Crystallization of Ethylene Plant Hormone Receptor—Screening for Structure
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
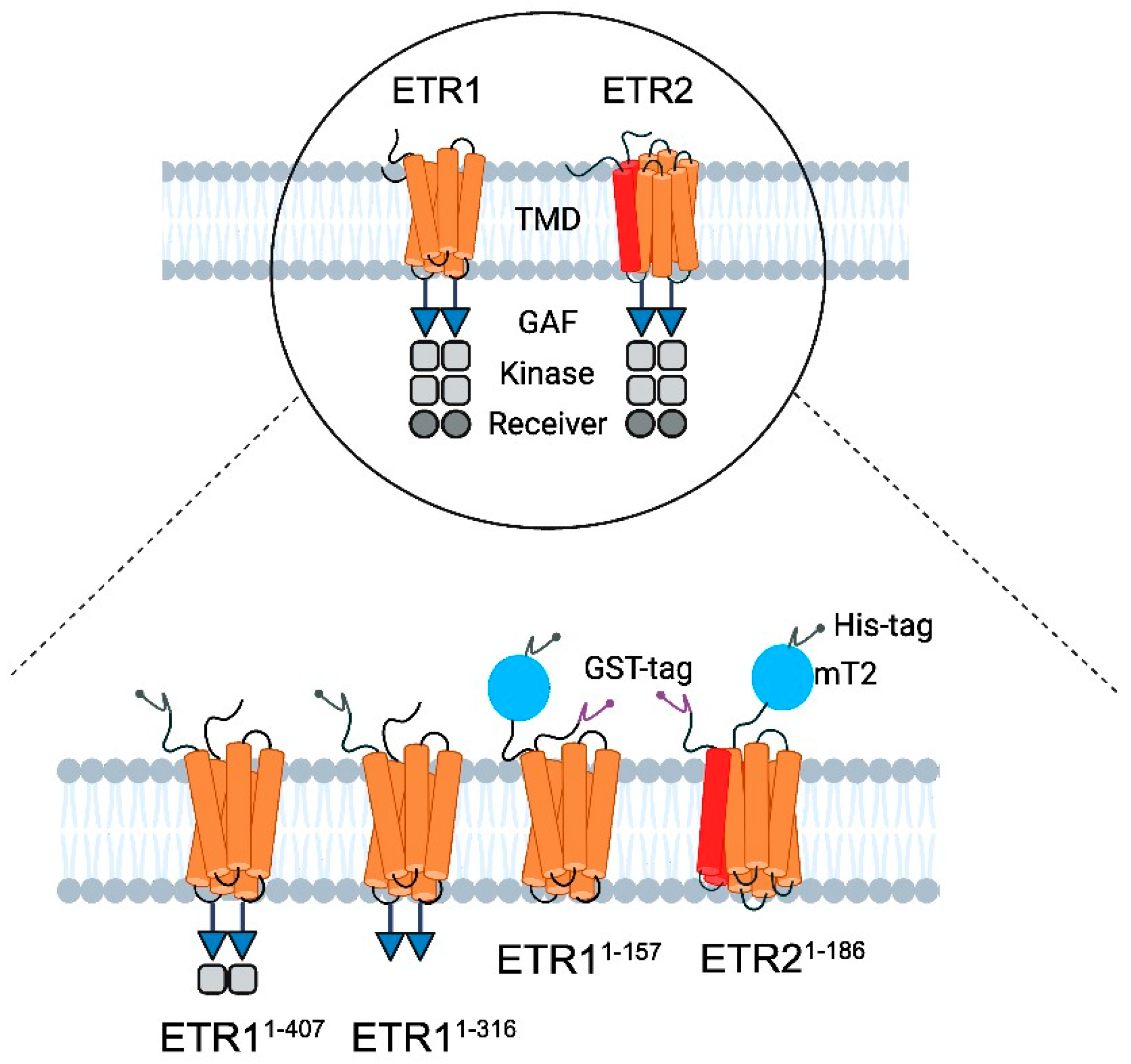
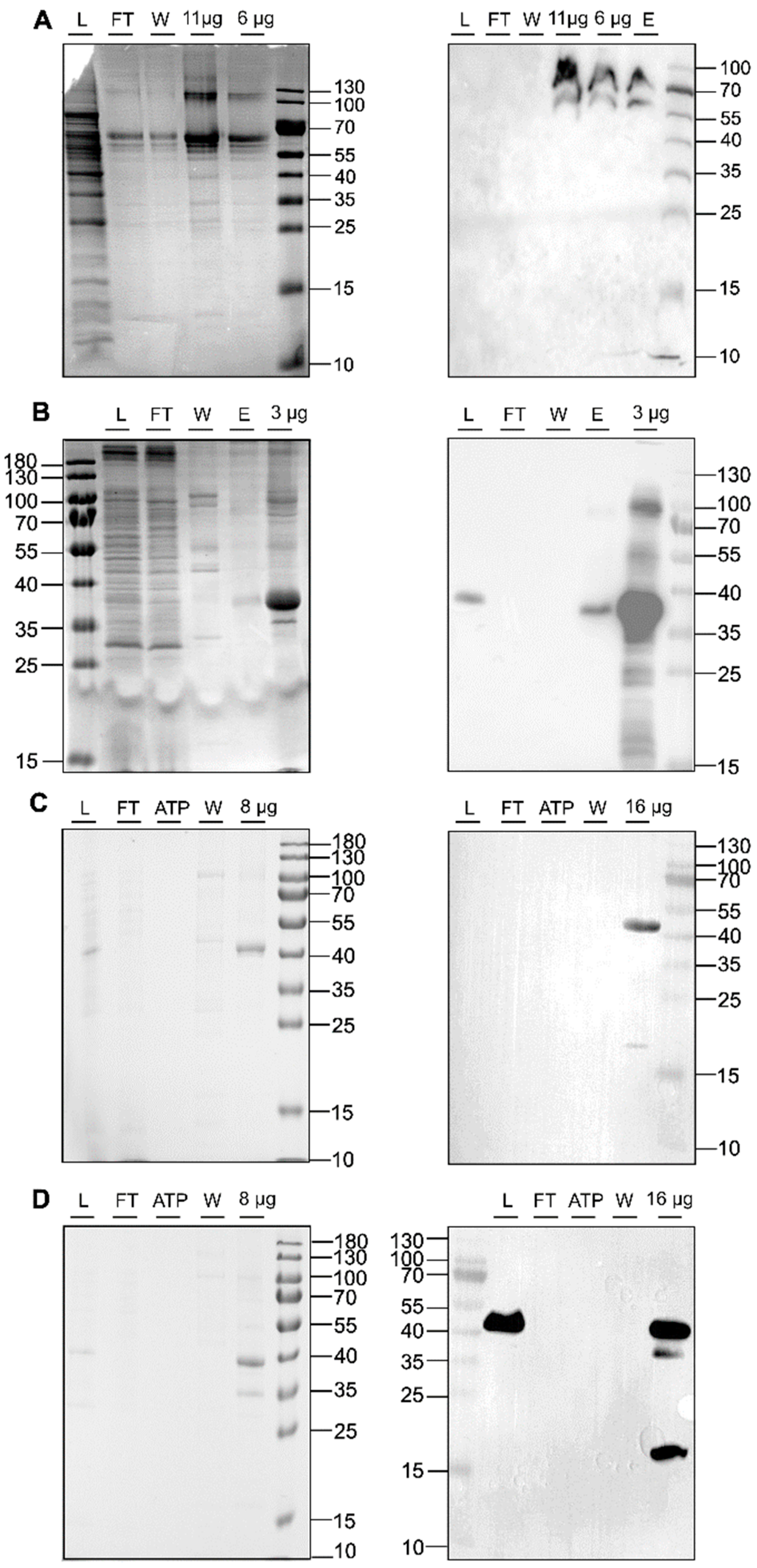
2.2.1. Cloning, Heterologous Expression, and Purification
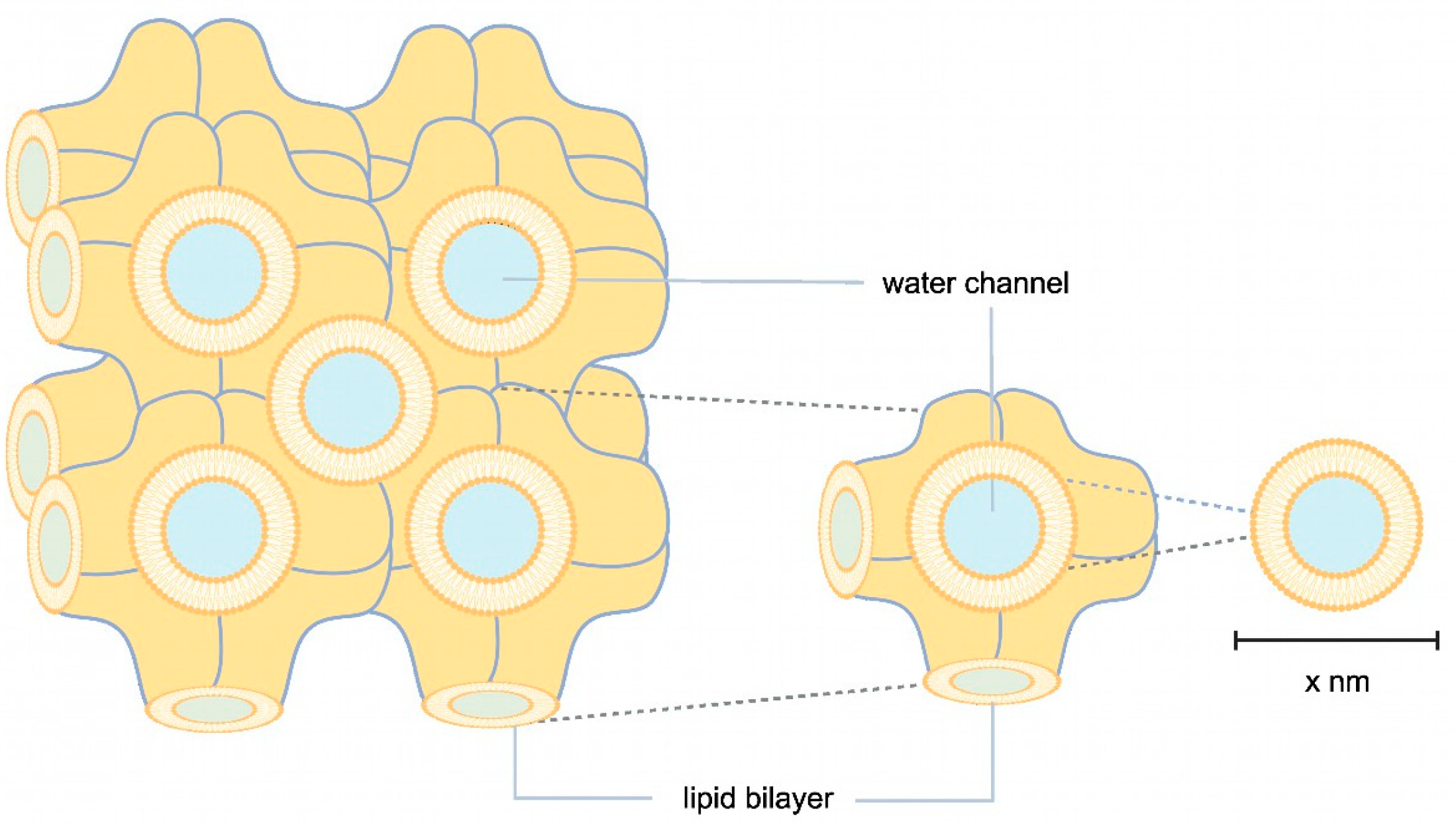
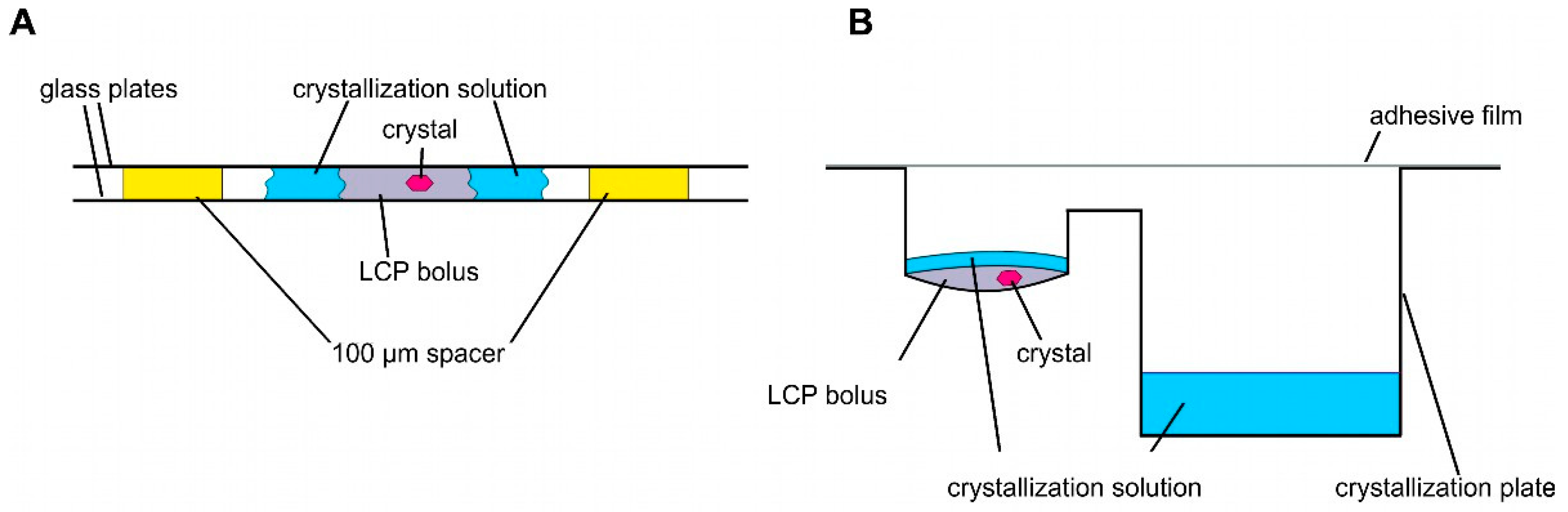
2.2.2. LCP Crystallization
2.2.3. LCP—Lipid Mixture Preparation
2.2.4. LCP—Sample Preparation
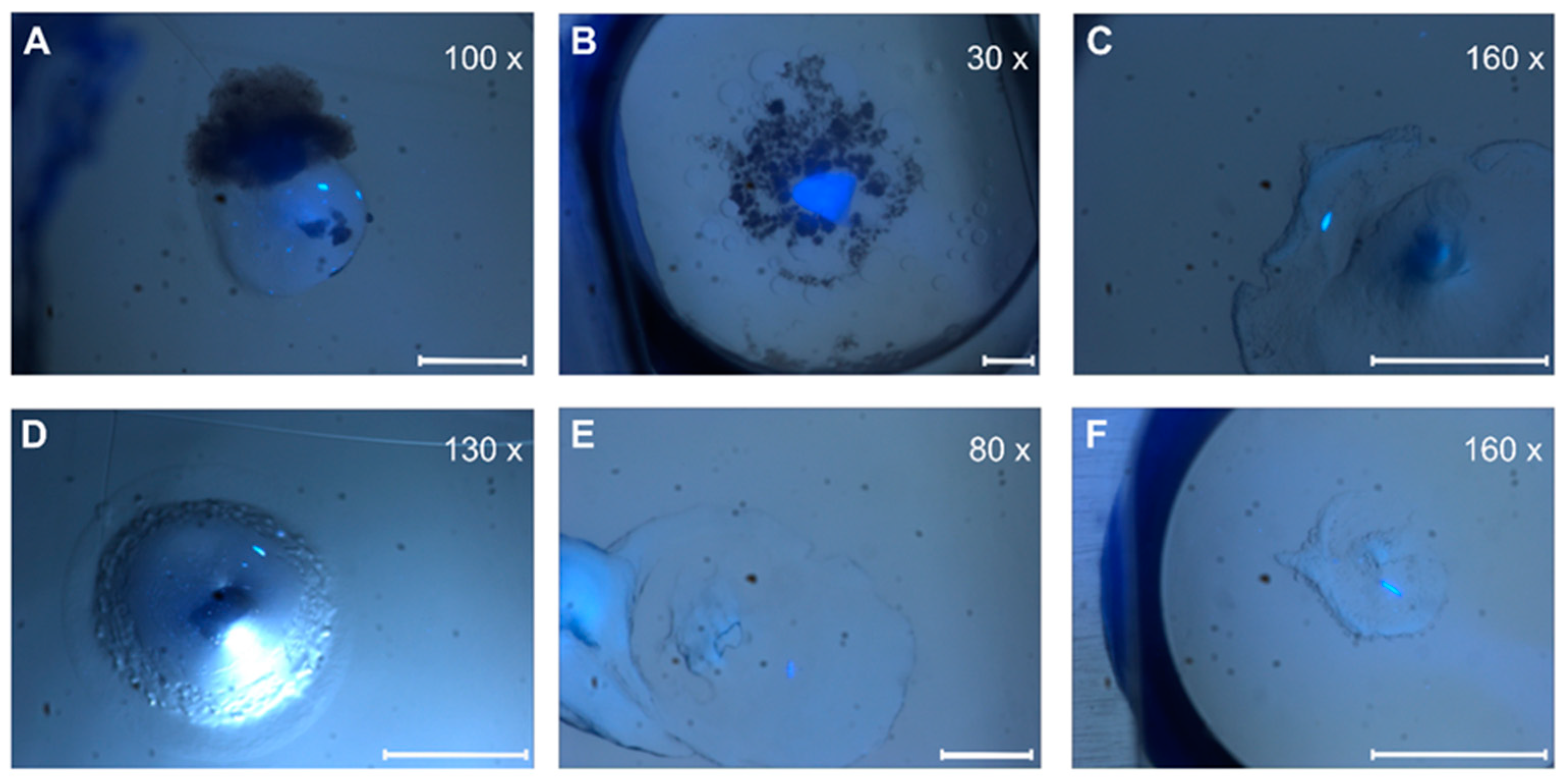
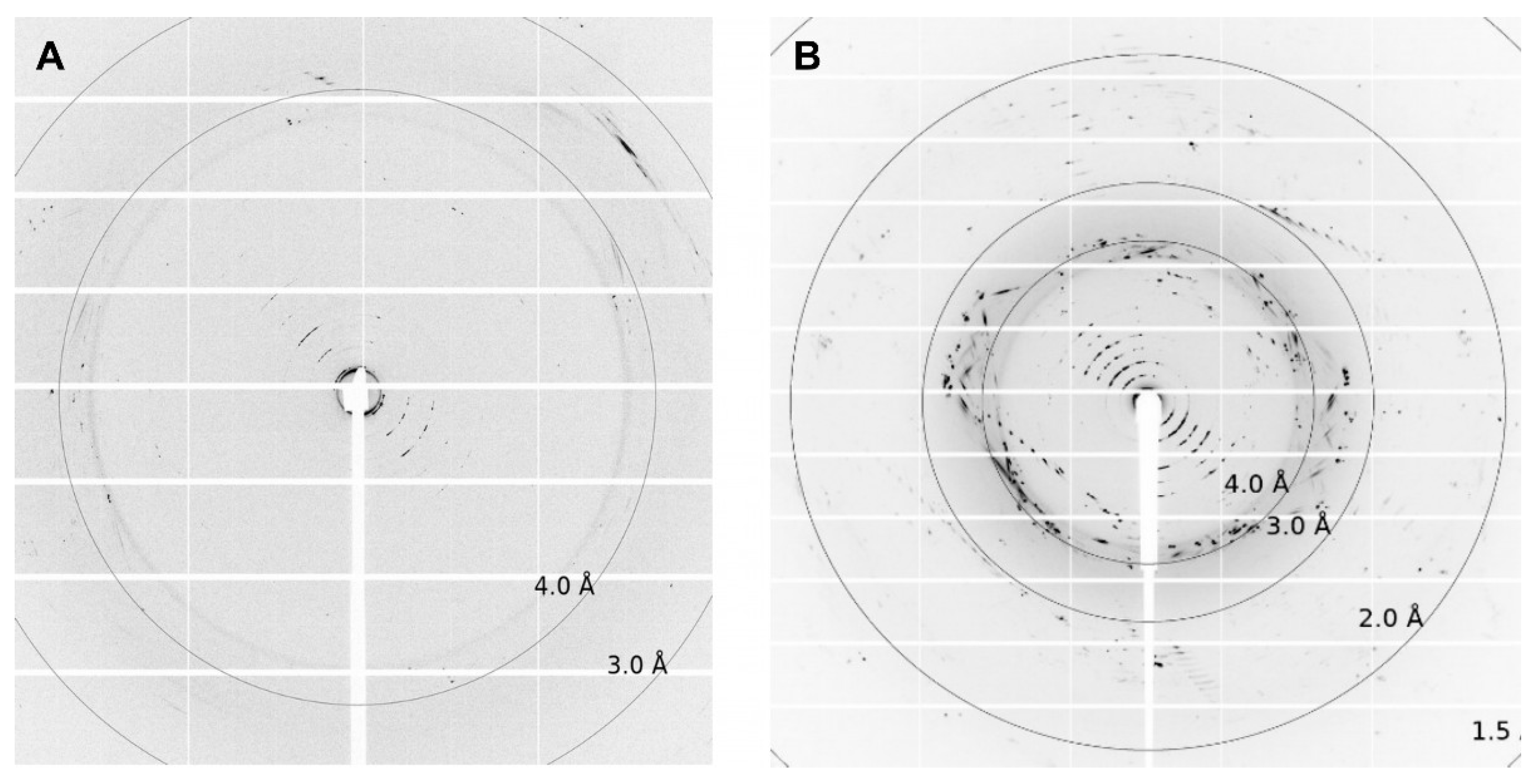
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef]
- Ju, C.; Chang, C. Advances in ethylene signalling: Protein complexes at the endoplasmic reticulum membrane. AoB Plants 2012, 2012, pls031. [Google Scholar] [CrossRef]
- Cho, W.; Stahelin, R.V. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 119–151. [Google Scholar] [CrossRef]
- Bleecker, A.B. Ethylene perception and signaling: An evolutionary perspective. Trends Plant Sci. 1999, 4, 269–274. [Google Scholar] [CrossRef]
- Bleecker, A.B.; Kende, H. Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 2000, 16, 1–18. [Google Scholar] [CrossRef]
- Ju, C.; van de Poel, B.; Cooper, E.D.; Thierer, J.H.; Gibbons, T.R.; Delwiche, C.F.; Chang, C. Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat. Plants 2015, 1, 14004. [Google Scholar] [CrossRef]
- Lacey, R.F.; Binder, B.M. How plants sense ethylene gas—The ethylene receptors. J. Inorg. Biochem. 2014, 133, 58–62. [Google Scholar] [CrossRef]
- Grefen, C.; Harter, K. Plant two-component systems: Principles, functions, complexity and cross talk. Planta 2004, 219, 733–742. [Google Scholar] [CrossRef]
- Shakeel, S.N.; Gao, Z.; Amir, M.; Chen, Y.; Rai, M.I.; Haq, N.U.; Schaller, G.E. Ethylene Regulates Levels of Ethylene Receptor/CTR1 Signaling Complexes in Arabidopsis thaliana. J. Biol. Chem. 2015, 290, 12415–12424. [Google Scholar] [CrossRef]
- Hua, J.; Meyerowitz, E.M. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 1998, 94, 261–271. [Google Scholar] [CrossRef]
- Hua, J.; Sakai, H.; Nourizadeh, S.; Qianhong, G.C.; Bleecker, A.B.; Ecker, J.R.; Meyerowitz, E.M. EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 1998, 10, 1321–1332. [Google Scholar] [CrossRef]
- Hall, A.E.; Findell, J.L.; Schaller, G.E.; Sisler, E.C.; Bleecker, A.B. Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiol. 2000, 123, 1449–1458. [Google Scholar] [CrossRef]
- Berleth, M.; Berleth, N.; Minges, A.; Hänsch, S.; Burkart, R.C.; Stork, B.; Stahl, Y.; Weidtkamp-Peters, S.; Simon, R.; Groth, G. Molecular Analysis of Protein-Protein Interactions in the Ethylene Pathway in the Different Ethylene Receptor Subfamilies. Front. Plant Sci. 2019, 10, 726. [Google Scholar] [CrossRef]
- Schaller, G.E.; Bleecker, A.B. Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 1995, 270, 1809–1811. [Google Scholar] [CrossRef]
- Gao, Z.; Wen, C.-K.; Binder, B.M.; Chen, Y.; Chang, J.; Chiang, Y.; Robert, J.K.R., III; Chang, C.; Schaller, G.E. Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis. J. Biol. Chem. 2008, 283, 23801–23810. [Google Scholar] [CrossRef]
- Chang, C.; Kwok, S.F.; Bleecker, A.B.; Meyerowitz, E.M. Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science 1993, 262, 539–544. [Google Scholar] [CrossRef]
- Mayerhofer, H.; Panneerselvam, S.; Kaljunen, H.; Tuukkanen, A.; Mertens, H.D.T.; Mueller-Dieckmann, J. Structural model of the cytosolic domain of the plant ethylene receptor 1 (ETR1). J. Biol. Chem. 2015, 290, 2644–2658. [Google Scholar] [CrossRef]
- Grantz, A.A.; Muller-Dieckmann, H.J.; Kim, S.-H. Subcloning, crystallization and preliminary X-ray analysis of the signal receiver domain of ETR1, an ethylene receptor from Arabidopsis thaliana. Acta Crystallogr. D Biol. Crystallogr. 1998, 54, 690–692. [Google Scholar] [CrossRef]
- Panneerselvam, S.; Kaljunen, H.; Mueller-Dieckmann, J. Cloning, overexpression, purification and preliminary X-ray analysis of the catalytic domain of the ethylene receptor ETR1 from Arabidopsis thaliana. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 1307–1309. [Google Scholar] [CrossRef]
- Birch, J.; Axford, D.; Foadi, J.; Meyer, A.; Eckhardt, A.; Thielmann, Y.; Moraes, I. The fine art of integral membrane protein crystallisation. Methods 2018, 147, 150–162. [Google Scholar] [CrossRef]
- Landau, E.M.; Rosenbusch, J.P. Lipidic cubic phases: A novel concept for the crystallization of membrane proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 14532–14535. [Google Scholar] [CrossRef]
- Caffrey, M.; Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 2009, 4, 706–731. [Google Scholar] [CrossRef]
- Wadsten, P.; Wöhri, A.B.; Snijder, A.; Katona, G.; Gardiner, A.T.; Cogdell, R.J.; Neutze, R.; Engström, S. Lipidic sponge phase crystallization of membrane proteins. J. Mol. Biol. 2006, 364, 44–53. [Google Scholar] [CrossRef]
- Zabara, A.; Meikle, T.G.; Newman, J.; Peat, T.S.; Conn, C.E.; Drummond, C.J. The nanoscience behind the art of in-meso crystallization of membrane proteins. Nanoscale 2017, 9, 754–763. [Google Scholar] [CrossRef]
- Cherezov, V.; Clogston, J.; Misquitta, Y.; Abdel-Gawad, W.; Caffrey, M. Membrane protein crystallization in meso: Lipid type-tailoring of the cubic phase. Biophys. J. 2002, 83, 3393–3407. [Google Scholar] [CrossRef]
- Zabara, A.; Chong, J.T.Y.; Martiel, I.; Stark, L.; Cromer, B.A.; Speziale, C.; Drummond, C.J.; Mezzenga, R. Design of ultra-swollen lipidic mesophases for the crystallization of membrane proteins with large extracellular domains. Nat. Commun. 2018, 9, 544. [Google Scholar] [CrossRef]
- Wu, H.; Wacker, D.; Mileni, M.; Katritch, V.; Han, G.W.; Vardy, E.; Liu, W.; Thompson, A.A.; Huang, X.; Carrol, F.I.; et al. Structure of the human κ-opioid receptor in complex with JDTic. Nature 2012, 485, 327–332. [Google Scholar] [CrossRef]
- Thomaston, J.L.; Alfonso-Prieto, M.; Woldeyes, R.A.; DeGrado, W.F. High-resolution structures of the M2 channel from influenza A virus reveal dynamic pathways for proton stabilization and transduction. Proc. Natl. Acad. Sci. USA 2015, 112, 14260–14265. [Google Scholar] [CrossRef]
- Siu, F.Y.; He, M.; de Graaf, C.; Han, G.W.; Yang, D.; Zhang, Z.; Zhou, C.; Xu, Q.; Wacker, D.; Joseph, J.S.; et al. Structure of the human glucagon class B G-protein-coupled receptor. Nature 2013, 499, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Shimamura, T.; Shiroishi, M.; Weyand, S.; Tsujimoto, H.; Winter, G.; Katritch, V.; Abagyan, R.; Cherezov, V.; Liu, W.; Han, G.W.; et al. Structure of the human histamine H1 receptor complex with doxepin. Nature 2011, 475, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Schöppe, J.; Ehrenmann, J.; Waltenspühl, Y.; Plückthun, A. Universal platform for the generation of thermostabilized GPCRs that crystallize in LCP. Nat. Protoc. 2022, 17, 698–726. [Google Scholar] [CrossRef] [PubMed]
- Caffrey, M. A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallogr. F Struct. Biol. Commun. 2015, 71, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, J.; Fellner, M.; Zhang, C.; Sui, D.; Hu, J. Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway. Sci. Adv. 2017, 3, e1700344. [Google Scholar] [CrossRef] [PubMed]
- Kusakizako, T.; Tanaka, Y.; Hipolito, C.J.; Kuroda, T.; Ishitani, R.; Suga, H.; Nureki, O. LCP crystallization and X-ray diffraction analysis of VcmN, a MATE transporter from Vibrio cholerae. Acta Crystallogr. F Struct. Biol. Commun. 2016, 72, 552–557. [Google Scholar] [CrossRef]
- Cherezov, V. Lipidic cubic phase technologies for membrane protein structural studies. Curr. Opin. Struct. Biol. 2011, 21, 559–566. [Google Scholar] [CrossRef]
- Goedhart, J.; von Stetten, D.; Noirclerc-Savoye, M.; Lelimousin, M.; Joosen, L.; Hink, M.A.; von Weeren, L.; Gadella, T.W.J., Jr.; Royant, A. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat. Commun. 2012, 3, 751. [Google Scholar] [CrossRef]
- Milić, D.; Dick, M.; Mulnaes, D.; Pfleger, C.; Kinnen, A.; Gohlke, H.; Groth, G. Recognition motif and mechanism of ripening inhibitory peptides in plant hormone receptor ETR1. Sci. Rep. 2018, 8, 3890. [Google Scholar] [CrossRef]
- Kanonenberg, K.; Royes, J.; Kedrov, A.; Poschmann, G.; Angius, F.; Solgadi, A.; Spitz, O.; Kleinschrodt, K.; Stühler, K.; Miroux, B.; et al. Shaping the lipid composition of bacterial membranes for membrane protein production. Microb. Cell Factories 2019, 18, 131. [Google Scholar] [CrossRef]
- Schott-Verdugo, S.; Müller, L.; Classen, E.; Gohlke, H.; Groth, G. Structural Model of the ETR1 Ethylene Receptor Transmembrane Sensor Domain. Sci. Rep. 2019, 9, 8869. [Google Scholar] [CrossRef]
- Lemke, M.; Reiners, J.; Smits, S.H.J.; Lakomek, N.; Groth, G. Functional reconstitution and structural characterization of the plant hormone receptor ETR1 in lipid nanodiscs. Chem. Commun. 2023, 59, 9344–9347. [Google Scholar] [CrossRef]
- Grefen, C.; Städele, K.; Růzicka, K.; Petr, O.; Klaus, H.; Jakub, H. Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Mol. Plant 2008, 1, 308–320. [Google Scholar] [CrossRef]
- Moussatche, P.; Klee, H.J. Autophosphorylation activity of the Arabidopsis ethylene receptor multigene family. J. Biol. Chem. 2004, 279, 48734–48741. [Google Scholar] [CrossRef]
- Rathmann, B.; Pais, D.Q.; Thielmann, Y. MPI tray: A versatile crystallization plate for membrane proteins. J. Appl. Crystallogr. 2017, 50, 327–330. [Google Scholar] [CrossRef]
- Zhao, J.; Xie, H.; Mehdipour, A.R.; Safarian, S.; Ermler, U.; Münke, C.; Thielmann, Y.; Hummer, G.; Ebersberger, I.; Wang, J.; et al. The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily. Proc. Natl. Acad. Sci. USA 2021, 118, e2107335118. [Google Scholar] [CrossRef]
- Zakrzewska, S.; Mehdipour, A.R.; Malviya, V.N.; Nonaka, T.; Köpke, J.; Münke, C.; Hausner, W.; Hummer, G.; Safarian, S.; Michel, H. Inward-facing conformation of a multidrug resistance MATE family transporter. Proc. Natl. Acad. Sci. USA 2019, 116, 12275–12284. [Google Scholar] [CrossRef]
- Joseph, J.S.; Liu, W.; Kunken, J.; Weiss, T.M.; Tsuruta, H.; Cherezov, V. Characterization of lipid matrices for membrane protein crystallization by high-throughput small angle X-ray scattering. Methods 2011, 55, 342–349. [Google Scholar] [CrossRef]
- Bisson, M.M.A.; Groth, G. Targeting Plant Ethylene Responses by Controlling Essential Protein-Protein Interactions in the Ethylene Pathway. Mol. Plant 2015, 8, 1165–1174. [Google Scholar] [CrossRef] [PubMed]
- Bisson, M.M.A.; Kessenbrock, M.; Müller, L.; Hofmann, A.; Schmitz, F.; Cristescu, S.M.; Groth, G. Peptides interfering with protein-protein interactions in the ethylene signaling pathway delay tomato fruit ripening. Sci. Rep. 2016, 6, 30634. [Google Scholar] [CrossRef] [PubMed]
- Zabara, A.; Meikle, T.G.; Trenker, R.; Yao, S.; Newman, J.; Peat, T.S.; Separovic, F.; Conn, C.E.; Call, M.J.; Call, M.E.; et al. Lipidic Cubic Phase-Induced Membrane Protein Crystallization: Interplay Between Lipid Molecular Structure, Mesophase Structure and Properties, and Crystallogenesis. Cryst. Growth Des. 2017, 17, 5667–5674. [Google Scholar] [CrossRef]
- Zabara, A.; Mezzenga, R. Plenty of room to crystallize: Swollen lipidic mesophases for improved and controlled in-meso protein crystallization. Soft Matter 2012, 8, 6535–6541. [Google Scholar] [CrossRef]
- Engblom, J.; Miezis, Y.; Nylander, T.; Razumas, V.; Larsson, K. On the swelling of monoolein liquid-crystalline aqueous phases in the presence of distearoylphosphatidylglycerol. In Surface and Colloid Science; Razumas, V., Lindman, B., Nylander, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2001; pp. 9–15. Volume 116. [Google Scholar]
- Cecchetti, C.; Strauss, J.; Stohrer, C.; Naylor, C.; Pryor, E.; Hobbs, J.; Tanley, S.; Goldman, A.; Byrne, B. A novel high-throughput screen for identifying lipids that stabilise membrane proteins in detergent based solution. PLoS ONE 2021, 16, e0254118. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Howe, N.; Dukkipati, A.; Shah, S.T.A.; Bax, B.D.; Edge, C.; Bridges, A.; Hardwicke, P.; Singh, O.M.P.; Giblin, G.; et al. Crystallizing Membrane Proteins in the Lipidic Mesophase. Experience with Human Prostaglandin E2 Synthase 1 and an Evolving Strategy. Cryst. Growth Des. 2014, 14, 2034–2047. [Google Scholar] [CrossRef] [PubMed]
- Reszczyńska, E.; Hanaka, A. Lipids Composition in Plant Membranes. Cell Biochem. Biophys. 2020, 78, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.J.; Dupont, F.M. Lipid Composition of Plasma Membranes and Endomembranes Prepared from Roots of Barley (Hordeum vulgare L.): Effects of Salt. Plant Physiol. 1989, 90, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, R.P.; Beevers, H. Lipid composition of organelles from germinating castor bean endosperm. Plant Physiol. 1977, 59, 259–263. [Google Scholar] [CrossRef]
- Gushchin, I.; Melnikov, I.; Polovinkin, V.; Ishchenko, A.; Yuzhakova, A.; Buslaev, P.; Bourenkov, G.; Grudinin, S.; Round, E.; Balandin, T.; et al. Mechanism of transmembrane signaling by sensor histidine kinases. Science 2017, 356. [Google Scholar] [CrossRef]
- Gordeliy, V.I.; Labahn, J.; Moukhametzianov, R.; Efremov, R.; Granzin, J.; Schlesinger, R.; Büldt, G.; Savopol, T.; Scheidig, A.J.; Klare, J.P.; et al. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature 2002, 419, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Gudipaty, S.A.; McEvoy, M.M. The histidine kinase CusS senses silver ions through direct binding by its sensor domain. Biochim. Biophys. Acta 2014, 1844, 1656–1661. [Google Scholar] [CrossRef]
- Cheung, J.; Hendrickson, W.A. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure 2009, 17, 190–201. [Google Scholar] [CrossRef]
- Sevvana, M.; Vijayan, V.; Zweckstetter, M.; Reinelt, S.; Madden, D.R.; Herbst-Irmer, R.; Sheldrick, G.M.; Bott, M.; Griesinger, C.; Becker, S. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J. Mol. Biol. 2008, 377, 512–523. [Google Scholar] [CrossRef]
- Schaller, G.E.; Ladd, A.N.; Lanahan, M.B.; Spanbauer, J.M.; Bleecker, A.B. The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J. Biol. Chem. 1995, 270, 12526–12530. [Google Scholar] [CrossRef]
- Kugele, A.; Uzun, B.; Müller, L.; Schott-Verdugo, S.; Gohlke, H.; Groth, G.; Drescher, M. Mapping the helix arrangement of the reconstituted ETR1 ethylene receptor transmembrane domain by EPR spectroscopy. RSC Adv. 2022, 12, 7352–7356. [Google Scholar] [CrossRef]
- Müller, L. Kupfertransport und -koordinationschemie der Kupferbindenden Domäne des Ethylenrezeptors ETR1. Ph.D. Thesis, Universitäts und Landesbibliothek, Düsseldorf, Germany, 2019. Available online: https://docserv.uni-duesseldorf.de/servlets/DocumentServlet?id=52654 (accessed on 12 January 2024).
- Andersson, M.; Mattle, D.; Sitsel, O.; Klymchuk, T.; Nielsen, A.M.; Møller, L.B.; White, S.H.; Nissen, P.; Gourdon, P. Copper-transporting P-type ATPases use a unique ion-release pathway. Nat. Struct. Mol. Biol. 2014, 21, 43–48. [Google Scholar] [CrossRef]
- Mangini, V.; Belviso, B.D.; Nardella, M.I.; Natile, G.; Arnesano, F.; Caliandro, R. Crystal Structure of the Human Copper Chaperone ATOX1 Bound to Zinc Ion. Biomolecules 2022, 12, 1494. [Google Scholar] [CrossRef]
- Badarau, A.; Baslé, A.; Firbank, S.J.; Dennison, C. Investigating the role of zinc and copper binding motifs of trafficking sites in the cyanobacterium Synechocystis PCC 6803. Biochemistry 2013, 52, 6816–6823. [Google Scholar] [CrossRef]
- Badarau, A.; Baslé, A.; Firbank, S.J.; Dennison, C. Crosstalk between Cu(I) and Zn(II) homeostasis via Atx1 and cognate domains. Chem. Commun. 2013, 49, 8000–8002. [Google Scholar] [CrossRef]
- Hoppen, C.; Müller, L.; Albrecht, A.C.; Groth, G. The NOP-1 peptide derived from the central regulator of ethylene signaling EIN2 delays floral senescence in cut flowers. Sci. Rep. 2019, 9, 1287. [Google Scholar] [CrossRef] [PubMed]
- Kessenbrock, M.; Klein, S.M.; Müller, L.; Hunsche, M.; Noga, G.; Groth, G. Novel Protein-Protein Inhibitor Based Approach to Control Plant Ethylene Responses: Synthetic Peptides for Ripening Control. Front. Plant Sci. 2017, 8, 1528. [Google Scholar] [CrossRef] [PubMed]
- Aghdam, M.S.; Razavi, F. Octapeptide NOP-1 treatment delays yellowing in broccoli floret during low temperature storage. Postharvest Biol. Technol. 2021, 180, 111628. [Google Scholar] [CrossRef]
- Klein, S.; Fiebig, A.; Neuwald, D.; Dluhosch, D.; Müller, L.; Groth, G.; Noga, G.; Hunsche, M. Influence of the ethylene-related signal-inhibiting octapeptide NOP-1 on postharvest ripening and quality of ‘Golden Delicious’ apples. J. Sci. Food Agric. 2019, 99, 3903–3909. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, S.; Krezel, A.M.; Li, W. Stabilization and structure determination of integral membrane proteins by termini restraining. Nat. Protoc. 2022, 17, 540–565. [Google Scholar] [CrossRef] [PubMed]
- Kastner, B.; Fischer, N.; Golas, M.M.; Sander, B.; Dube, P.; Böhringer, D.; Hartmuth, K.; Deckert, J.; Hauer, F.; Wolf, E.; et al. GraFix: Sample preparation for single-particle electron cryomicroscopy. Nat. Methods 2008, 5, 53–55. [Google Scholar] [CrossRef] [PubMed]
- Lusty, C.J. A gentle vapor-diffusion technique for cross-linking of protein crystals for cryocrystallography. J. Appl. Crystallogr. 1999, 32, 106–112. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rüffer, B.; Thielmann, Y.; Lemke, M.; Minges, A.; Groth, G. Crystallization of Ethylene Plant Hormone Receptor—Screening for Structure. Biomolecules 2024, 14, 375. https://doi.org/10.3390/biom14030375
Rüffer B, Thielmann Y, Lemke M, Minges A, Groth G. Crystallization of Ethylene Plant Hormone Receptor—Screening for Structure. Biomolecules. 2024; 14(3):375. https://doi.org/10.3390/biom14030375
Chicago/Turabian StyleRüffer, Buket, Yvonne Thielmann, Moritz Lemke, Alexander Minges, and Georg Groth. 2024. "Crystallization of Ethylene Plant Hormone Receptor—Screening for Structure" Biomolecules 14, no. 3: 375. https://doi.org/10.3390/biom14030375
APA StyleRüffer, B., Thielmann, Y., Lemke, M., Minges, A., & Groth, G. (2024). Crystallization of Ethylene Plant Hormone Receptor—Screening for Structure. Biomolecules, 14(3), 375. https://doi.org/10.3390/biom14030375





