Insights into the Allosteric Regulation of Human Hsp90 Revealed by NMR Spectroscopy
Abstract
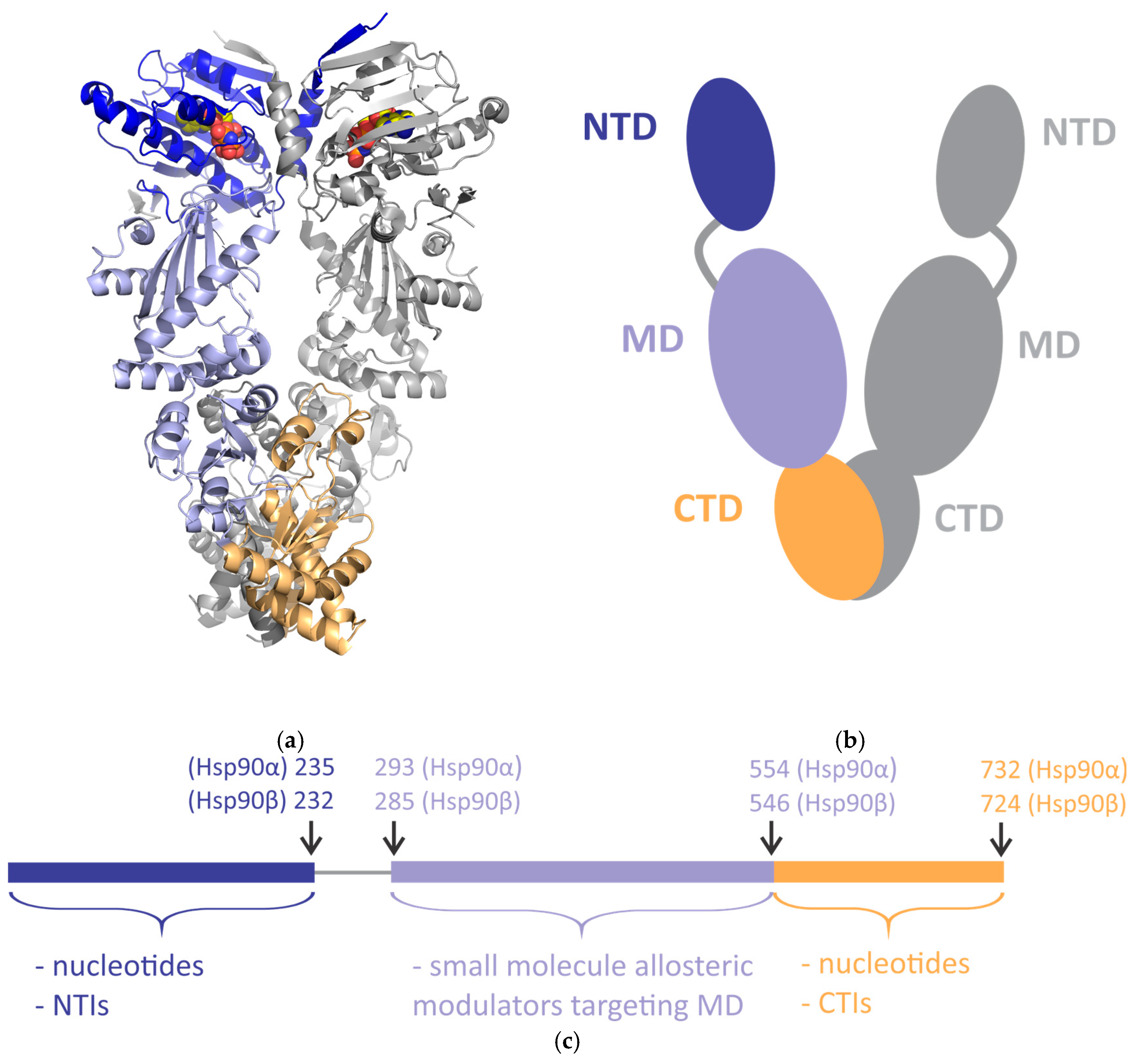
1. Introduction
2. Allosteric Regulation of Human Hsp90 Investigated by NMR Spectroscopy
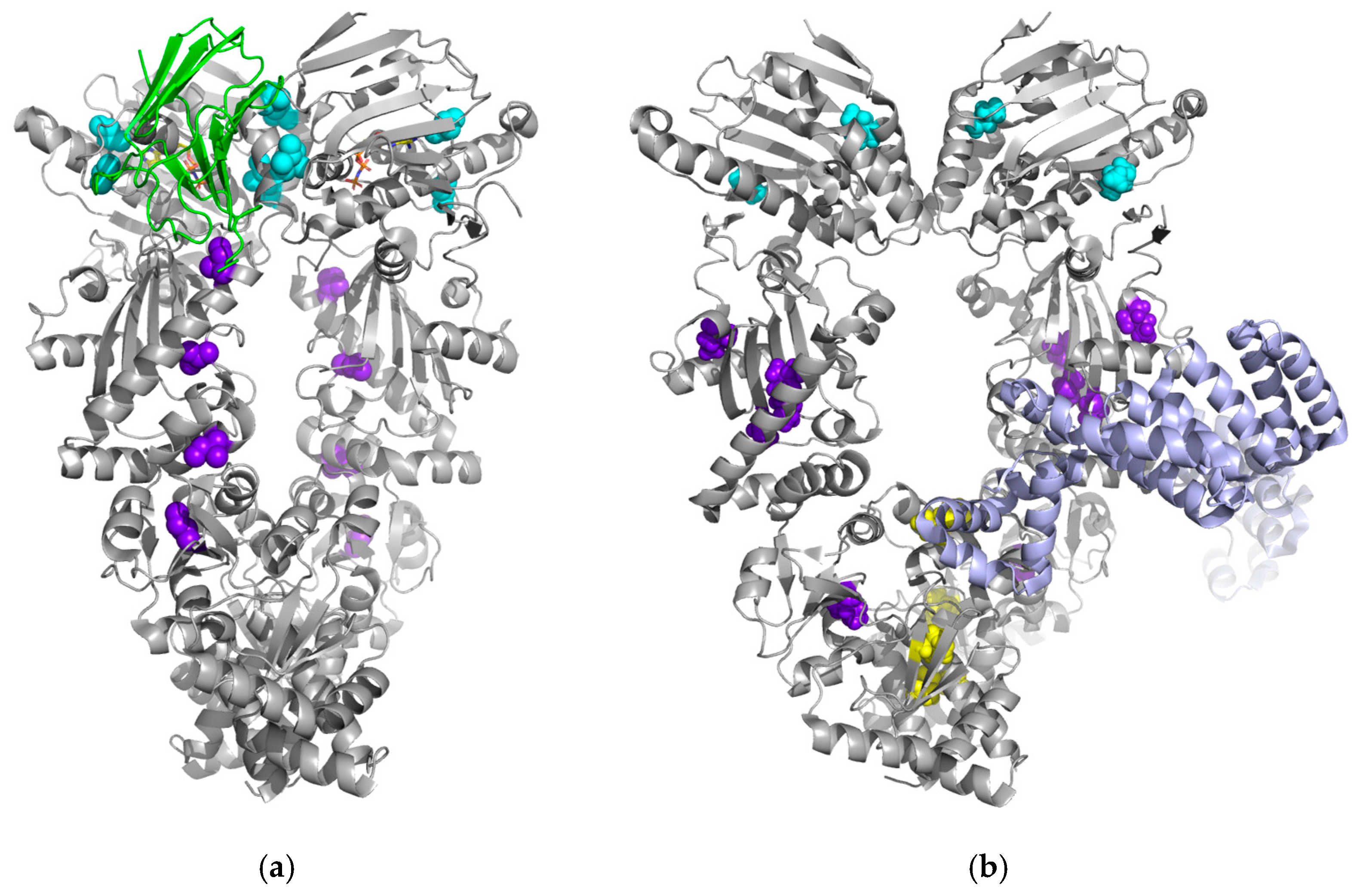
2.1. Allosteric Regulation of Human Hsp90 with CTIs

2.2. Allosteric Regulation of Human Hsp90 with the Synthetic Small-Molecule Allosteric Modulator Targeting the MD
2.3. Allosteric Regulation of Human Hsp90 with Co-Chaperones and Nucleotides
2.3.1. Allosteric Regulation of Hsp90β with p23 and ATP
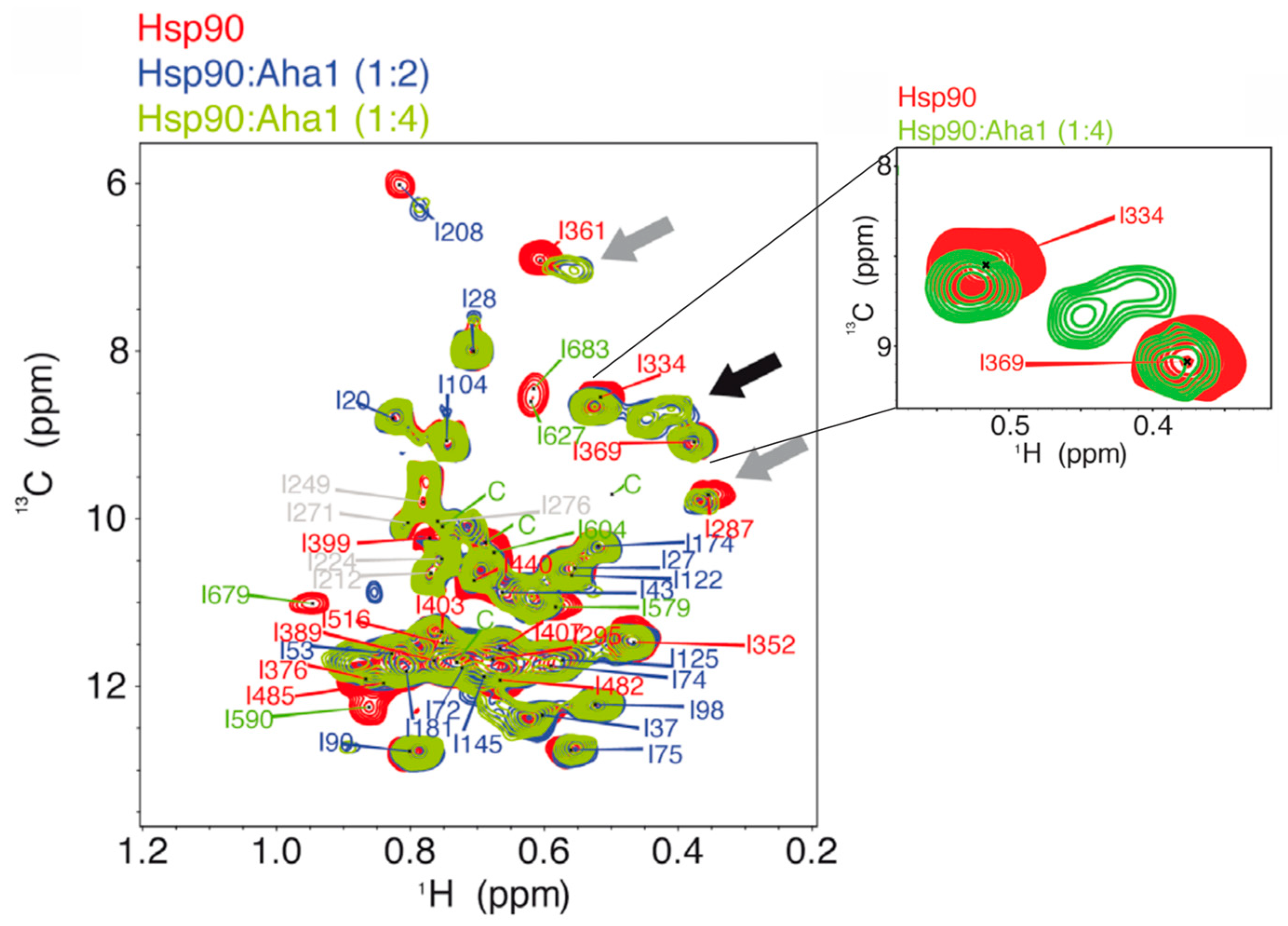
2.3.2. Allosteric Regulation of Hsp90β with Aha1
2.3.3. Allosteric Regulation of Hsp90β with Hop
2.4. Allosteric Regulation of Human Hsp90 with the Client Protein
3. Discussion
4. Conclusions and Outlook
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Grammatikakis, N.; Lin, J.-H.; Grammatikakis, A.; Tsichlis, P.N.; Cochran, B.H. p50cdc37 Acting in Concert with Hsp90 Is Required for Raf-1 Function. Mol. Cell Biol. 1999, 19, 1661–1672. [Google Scholar] [CrossRef]
- Boczek, E.E.; Reefschläger, L.G.; Dehling, M.; Struller, T.J.; Häusler, E.; Seidl, A.; Kaila, V.R.I.; Buchner, J. Conformational processing of oncogenic v-Src kinase by the molecular chaperone Hsp90. Proc. Natl. Acad. Sci. USA 2015, 112, E3189–E3198. [Google Scholar] [CrossRef] [PubMed]
- Eckl, J.M.; Daake, M.; Schwartz, S.; Richter, K. Nucleotide-Free sB-Raf is Preferentially Bound by Hsp90 and Cdc37 In Vitro. J. Mol. Biol. 2016, 428, 4185–4196. [Google Scholar] [CrossRef] [PubMed]
- Biebl, M.M.; Buchner, J. Structure; function, and regulation of the hsp90 machinery. Cold Spring Harb. Perspect. Biol. 2019, 11, a034017. [Google Scholar] [CrossRef] [PubMed]
- Didenko, T.; Duarte, A.M.S.; Karagöz, G.E.; Rüdiger, S.G.D. Hsp90 structure and function studied by NMR spectroscopy. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 636–647. [Google Scholar] [CrossRef]
- Ali, M.M.U.; Roe, S.M.; Vaughan, C.K.; Meyer, P.; Panaretou, B.; Piper, P.W.; Prodromou, C.; Pearl, L.H. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 2006, 440, 1013–1017. [Google Scholar] [CrossRef] [PubMed]
- Scheufler, C.; Brinker, A.; Bourenkov, G.; Pegoraro, S.; Moroder, L.; Bartunik, H.; Hartl, F.U.; Moarefi, I. Structure of TPR domain-peptide complexes: Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 2000, 101, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Roe, S.M.; Ali, M.M.U.; Meyer, P.; Vaughan, C.K.; Panaretou, B.; Piper, P.W.; Prodromou, C.; Pearl, L.H. The Mechanism of Hsp90 Regulation by the Protein Kinase-Specific Cochaperone p50cdc37. Cell 2004, 116, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Karagöz, G.E.; Duarte, A.M.S.; Ippel, H.; Uetrecht, C.; Sinnige, T.; Van Rosmalen, M.; Hausmann, J.; Heck, A.J.R.; Boelens, R.; Rüdiger, S.G.D. N-terminal domain of human Hsp90 triggers binding to the cochaperone p23. Proc. Natl. Acad. Sci. USA 2011, 108, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Nathan, D.F.; Lindquist, S. Mutational Analysis of Hsp90 Function: Interactions with a Steroid Receptor and a Protein Kinase. Mol. Cell Biol. 1995, 15, 3917–3925. [Google Scholar] [CrossRef] [PubMed]
- Verba, K.A.; Wang, R.Y.R.; Arakawa, A.; Liu, Y.; Shirouzu, M.; Yokoyama, S.; Agard, D.A. Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science 2016, 352, 1542–1547. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Borin, B.N.; Martinez-Yamout, M.A.; Dyson, H.J. The client protein p53 adopts a molten globule-like state in the presence of Hsp90. Nat. Struct. Mol. Biol. 2011, 18, 537–541. [Google Scholar] [CrossRef]
- Karagöz, G.E.; Duarte, A.M.S.; Akoury, E.; Ippel, H.; Biernat, J.; Luengo, T.M.; Radli, M.; Didenko, T.; Nordhues, B.A.; Veprintsev, D.B.; et al. Hsp90-tau complex reveals molecular basis for specificity in chaperone action. Cell 2014, 156, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Kostic, M.; Dyson, H.J. Dynamic interaction of Hsp90 with its client protein p53. J. Mol. Biol. 2011, 411, 158–173. [Google Scholar] [CrossRef] [PubMed]
- Young, J.C.; Schneider, C.; Hartl, F.U. In vitro evidence that hsp90 contains two independent chaperone sites. FEBS Lett. 1997, 418, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Hagn, F.; Lagleder, S.; Retzlaff, M.; Rohrberg, J.; Demmer, O.; Richter, K.; Buchner, J.; Kessler, H. Structural analysis of the interaction between Hsp90 and the tumor suppressor protein p53. Nat. Struct. Mol. Biol. 2010, 18, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Marcu, M.G.; Chadli, A.; Bouhouche, I.; Catelli, M.; Neckers, L.M. The heat shock protein 90 antagonist novobiocin interacts with a previously unrecognized ATP-binding domain in the carboxyl terminus of the chaperone. J. Biol. Chem. 2000, 275, 37181–37186. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Yadav, R.P.; Singh, S.; Boyd, K.; Artemyev, N.O. Unique interface and dynamics of the complex of HSP90 with a specialized cochaperone AIPL1. Structure 2023, 31, 309–317.e5. [Google Scholar] [CrossRef]
- Dernovšek, J.; Tomašič, T. Following the design path of isoform-selective Hsp90 inhibitors: Small differences, great opportunities. Pharmacol. Ther. 2023, 245, 108396. [Google Scholar] [CrossRef]
- Oroz, J.; Blair, L.J.; Zweckstetter, M. Dynamic Aha1 co-chaperone binding to human Hsp90. Protein Sci. 2019, 28, 1545–1551. [Google Scholar] [CrossRef] [PubMed]
- Lott, A.; Oroz, J.; Zweckstetter, M. Molecular basis of the interaction of Hsp90 with its co-chaperone Hop. Protein Sci. 2020, 29, 2422–2432. [Google Scholar] [CrossRef] [PubMed]
- Lopez, A.; Dahiya, V.; Delhommel, F.; Freiburger, L.; Stehle, R.; Asami, S.; Rutz, D.; Blair, L.; Buchner, J.; Sattler, M. Client binding shifts the populations of dynamic Hsp90 conformations through an allosteric network. Sci. Adv. 2021, 7, eabl7295. [Google Scholar] [CrossRef] [PubMed]
- Amatya, E.; Blagg, B.S.J. Recent advances toward the development of Hsp90 C-terminal inhibitors. Bioorg. Med. Chem. Lett. 2023, 80, 129111. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, S.; Joshi, A.; Sato, N.; Lee, S.; Lee, M.J.; Trepel, J.B.; Neckers, L. An update on the status of HSP90 inhibitors in cancer clinical trials. Cell Stress. Chaperones 2024, 29, 519–539. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, Y.; Honma, Y.; Sawaki, A.; Naito, Y.; Iwagami, S.; Komatsu, Y.; Takahashi, T.; Nishida, T.; Doi, T. Pimitespib in patients with advanced gastrointestinal stromal tumor (CHAPTER-GIST-301): A randomized, double-blind, placebo-controlled phase III trial. Ann. Oncol. 2022, 33, 959–967. [Google Scholar] [CrossRef] [PubMed]
- MV, V.K.; Noor, R.E.; Davis, R.E.; Zhang, Z.; Sipavicius, E.; Keramisanou, D.; Blagg, B.S.J.; Gelis, I. Molecular insights into the interaction of Hsp90 with allosteric inhibitors targeting the C-terminal domain. Medchemcomm 2018, 9, 1323–1331. [Google Scholar] [CrossRef]
- Zhou, C.; Zhang, C.; Zhu, H.; Liu, Z.; Su, H.; Zhang, X.; Chen, T.; Zhong, Y.; Hu, H.; Xiong, M.; et al. Allosteric Regulation of Hsp90α’s Activity by Small Molecules Targeting the Middle Domain of the Chaperone. IScience 2020, 23, 100857. [Google Scholar] [CrossRef] [PubMed]
- Kerfah, R.; Plevin, M.J.; Sounier, R.; Gans, P.; Boisbouvier, J. Methyl-specific isotopic labeling: A molecular tool box for solution NMR studies of large proteins. Curr. Opin. Struct. Biol. 2015, 32, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Ruschak, A.M.; Kay, L.E. Methyl groups as probes of supra-molecular structure, dynamics and function. J. Biomol. NMR 2010, 46, 75–87. [Google Scholar] [CrossRef]
- Fiaux, J.; Bertelsen, E.B.; Horwich, A.L.; Wütrich, K. NMR analysis of a 900K GroEL-GroES complex. Nature 2002, 418, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Sprangers, R.; Kay, L.E. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature 2007, 445, 618–622. [Google Scholar] [CrossRef]
- Jiang, Y.; Kalodimos, C.G. NMR Studies of Large Proteins. J. Mol. Biol. 2017, 429, 2667–2676. [Google Scholar] [CrossRef]
- Pervushin, K.; Riek, R.; Wider, G.; Wüthrich, K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Biophysics 1997, 94, 12366–12371. [Google Scholar] [CrossRef]
- Riek, R.; Pervushin, K.; Wüthrich, K. TROSY and CRINEPT: NMR with large molecular and supramolecular structures in solution. Trends Biochem. Sci. 2000, 25, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Riek, R.; Wider, G.; Pervushin, K.; Wüthrich, K. Polarization transfer by cross-correlated relaxation in solution NMR with very large molecules. Biophysics 1999, 96, 4918–4923. [Google Scholar] [CrossRef] [PubMed]
- Tugarinov, V.; Kay, L.E. Ile, Leu, and Val Methyl Assignments of the 723-Residue Malate Synthase G Using a New Labeling Strategy and Novel NMR Methods. J. Am. Chem. Soc. 2003, 125, 13868–13878. [Google Scholar] [CrossRef] [PubMed]
- Schütz, S.; Sprangers, R. Methyl TROSY spectroscopy: A versatile NMR approach to study challenging biological systems. Prog. Nucl. Magn. Reson. Spectrosc. 2020, 116, 56–84. [Google Scholar] [CrossRef] [PubMed]
- Oroz, J.; Chang, B.J.; Wysoczanski, P.; Lee, C.T.; Pérez-Lara, Á.; Chakraborty, P.; Hofele, R.V.; Baker, J.D.; Blair, L.J.; Biernat, J.; et al. Structure and pro-toxic mechanism of the human Hsp90/PPIase/Tau complex. Nat. Commun. 2018, 9, 4532. [Google Scholar] [CrossRef]
- Henot, F.; Crublet, E.; Frech, M.; Boisbouvier, J. NMR assignment of human HSP90 N-terminal domain bound to a long residence time resorcinol ligand. Biomol. NMR Assign. 2022, 16, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Henot, F.; Kerfah, R.; Törner, R.; Macek, P.; Crublet, E.; Gans, P.; Frech, M.; Hamelin, O.; Boisbouvier, J. Optimized precursor to simplify assignment transfer between backbone resonances and stereospecifically labelled valine and leucine methyl groups: Application to human Hsp90 N-terminal domain. J. Biomol. NMR 2021, 75, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, D.M.; Langer, T.; Elshorst, B.; Saxena, K.; Fiebig, K.M.; Vogtherr, M.; Schwalbe, H. NMR Backbone Assignment of the N-terminal Domain of Human HSP90. J. Biomol. NMR 2006, 36, 52. [Google Scholar] [CrossRef] [PubMed]
- Oroz, J.; Kim, J.H.; Chang, B.J.; Zweckstetter, M. Mechanistic basis for the recognition of a misfolded protein by the molecular chaperone Hsp90. Nat. Struct. Mol. Biol. 2017, 24, 407–413. [Google Scholar] [CrossRef]
- Söti, C.; Rácz, A.; Csermely, P. A nucleotide-dependent molecular switch controls ATP binding at the C-terminal domain of Hsp90. N-terminal nucleotide binding unmasks a C-terminal binding pocket. J. Biol. Chem. 2002, 277, 7066–7075. [Google Scholar] [CrossRef] [PubMed]
- Garnier, C.; Lafitte, D.; Tsvetkov, P.O.; Barbier, P.; Leclerc-Devin, J.; Millot, J.M.; Briand, C.; Makarov, A.A.; Catelli, M.G.; Peyrot, V. Binding of ATP to heat shock protein 90: Evidence for an ATP-binding site in the C-terminal domain. J. Biol. Chem. 2002, 277, 12208–12214. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, B.K.; Jayaraj, A.; Kumar, V.; Blagg, B.; Davis, R.E.; Jayaram, B.; Deep, S.; Chaudhuri, T.K. Stimulation of heat shock protein 90 chaperone function through binding of a novobiocin analog KU-32. J. Biol. Chem. 2019, 294, 6450–6467. [Google Scholar] [CrossRef]
- Mayer, M.; Meyer, B. Characterization of Ligand Binding by Saturation Transfer Difference NMR Spectroscopy. Angew. Chem. Int. Ed. 1999, 38, 1784–1788. [Google Scholar] [CrossRef]
- Cloreand, G.M.; Gronenborn, A.M. Theory and Applications of the Transferred Nuclear Overhauser Effect to the Study of the Conformations of Small Ligands Bound to Proteins. J. Magn. Reson. 1982, 48, 402–417. [Google Scholar] [CrossRef]
- Dernovšek, J.; Gradišek, N.; Zajec, Ž.; Urbančič, D.; Cingl, J.; Goričan, T.; Grdadolnik, S.G.; Tomašič, T. Discovery of new Hsp90-Cdc37 protein-protein interaction inhibitors: In silico screening and optimization of anticancer activity. RSC Adv. 2024, 14, 28347–28375. [Google Scholar] [CrossRef] [PubMed]
- Dernovšek, J.; Zajec, Ž.; Poje, G.; Urbančič, D.; Sturtzel, C.; Goričan, T.; Grissenberger, S.; Ciura, K.; Woziński, M.; Gedgaudas, M.; et al. Exploration and optimisation of structure-activity relationships of new triazole-based C-terminal Hsp90 inhibitors towards in vivo anticancer potency. Biomed. Pharmacother. 2024, 177, 116941. [Google Scholar] [CrossRef]
- Zajec, Ž.; Dernovšek, J.; Cingl, J.; Ogris, I.; Gedgaudas, M.; Zubrienė, A.; Mitrović, A.; Grdadolnik, S.G.; Gobec, M.; Tomašič, T. New Class of Hsp90 C-Terminal Domain Inhibitors with Anti-tumor Properties against Triple-Negative Breast Cancer. J. Med. Chem. 2024, 67, 12984–13018. [Google Scholar] [CrossRef]
- Dernovšek, J.; Urbančič, D.; Zajec, Ž.; Sturtzel, C.; Grissenberger, S.; Wenninger-Weinzierl, A.; Gedgaudas, M.; Zubrienė, A.; Goričan, T.; Grdadolnik, S.G.; et al. First dual inhibitors of human topoisomerase IIα and Hsp90 C-terminal domain inhibit the growth of Ewing sarcoma in vitro and in vivo. Bioorg. Chem. 2024, 153, 107850. [Google Scholar] [CrossRef] [PubMed]
- Dernovšek, J.; Goričan, T.; Gedgaudas, M.; Zajec, Ž.; Urbančič, D.; Jug, A.; Skok, Ž.; Sturtzel, C.; Distel, M.; Grdadolnik, S.G.; et al. Hiding in plain sight: Optimizing topoisomerase IIα inhibitors into Hsp90β selective binders. Eur. J. Med. Chem. 2024, 280, 116934. [Google Scholar] [CrossRef] [PubMed]
- Di Carluccio, C.; Forgione, M.C.; Martini, S.; Berti, F.; Molinaro, A.; Marchetti, R.; Silipo, A. Investigation of protein-ligand complexes by ligand-based NMR methods. Carbohydr. Res. 2021, 503, 108313. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.; Meyer, B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 2001, 123, 6108–6117. [Google Scholar] [CrossRef] [PubMed]
- Jahnke, W.; Floersheim, P.; Ostermeier, C.; Zhang, X.; Hemmig, R.; Hurth, K.; Uzunov, D.P. NMR reporter screening for the detection of high-affinity ligands. Angew. Chem.-Int. Ed. 2002, 41, 3420–3423. [Google Scholar] [CrossRef]
- Cheng, Y.; Chen, Y.; Li, K.; Liu, S.; Pang, C.; Gao, L.; Xie, J.; Wenjing, L.V.; Yu, H.; Deng, B. How inflammation dictates diabetic peripheral neuropathy: An enlightening review. CNS Neurosci. Ther. 2024, 30, e14477. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.J.; Li, C.; Yu, C.; Lu, Y.; Krise, J.M.; Mcintosh, M.P.; Rajewski, R.A.; Blagg, B.S.J.; Dobrowsky, R.T. Inhibiting heat-shock protein 90 reverses sensory hypoalgesia in diabetic mice. ASN Neuro 2010, 2, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.J.; Pan, P.; Farmer, K.L.; Zhao, H.; Blagg, B.S.J.; Dobrowsky, R.T. Modulating molecular chaperones improves sensory fiber recovery and mitochondrial function in diabetic peripheral neuropathy. Exp. Neurol. 2012, 235, 388–396. [Google Scholar] [CrossRef]
- Southworth, D.R.; Agard, D.A. Species-Dependent Ensembles of Conserved Conformational States Define the Hsp90 Chaperone ATPase Cycle. Mol. Cell 2008, 32, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Thwin, A.C.; Nadel, C.M.; Tse, E.; Gates, S.N.; Gestwicki, J.E.; Southworth, D.R. The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state. Mol. Cell 2021, 81, 3496–3508. [Google Scholar] [CrossRef] [PubMed]
- Meyer, P.; Prodromou, C.; Liao, C.; Hu, B.; Roe, S.M.; Vaughan, C.K.; Vlasic, I.; Panaretou, B.; Piper, P.W.; Pear, L.H. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 2004, 23, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Krukenberg, K.A.; Street, T.O.; Lavery, L.A.; Agard, D.A. Conformational dynamics of the molecular chaperone Hsp90. Q. Rev. Biophys. 2011, 44, 229–255. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Richter, K.; Reinstein, J.; Buchner, J. Integration of the accelerator Aha1 in the Hsp90 co-chaperone cycle. Nat. Struct. Mol. Biol. 2013, 20, 326–331. [Google Scholar] [CrossRef]
- Hessling, M.; Richter, K.; Buchner, J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat. Struct. Mol. Biol. 2009, 16, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Siligardi, G.; Hu, B.; Panaretou, B.; Piper, P.W.; Pearl, L.H.; Prodromou, C. Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J. Biol. Chem. 2004, 279, 51989–51998. [Google Scholar] [CrossRef] [PubMed]
- Schmid, A.B.; Lagleder, S.; Gräwert, M.A.; Röhl, A.; Hagn, F.; Wandinger, S.K.; Cox, M.B.; Demmer, O.; Richter, K.; Groll, M.; et al. The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop. EMBO J. 2012, 31, 1506–1517. [Google Scholar] [CrossRef]
- Wang, R.Y.R.; Noddings, C.M.; Kirschke, E.; Myasnikov, A.G.; Johnson, J.L.; Agard, D.A. Structure of Hsp90–Hsp70–Hop–GR reveals the Hsp90 client-loading mechanism. Nature 2022, 601, 460–464. [Google Scholar] [CrossRef] [PubMed]
- Southworth, D.R.; Agard, D.A. Client-Loading Conformation of the Hsp90 Molecular Chaperone Revealed in the Cryo-EM Structure of the Human Hsp90:Hop Complex. Mol. Cell 2011, 42, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Kirschke, E.; Goswami, D.; Southworth, D.; Griffin, P.R.; Agard, D.A. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 2014, 157, 1685–1697. [Google Scholar] [CrossRef]
- Alvira, S.; Cuéllar, J.; Röhl, A.; Yamamoto, S.; Itoh, H.; Alfonso, C.; Rivas, G.; Buchner, J.; Valpuesta, J.M. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nat. Commun. 2014, 5, 5484. [Google Scholar] [CrossRef]
- Gampp, O.; Kadavath, H.; Riek, R. NMR tools to detect protein allostery. Curr. Opin. Struct. Biol. 2024, 86, 102792. [Google Scholar] [CrossRef] [PubMed]
- Shukla, V.K.; Heller, G.T.; Hansen, D.F. Biomolecular NMR spectroscopy in the era of artificial intelligence. Structure 2023, 31, 1360–1374. [Google Scholar] [CrossRef]
- Zhu, K.F.; Yuan, C.; Du, Y.M.; Sun, K.L.; Zhang, X.K.; Vogel, H.; Jia, X.D.; Gao, Y.Z.; Zhang, Q.F.; Wang, D.P.; et al. Applications and prospects of cryo-EM in drug discovery. Mil. Med. Res. 2023, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Van Drie, J.H.; Tong, L. Cryo-EM as a powerful tool for drug discovery. Bioorg. Med. Chem. Lett. 2020, 30, 127524. [Google Scholar] [CrossRef]
- Lees, J.A.; Dias, J.M.; Han, S. Applications of Cryo-EM in small molecule and biologics drug design. Biochem. Soc. Trans. 2021, 49, 2627–2638. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, T.M.; van Beek, L.; Shilliday, F.; Debreczeni, J.; Phillips, C. Cryo-EM: The Resolution Revolution and Drug Discovery. SLAS Discov. 2021, 26, 17–31. [Google Scholar] [CrossRef]
- Son, A.; Kim, W.; Park, J.; Lee, W.; Lee, Y.; Choi, S.; Kim, H. Utilizing Molecular Dynamics Simulations, Machine Learning, Cryo-EM, and NMR Spectroscopy to Predict and Validate Protein Dynamics. Int. J. Mol. Sci. 2024, 25, 9725. [Google Scholar] [CrossRef] [PubMed]
- Di Prisco, G. Effect of pH and ionic Strength on the Catalytic and Allosteric Properties of Native and Chemically Modified Ox Liver Mitochondrial Glutamate Dehydrogenase. Arch. Biochem. Biophys. 1975, 171, 604–612. [Google Scholar] [CrossRef]
- Nussinov, R.; Tsai, C.J. Unraveling structural mechanisms of allosteric drug action. Trends Pharmacol. Sci. 2014, 35, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Arntson, K.E.; Pomerantz, W.C.K. Protein-Observed Fluorine NMR: A Bioorthogonal Approach for Small Molecule Discovery. J. Med. Chem. 2016, 59, 5158–5171. [Google Scholar] [CrossRef] [PubMed]
- Hull, W.E.; Sykes, B.D. Fluorotyrosine Alkaline Phosphatase: Internal Mobility of Individual Tyrosines and the Role of Chemical Shift Anisotropy as a 19F Nuclear Spin Relaxation Mechanism in Proteins. J. Mol. Biol. 1975, 98, 121–153. [Google Scholar] [CrossRef] [PubMed]
- Magnan, B.; Chen, X.H.; Rashid, S.; Minard, A.; Chau, W.; Uyesugi, T.; Edwards, R.A.; Panigrahi, R.; Glover, J.N.M.; LaPointe, P.; et al. Asymmetric Dynamics Drive Catalytic Activation of the Hsp90 Chaperone. J. Phys. Chem. B 2024, 128, 8388–8399. [Google Scholar] [CrossRef] [PubMed]
- Pham, L.B.T.; Costantino, A.; Barbieri, L.; Calderone, V.; Luchinat, E.; Banci, L. Direct Expression of Fluorinated Proteins in Human Cells for 19F In-Cell NMR Spectroscopy. J. Am. Chem. Soc. 2023, 145, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Rashid, S.; Lee, B.L.; Wajda, B.; Spyracopoulos, L. Nucleotide Binding and Active Site Gate Dynamics for the Hsp90 Chaperone ATPase Domain from Benchtop and High Field 19F NMR Spectroscopy. J. Phys. Chem. B 2020, 124, 2984–2993. [Google Scholar] [CrossRef] [PubMed]
- Rashid, S.; Lee, B.L.; Wajda, B.; Spyracopoulos, L. Side-Chain Dynamics of the Trifluoroacetone Cysteine Derivative Characterized by 19 F NMR Relaxation and Molecular Dynamics Simulations. J. Phys. Chem. B 2019, 123, 3665–3671. [Google Scholar] [CrossRef]
- Lee, B.L.; Rashid, S.; Wajda, B.; Wolmarans, A.; Lapointe, P.; Spyracopoulos, L. The Hsp90 Chaperone: 1H and 19F Dynamic Nuclear Magnetic Resonance Spectroscopy Reveals a Perfect Enzyme. Biochemistry 2019, 58, 1869–1877. [Google Scholar] [CrossRef] [PubMed]

| Domain (+ligand)/Construct | Assignment | Reference | ||
|---|---|---|---|---|
| Resonances—Backbone/Methyl Groups | % of Assigned Residues | % of Unassigned Residues | ||
| NTD | 1H, 15N, 13C—backbone | 82% of backbone and Cβ resonances (9–223) 89% of Cα, 80% of Cβ (17–224) 81% of NH 1 | 18% of backbone and Cβ resonances (9–223) 11% of Cα, 20% of Cβ (17–224) 19% of NH 1 | BMRB: 7003, Jacobs et al., 2006 [41] BMRB: 19,560, Park et al., 2011 [14] BMRB: 50,786, Henot et al., 2021 [40] |
| 1H, 13C—methyl groups AILpro-SMTVpro-S | 100% of AILpro-SMTVpro-S | 0% of AILpro-SMTVpro-S | BMRB: 19,560, Park et al., 2011 [14] Karagöz et al., 2011 [9] BMRB: 50,786, Henot et al., 2021 [40] | |
| NTD + ligand | 1H, 15N, 13C—backbone | 96% of Cα, 95% of Cβ (non-proline residues) 92% of non-proline backbone (NH) 85% of NH 1 | 8% of non-proline backbone (NH) 4% of Cα, 5% of Cβ (non-proline residues) 15% of NH 1 | BMRB: 51,378, Henot et al., 2022 [39] |
| 1H, 13C—methyl groups AILpro-SMTVpro-S | 100% of AILpro-SMTVpro-S | 0% of AILpro-SMTVpro-S | ||
| MD | 1H, 15N, 13C—backbone 1H, 13C—methyl groups Ile-δ1 | 82% of NH 1 100% of all Ile-δ1 | 18% of NH 1 0% of all Ile-δ1 | BMRB: 19,560, Park et al., 2011 [14] Karagöz et al., 2011 [9] |
| NTD + MD | 1H, 15N—backbone 1H, 13C—methyl groups Ile-δ1 | / 2 95% of all Ile-δ1 | / 2 5% of all Ile-δ1 | BMRB: 19,560, Park et al., 2011 [14] |
| Hsp90Δ | 1H, 13C—methyl groups Ile-δ1 | / 2 | / 2 | BMRB: 19,560, Park et al., 2011 [14] |
| full-length | 1H, 13C—methyl groups Ile-δ1 and Met-ε | 100% of all Ile-δ1 35% of all Met-ε | 0% of all Ile-δ1 65% of all Met-ε | Karagöz et al., 2011 [9] Oroz et al., 2017–2019 [20,38,42] Lopez et al., 2021 [22] |
| ATP | ATP + p23 | ADP | Aha1 | Hop | ||
|---|---|---|---|---|---|---|
| NTD | I20 | X 1 | X | X | X 1 | X 1 |
| I27 | X 1 | X | X | X 1 | ||
| I28 | X 1 | X | X | X | ||
| I37 | X | X 1 | ||||
| I53 | X | |||||
| I75 | X | X 1 | X 1 | |||
| I90 | X | X | ||||
| I98 | X | X | ||||
| I122 | X | X | X | X 2 | ||
| I125 | X | X 2 | ||||
| I208 | X 1 | |||||
| MD | I287 | X | ||||
| I334 | X | |||||
| I361 | X | X | ||||
| I369 | X 1 | |||||
| I376 | X | |||||
| I389 | X | X | ||||
| I399 | X 2 | |||||
| I407 | X | |||||
| I440 | X 1 | |||||
| I482 | X 1 | |||||
| I485 | X | |||||
| CTD | I590 | X | ||||
| I604 | X | |||||
| I679 | X |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goričan, T.; Golič Grdadolnik, S. Insights into the Allosteric Regulation of Human Hsp90 Revealed by NMR Spectroscopy. Biomolecules 2025, 15, 37. https://doi.org/10.3390/biom15010037
Goričan T, Golič Grdadolnik S. Insights into the Allosteric Regulation of Human Hsp90 Revealed by NMR Spectroscopy. Biomolecules. 2025; 15(1):37. https://doi.org/10.3390/biom15010037
Chicago/Turabian StyleGoričan, Tjaša, and Simona Golič Grdadolnik. 2025. "Insights into the Allosteric Regulation of Human Hsp90 Revealed by NMR Spectroscopy" Biomolecules 15, no. 1: 37. https://doi.org/10.3390/biom15010037
APA StyleGoričan, T., & Golič Grdadolnik, S. (2025). Insights into the Allosteric Regulation of Human Hsp90 Revealed by NMR Spectroscopy. Biomolecules, 15(1), 37. https://doi.org/10.3390/biom15010037







