 ± 1.1 | 0.0 |
| Acetone | n.d. | n.d. | n.d. | n.d. | 28.2 ± 1.6 | 0.0 |
| DMSO | n.d. | n.d. | n.d. | n.d. | 9.4 ± 0.9 | 0.0 |
| pH 7.2 | ||||||
| Tween 20 | n.d. | 97.4 ± 0.5 | 90.7 ± 0.7 | n.d. | n.d. | n.d. |
| Tween 40 | n.d. | 99.8 ± 0.2 | 96.1 ± 1.6 | n.d. | n.d. | n.d. |
| Tween 60 | n.d. | 91.8 ± 0.9 | 77.7 ± 0.7 | n.d. | n.d. | n.d. |
| Tween 80 | n.d. | 94.2 ± 0.9 | 88.6 ± 0.2 | n.d. | n.d. | n.d. |
| Triton X-100 | n.d. | 96.0 ± 1.5 | 83.6 ± 4.0 | n.d. | n.d. | n.d. |
| SDS | n.d. | n.d | 53.2 ± 3.2 | 29.8 ± 1.0 | n.d. | n.d. |
| EDTA | 158.7 ± 3.8 | 160.8 ± 3.5 | 161.7 ± 2.3 | 179.2 ± 0.6 | n.d. | n.d. |
| CTAB | 14.2 ± 2.5 | 0.0 | 0.0 | 0.0 | n.d. | n.d. |
| Ethanol | n.d. | n.d. | n.d. | n.d. | 60.8 ± 3.4 | 40.8 ± 0.8 |
| Acetonitrile | n.d. | n.d. | n.d. | n.d. | 36.1 ± 2.4 | 0.0 |
| Acetone | n.d. | n.d. | n.d. | n.d. | 56.1 ± 1.4 | 17.1 ± 1.6 |
| DMSO | n.d | n.d | n.d | n.d | 38.8 ± 1.2 | 14.8 ± 0.3 |
| MW, g/mol | Mass Spectrum (Relative Intensity, %) | Linkage | Oligomericity | Catalyst (pH) | Reference |
|---|---|---|---|---|---|
| coniferyl alcohol (MW = 180 Da) | |||||
| 576 | [M+H]+577(70%), 549(25%), 506(100%), 464(15%), 393(25%) | No info | Trimer | Recombinant laccase or wild type laccase from P. roridum VKM F-3565 (pH 7.2) | present work, [41] |
| 432 | [M+H]+433(100%), 432(86%), 289(59%), 273(66%) | No info | Trimer | Laccase from Lentinus strigosus 1566 (pH 7.2) | [11] |
| 446 | [M+H]+447(31%), 307(11%), 303(17%), 289(100%), 274(12%), 141(20%) | No info | Trimer | ||
| 340 | [M+H]+341(100%), 323(42%), 311(17%), 208(76%) | No info | Dimer | Laccase from Curvularia geniculata VKM F-3561 (pH 7.2) | [31] |
| 340 | [M+H]+341(58%), 323(98%), 311(27%), 175(16%), 161(100%), 137(25%) | No info | Dimer | ||
| 588 | no info | β-5/β-O-4 | Trimer | Chemo-enzymatic pathway using laccase from Trametes versicolor (pH 4.5–5.0) | [61] |
| 358 | [M−H]−357(), 342(15%), 327(24%), 311(18%), 151(100%), 136(35%) | β-β | Dimer (pinoresinol) | Natural oligomers from wild-type poplar | [62] |
| 358 | [M−H]−357(), 342(25%), 339(100%), 327(25%), 221(22%), 203(15%), 191(5%) | β-5 | Dimer | ||
| 376 | [M−H]−375(), 357(6%), 327(98%), 195(15%), 179(3%), 165(4%) | β-O-4 | Dimer | ||
| 572 | [M−H]−571(), 553(12%), 523(100%), 391(29%), 375(4%), 343(20%), 327(3%), 195(), 179(11%) | β-O-4/β-O-4 | Trimer | ||
| 584 | [M−H]−583(), 565(17%), 535(100%), 387(2%), 369(53%), 357(23%), 195(3%) | β-5/β-O-4 | Trimer | ||
| 584 | [M−H]−583(), 565(23%), 535(100%), 387(42%), 373(11%), 357(14%), 343(5%), 195(4%) | β-β/β-O-4 | Trimer | ||
| 618 | [M−H]−617(), 587(5%), 569(100%), 491(18%), 403(35%), 391(5%), 195(1%) | β-5/β-O-4 | Trimer | ||
| ferulic acid (MW = 194 Da) | |||||
| 545 | [M+H]+546(54%), 528(100%), 510(17%), 366(18%) | No info | Dehydroderivative of trimer with [M+H]+548 | Recombinant laccase or wild type laccase from P. roridum VKM F-3565 (pH 7.2) | present work, [41] |
| 547 | [M+H]+548(65%), 530(100%), 368(18%), 277(9%), 259(11%) | No info | Trimer | ||
| 636 | [M+H]+637(50%), 581(100%) | No info | Trimer | Laccase from Curvularia geniculata VKM F-3561 (pH 7.2) | [31] |
| 735 | [M+H]+736(100%), 574(42%), 556(90%), 538(23%) | No info | Tetramer | ||
| 386 | [M−H]−385(100%) ([M+H]+387) | β-5 | Dimer | Laccase from Trametes pubescens strain CBS 696.94 (pH 5.0) | [63] |
| 386 | [M−H]−385(96%), 341(38%), 297(100%) | β-β | Dimer | ||
| 579 | no info | No info | Trimer | ||
| 769 | no info | No info | Tetramer | ||
| 324 | [M+H]+325(87%), 307(100%), 223(94%) | No info | No info | Laccase (culture liquid) of Myrothecium verrucaria VKM F-3851 (pH 7.2) | [49] |
| 386 (P1) | [M−H]+387(70%), 341(49%), 297(21%) | β-β | Dehydrodimer | Recombinant laccase from Myceliophthora thermophila (pH 7.5) | [64] |
| 386 (P2) | [M−H]+387(75%), 343(45%), 297(59%) | β-5 | Dehydrodimer | ||
| 386 (P3) | [M−H]+387(90%), 343(65%), 325(53%), 297(11%) | β-O-4 | Dehydrodimer | ||
| 340 (P4) | [M−H]+341(92%), 323(23%), 235(8%), 177(80%) | β-5 | Dehydrodimer | ||
| 403 (P5) | [M−H]+404(3%), 387(93%), 341(38%), 271(62%), 219(48%), 175(36%) | β-β | Dehydrodimer | ||
| 770 (P6) | [M−H]+771(92%), 753(100%), 717(27%) | No info | DehydroTetramer | ||
| 770 (P7) | [M−H]+771(100%), 735(45%), 689(20%) | No info | Dehydrotetramer | ||
| 298 | [M−H]−297.111(-), 146(-), 109(-) | No info | Double decarboxylated diferulic acid | Laccases from Pleurotus citrinopileatus LGAM 28,684 (pH 4.0) | [65] |
| 342 | [M−H]−341(-), 281(-), 267(-), 209(-), 159(-), 146(-) | No info | Decarboxylated Diferulic acid | ||
| 386 | [M−H]−385(-), 267(-), 239(-) | No info | Diferulic acid | ||
| 578 | [M−H]−577(-), 193(-) | No info | Triferulic acid | ||
| 534 | [M−H]−533(-), 193(-) | No info | Decarboxylated Triferulic acid | ||
| 566 | [M−H]−565(-), 193(-), 178(-), 149(-), 134(-) | No info | Triferulic acid | ||
| 552 | [M−H]−551(-), 193(-), 178(-), 149(-), 134(-) | No info | Decarboxylated Triferulic acid (water molecule missing) | ||
| 386 (P1) | [M−H]−385(43%), 341(100%), 326(24%), 282(8%), 193(16%) | β-4 | Dimer | Laccase from Pyricularia oryzae (pH 6.0) | [66] |
| 386 (P2) | [M−H]−385(16%), 341(68%), 297(7%), 281(15%), 173(42%), 159(73%), 123(100%) | β-O-4 | Dimer | ||
| 358 | no info | β-5 | Dimer | Natural compounds from maize bran fiber | [67,68] |
| 344 | no info | 5-5 | Dimer | ||
| 578 | no info | β-5/β-O-4 | Trimer | ||
| 596 | no info | 5-5/β-O-4 | Trimer | ||
| 578 | [M−H]−577 | 5-5/β-O-4 | Dehydrotrimer | ||
| 578 | [M−H]−577 | β-O-4/β-O-4 | Dehydrotrimer | Natural compound from saponified maize bran | [69] |
| 578 | [M−H]−577 | β-O-4/β-β(cyclic) | Dehydrotrimer | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Renfeld, Z.V.; Chernykh, A.M.; Gorina, S.Y.; Baskunov, B.P.; Moiseeva, O.V.; Trachtmann, N.V.; Validov, S.Z.; Kolomytseva, M.P. Specific Phenylpropanoid Oligomerization in a Neutral Environment by the Recombinant Alkaline Laccase from Paramyrothecium roridum VKM F-3565. Biomolecules 2025, 15, 1437. https://doi.org/10.3390/biom15101437
Renfeld ZV, Chernykh AM, Gorina SY, Baskunov BP, Moiseeva OV, Trachtmann NV, Validov SZ, Kolomytseva MP. Specific Phenylpropanoid Oligomerization in a Neutral Environment by the Recombinant Alkaline Laccase from Paramyrothecium roridum VKM F-3565. Biomolecules. 2025; 15(10):1437. https://doi.org/10.3390/biom15101437
Chicago/Turabian StyleRenfeld, Zhanna V., Alexey M. Chernykh, Sofia Yu. Gorina, Boris P. Baskunov, Olga V. Moiseeva, Natalia V. Trachtmann, Shamil Z. Validov, and Marina P. Kolomytseva. 2025. "Specific Phenylpropanoid Oligomerization in a Neutral Environment by the Recombinant Alkaline Laccase from Paramyrothecium roridum VKM F-3565" Biomolecules 15, no. 10: 1437. https://doi.org/10.3390/biom15101437
APA StyleRenfeld, Z. V., Chernykh, A. M., Gorina, S. Y., Baskunov, B. P., Moiseeva, O. V., Trachtmann, N. V., Validov, S. Z., & Kolomytseva, M. P. (2025). Specific Phenylpropanoid Oligomerization in a Neutral Environment by the Recombinant Alkaline Laccase from Paramyrothecium roridum VKM F-3565. Biomolecules, 15(10), 1437. https://doi.org/10.3390/biom15101437







