Abstract
Collagen VI-related dystrophies (COL6RD) are a group of rare muscle disorders caused by mutations in specific genes responsible for type VI collagen production. It affects muscles, joints, and connective tissues, leading to weakness, joint problems, and structural issues. Currently, there is no effective treatment for COL6RD; its management typically addresses symptoms and complications. Therefore, it is essential to decipher the disease’s molecular mechanisms, identify drug targets, and develop effective treatment strategies to treat COL6RD. In this study, we employed differential gene expression analysis, weighted gene co-expression network analysis, and genome-scale metabolic modeling to investigate gene expression patterns in COL6RD patients, uncovering key genes, significant metabolites, and disease-related pathophysiological pathways. First, we performed differential gene expression and weighted gene co-expression network analyses, which led to the identification of 12 genes (CHCHD10, MRPS24, TRIP10, RNF123, MRPS15, NDUFB4, COX10, FUNDC2, MDH2, RPL3L, NDUFB11, PARVB) as potential hub genes involved in the disease. Second, we utilized a drug repurposing strategy to identify pharmaceutical candidates that could potentially modulate these genes and be effective in the treatment. Next, we utilized context-specific genome-scale metabolic models to compare metabolic variations between healthy individuals and COL6RD patients. Finally, we conducted reporter metabolite analysis to identify reporter metabolites (e.g., phosphatidates, nicotinate ribonucleotide, ubiquinol, ferricytochrome C). In summary, our analysis revealed critical genes and pathways associated with COL6RD and identified potential targets, reporter metabolites, and candidate drugs for therapeutic interventions.
1. Introduction
Muscular dystrophies are a group of congenital disorders resulting from gene mutations that play an essential role in muscle function and structure. Their clinical symptoms are progressive, characterized by muscle weakness and disability [1,2]. There are various types of muscular dystrophy, each with distinct onset times, severity levels, and life expectancies. COL6RD represents a subgroup of muscular dystrophies, extending from the less severe Bethlem muscular dystrophy to the more severe Ullrich muscular dystrophy [3]. These conditions result from genetic mutations affecting the genes responsible for producing the three primary α-chains of collagen type VI: COL6A1, COL6A2, and COL6A3 [4]. COL6RD is associated with multiple metabolic dysfunctions, especially those impacting mitochondria and cellular energy production [4,5]. Unfortunately, there is not an FDA-approved drug specifically developed for COL6RD [4]. The treatment primarily aims at managing symptoms and complications, including physical therapy, respiratory support, orthopedic care, nutritional assistance, and cardiac evaluations [4].
Systems biology is an interdisciplinary field that explores the interactions of biological components as an integrated system, rather than analyzing them individually. The development of high-throughput technologies and enhanced computational capabilities has given rise to systems biology as a novel field of research [6]. Its methodologies are employed to comprehend complex diseases, uncover molecular mechanisms, and accelerate biomarker and drug discovery [7,8,9,10,11,12,13]. It combines experiments and computational analysis to explore how biological components operate, employing diverse methods such as bioinformatic tools, network modeling, and computational modeling [14,15]. These approaches are substantial for gaining a comprehensive understanding of biology. For example, weighted gene co-expression network analysis (WGCNA) has been effectively used in previous studies to build gene networks in diverse diseases, identifying hub genes as potential biomarkers and therapeutic targets [16,17]. Furthermore, genome-scale metabolic models (GEMs) have been applied in various contexts, they offer detailed insight into the connections between metabolites, genes, and reactions in the organisms by consolidating current literature knowledge and omics data [18,19,20].
Drug repurposing, exploring new uses for existing medications, is gaining popularity due to the high costs and low success rates of new drug development [21]. This method utilizes approved drugs or well-characterized drug candidates, potentially cutting down costs and development time [22]. Approaches include experimental and computational methods such as analyzing disease pathways, drug interactions, multi-omics data integration, and clinical data mining [23]. As a successful example, combined metabolic activators (CMAs) have been repurposed for treating mitochondrial dysfunction-associated diseases, including neurodegenerative diseases, nonalcoholic fatty liver disease, and COVID-19; their effects have been demonstrated in preclinical and clinical studies [24,25,26,27,28,29]. Previously, anti-TGFβ antibodies [30], SB431542 [30], losartan [30], and cyclosporin A [31,32] have been suggested for treating COL6RD based on previous studies, but further research and clinical trials are necessary to assess their safety and effectiveness. Within this context, employing a systems biology approach could assist in repurposing current medications and innovating new treatment options for COL6RD.
In this study, we employed systems biology methodologies to understand the molecular pathophysiology of COL6RD, decipher the underlying molecular mechanisms of the disease, and identify potential drug targets that could aid in developing more effective treatment options. First, we performed differential expression analysis and gene set enrichment analyses and identified the upregulated and downregulated genes and their associated pathways along two conditions. Secondly, we generated a weighted gene co-expression network to reveal the genes that work collaboratively in healthy and diseased states. It enabled the identification of the hub genes that have important roles in the occurrence and progression of the disease. Thirdly, we applied a pathway-based drug repurposing method to discover potent drug candidates by calculating the correlation between gene perturbation profiles and functional enrichment results of the target genes. Finally, we reconstructed context-specific genome-scale metabolic models for both COL6RD patients and the control group. This allowed us to identify reporter metabolites and present the global changes at the metabolite level. The general outflow of the study is represented in Figure 1.
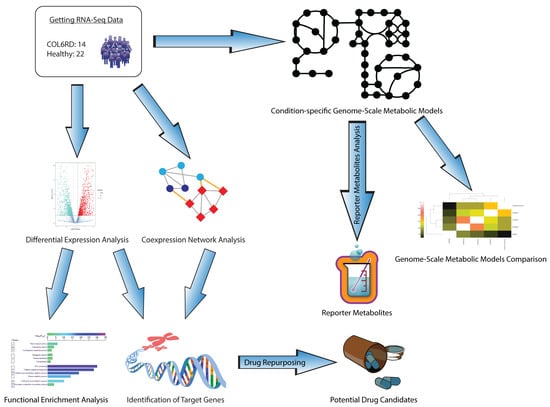
Figure 1.
A blueprint illustrating a systematic approach for our research. Initially, we analyzed RNA sequencing data from a prior cohort to pinpoint gene targets. This involved conducting differential expression, functional enrichment, and gene co-expression network analysis, then integrating their findings. Subsequently, we employed a pathway-based method for drug repurposing to identify drugs that can activate these genes in COL6RD. Additionally, we developed condition-specific GEMs to highlight metabolic distinctions between COL6RD patients and healthy controls. Finally, we utilized reporter metabolite analysis to propose potential biomarkers for COL6RD.
2. Materials and Methods
2.1. Transcriptomics Data and Pre-Processing
In this study, we employed previously generated global RNA-sequencing (transcriptomics) data of muscle tissues obtained from 22 patients with COL6RD and 14 healthy controls [33]. First, raw fastq files were obtained from the Gene Expression Omnibus database (accession number: GSE103608, access date: 8 April 2024) with the aid of “prefetch” and “fasterq-dump” functions of the SRA Toolkit [34]. Additionally, we downloaded the latest human reference genome (GRCh38.p14) from the Ensembl database [35]. First, an index file was created using the “index” function. Then, transcript abundances were calculated using the “quant” function of Kallisto (version 0.46.1) [36]. The expression level of each gene was calculated by summing the values of all its transcripts. We selected only protein-coding genes annotated in the BioMart data mining tool [37] because a core objective of our study is to identify protein targets and modulate their activity with drugs. The metadata of the samples (age, sex, clinical phenotype, etc.) can be found in the Supplementary Data.
2.2. Differential Gene Expression and Gene Set Enrichment Analyses
We used the “DESeq2” package in R to conduct the differential gene expression analysis [38]. Initially, a matrix was generated by correlating clinical data with raw count numbers from COL6RD patients and the control group. Later, we eliminated genes with total counts below 10 across all samples to achieve more reliable outcomes and minimize noise. Subsequently, the analysis was run by choosing the healthy control group as the baseline condition to detect differentially expressed genes (DEGs). DEGs with an adjusted p-value smaller than 10−5 were considered statistically significant for our downstream analysis. DEGs with a positive log2FoldChange value were labeled as upregulated, while those with a negative log2FoldChange value were labeled as downregulated. We utilized the “ggplot” package in R to visualize the results of the differential gene expression analysis [39].
For gene set enrichment analyses, the “enrichGo” function of the clusterProfiler package in R was employed to identify significantly enriched genes utilizing the hypergeometric distribution for biological process pathways [40]. A p-value < 0.05 was employed as a cut-off to pinpoint significantly enriched pathways. The “org.Hs.eg.db” package was used for human gene annotation and only the biological process (BP) ontology parameter was selected for the analyses [41]. The “enrichplot” package was used for illustration of the gene set enrichment outcomes.
2.3. Weighted Gene Co-Expression Network Analysis
Weighted gene co-expression network analysis was used to identify co-expression patterns among genes and their relationship with certain conditions. To build co-expression networks, we employed the WGCNA package in R [42]. As a first step, we undertook quality control utilizing the “goodSamplesGenes” function that helped to exclude genes with many incomplete records. By this means, we ensured that the analysis was conducted using reliable data. Later, a cluster tree map was created to find and exclude outlier samples. Next, genes with low counts (fewer than 15 in 75% of samples) were removed to ensure that the remaining genes had sufficient expression levels in most samples. Thus, we enabled the detection of more robust co-expression patterns.
“DESeq2” was employed for variance stabilization and normalization of the count data, in order to maintain comparable expression levels across samples. Next, correlation coefficients were calculated between every pair of genes to generate a gene expression similarity matrix, representing the strength of their co-expression relationships. The package’s “pickSoftThreshold” function was used to select the optimal soft threshold value, aiming to achieve a scale-free network that has a minimum scale independence of 0.8. Subsequently, the adjacency matrix was derived from the gene expression similarity matrix. Then, it was converted into a topological overlap matrix that captures both direct and indirect relationships between genes using hierarchical clustering. Dynamic cut tree methods were used to detect gene co-expression network modules (with the “minCoreKME” parameter set to 0.5 and the “mergeCutHeight” parameter set to 0.25). The topological overlap matrix (TOM) was designated as “signed” to identify gene modules that were positively correlated, indicating they have similar biological functions.
After building modules, we calculated their eigengenes, which represent the dominant expression pattern shared by genes within the modules. Following, the correlation between module eigengenes and disease state was measured with the help of the Pearson correlation coefficient [43]. Modules with a p-value below 0.05 and a positive correlation with COL6RD were termed upregulated modules, while those with a negative correlation were termed downregulated modules. Last, we employed intramodular analysis to pinpoint hub genes by measuring module membership parameters along with p-values.
2.4. Defining Gene Targets
Genes in each module were ranked in decreasing order according to their module membership scores. Subsequently, the top 1% of genes from every module were identified and extracted. Initially, we examined the overlap between upregulated and downregulated DEGs and the top 1% of upregulated and downregulated modules. Next, a more detailed investigation of these genes within the Human Protein Atlas Database was conducted [44]. We selected genes with certain RNA expression levels in muscle tissue (tissue-enriched or tissue-enhanced) as our targets. Since COL6RD primarily affects muscles, we hypothesized that targeting muscle-specific genes would reduce systemic side effects and increase the therapy’s success rate when we manipulated these genes with drugs.
2.5. Pathway-Based Drug Repositioning
In this study, we used a web-based tool known as Gene2drug, which leverages gene expression data sourced from the Connectivity Map [45,46]. These data were transformed into pathway expression profiles. Then, they were sorted based on the p-value of the Kolmogorov–Smirnov statistic. Subsequently, when a collection of pathways was provided, the Kolmogorov–Smirnov statistic was reapplied to identify drugs that consistently induce upregulation or downregulation across most pathways within the set.
We repeated gene set enrichment analyses on our specified gene targets from the previous step. Following this, we integrated biological process gene ontology results into the Gene2drug tool and downloaded all the findings in an Excel file. We selected only drugs with positive enrichment scores that indicate pathway upregulation and p-values below 0.05. We excluded antineoplastic agents due to their cytotoxic effects.
2.6. Genome-Scale Metabolic Modeling and Reporter Metabolite Analysis
We separately computed the average transcripts per million (TPM) values for each gene in the COL6RD patients and the healthy control group. Genes with an average TPM of at least 1 were removed to reduce noise from low-expression genes in the cohort. Subsequently, we used these values as the starting point for creating specialized GEMs with the help of a task-driven integrative network inference algorithm called tINIT using Human-GEM v1.18 as the reference model [8,47]. This algorithm automated the creation of GEMs by incorporating omics data. Modeling was conducted in MATLAB R2023a (MathWorks, Natick, MA, USA) with the RAVEN v2.8.6 toolbox for constructing the models and the Gurobi 11.0 (Gurobi Optimization LLC, Beaverton, OR, USA) solver for optimization [48]. Before advancing in our pipeline, we assessed the models to determine if they could successfully execute 57 predefined metabolic tasks, referred to as essential tasks in the reference model [8]. These tasks encompassed vital cellular functions, e.g., protein construction, ATP production, and nucleotide synthesis. Five GEMs were built to represent healthy controls, COL6RD patients with 3 different histological states, and a common COL6RD model to describe all patients’ average expression profiles.
The reporter metabolites analysis (RMA) was carried out employing the “reporterMetabolites” function within the RAVEN Toolbox. This worked through results integration from the differential gene expression analysis into the common COL6RD model. The analysis pinpointed metabolites that were influenced by alterations in metabolic networks and gene expression levels in COL6RD patients. Before running the RMA, 20 currency metabolites (“H2O”, “CO2”, “O2”, “H+”, “HCO3−”, “Na+”, “CoA”, “Pi”, “PPi”, “AMP”, “ADP”, “ATP”, “NAD+”, “NADH”, “NADP+”, “NADPH”, “PAP”, “PAPS”, “FAD”, and “FADH2”) were excluded from the model because their widespread involvement in reactions can obscure specific changes, add noise, and reduce clarity. Their removal highlighted relevant pathway-specific changes and improved statistical power. Our primary focus was on metabolites exhibiting lower p-values than 0.05, and RMA was conducted separately for those linked to downregulated and upregulated genes.
To investigate diversities between common COL6RD and the healthy GEMs, the hamming distance was computed for each couple of models, based on the count of distinct reactions present exclusively in one of the models. Additionally, the findings were visually represented through a cluster gram. Furthermore, differences in subsystem coverage were demonstrated for subsystems showing at least a 25% variation in one or more GEMs, emphasizing significant alterations in metabolic processes under varying conditions. Also, we created a dot plot to illustrate the diversity in metabolic networks among various models. The diversity was quantified using the “compareMultipleModels” function from the RAVEN Toolbox. For this analysis, we utilized an extensive metabolic task file containing 256 tasks, named “metabolicTasks_Full.xlsx”, which was in the Human-GEM repository.
3. Results
3.1. Analysis of Transcriptomic Data from COL6RD Patients Reveals Distinctive Gene Signatures
We performed the differential gene expression analysis for transcriptomics data of COL6RD patients versus healthy controls to identify the changes in gene signatures at the disease state. The cohort included samples from 22 muscular dystrophy patients and 14 healthy individuals. Consequently, we detected 2528 DEGs within the cohort (with a p-adjusted value < 10−5), comprising 1679 upregulated genes and 849 downregulated genes in COL6RD samples relative to the control samples (Figure 2A).
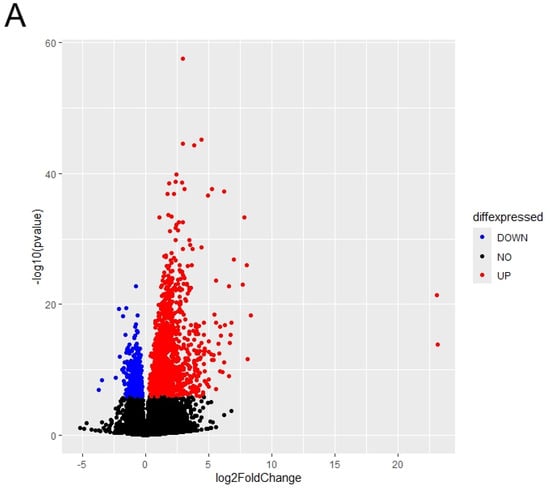
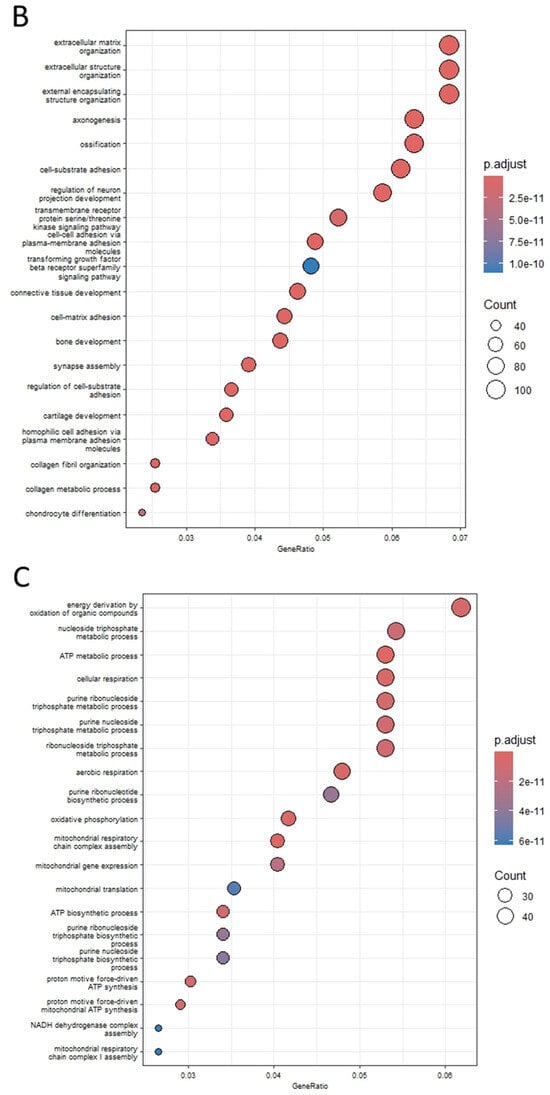
Figure 2.
(A) The distribution of differentially expressed genes is depicted. Genes exhibiting significantly upregulated expressions (FDR < 10−5) are highlighted in red, while those with significantly downregulated expressions (FDR < 10−5) are depicted in blue. Genes with FDR scores exceeding 10−5 are illustrated in black. (B,C) The top 20 enriched pathways of upregulated DEGs are presented in (B), while the top 20 enriched pathways of downregulated DEGs are displayed in (C), based on the p-adjusted score. In this representation, a smaller p-adjusted value is denoted by the color red, while blue indicates a higher value. The size of the dots reflects the number of genes associated with a specific pathway.
Functional enrichment analysis revealed that upregulated DEGs were significantly enriched in extracellular matrix and structure organization, cell-substrate adhesion, bone development, collagen fibril organization, axonogenesis, cell–cell adhesion via plasma membrane adhesion molecules, and ossification (Figure 2B). On the other hand, downregulated DEGs were predominantly enriched in aerobic respiration and its associated processes, including mitochondrial respiratory chain complex assembly, oxidative phosphorylation, proton motive force-driven mitochondrial ATP synthesis, ATP metabolic process, and mitochondrial gene expression (Figure 2C). Likewise, the initial research paper emphasized the increased activity of extracellular matrix and structural organization pathways while noting a decrease in mitochondrial pathways [33]. However, there were notable differences. For instance, we identified 2528 DEGs, whereas the original study reported only 248. Additionally, our gene ontology analysis revealed oxidative phosphorylation and mitochondrial energy metabolism as significant pathways for downregulated DEGs, contrasting with the original study’s emphasis on fiber contraction and metabolic processes.
3.2. Weighted Gene Co-Expression Network Analysis Reveals the Gene Modules for COL6RD
We generated a gene co-expression network matrix using the raw count from the RNA-seq data to investigate functional gene modules. During the initial quality control phase, 1361 genes were eliminated because of either having excess missing values or zero variance. Later, the hierarchical clustering method detected two outlier samples (SRR6015106, SRR6015079) and omitted them from the analysis (Figure 3A). Following that, 7886 genes that had less than 15 counts in 75% of samples were eliminated for the downstream pipeline. After normalizing the counts using the Deseq2 package, the soft threshold power of 9 was selected by reason of yielding high R2 results and lower mean connectivity (Figure 3B). Subsequently, an adjacency matrix was constructed, and the topological overlap measure along with dynamic tree cutting using a cut-off value of 0.25 was applied to generate clusters (Figure 3C). This process resulted in the identification of 12 modules, with module sizes varying from 24 to 4872.
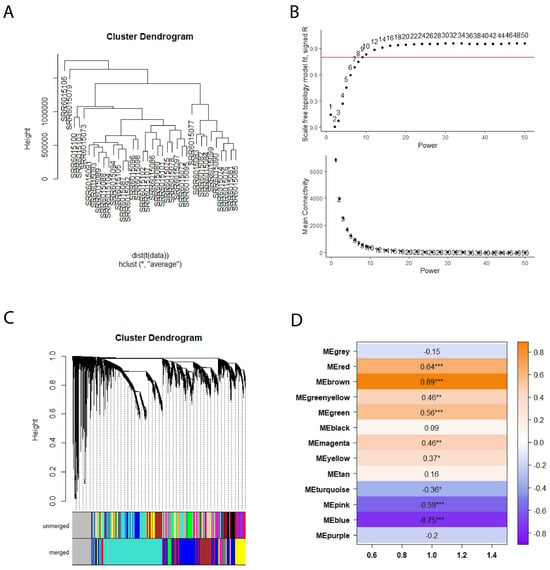
Figure 3.
(A) The dendrogram illustrating the hierarchical clustering of the COL6RD cohort’s samples was generated, but two outliers (SRR6015106 and SRR6015079) were excluded. (B) Graphs were created to depict the mean connectivity and fit of the scale-free topology model, with the y-intercept set at 0.8. Based on a high R2 value and reduced mean connectivity, a soft threshold power of 9 was chosen. (C) Profiles of cluster dendrograms and module detection are displayed, with module colors shown before and after merging below the dendrogram. (D) The heatmap shows Pearson correlations between module eigengenes and COL6RD disease state, with positive correlations in brown and negative in purple. Asterisks indicate significance levels: a single asterisk (*) denotes a p-value below 0.05, a double asterisk (**) denotes a p-value below 0.01, and a triple asterisk (***) denotes a p-value below 0.001. Genes that are not part of any module are shown in grey.
Following the computation of module eigengenes for each module, the Pearson correlation coefficient was determined between module eigengenes and the disease state. Among the 12 modules tested, 9 displayed statistically significant results (p < 0.05). Within these modules, six were classified as upregulated (yellow, green-yellow, magenta, green, red, brown), while three were categorized as downregulated (blue, pink, turquoise) (Figure 3D). When examining gene set enrichment analyses of upregulated modules, various distinct patterns emerge. For instance, the yellow module was enriched in processes such as ncRNA processing and endosomal transport, while the green-yellow module shows enrichment in the planar cell polarity pathway and epithelial tube morphogenesis. The green module exhibits enrichment in pathways related to immune response activation via cell surface receptors and myeloid leukocyte activation. In contrast, the red module demonstrates enrichment in lipid catabolism and alcohol metabolism, and the brown module was enriched in extracellular matrix organization. Interestingly, the magenta module did not show enrichment in any specific biological pathways. Similarly, downregulated modules also displayed unique enrichment patterns. For example, the blue module was enriched in aerobic and cellular respiration as well as oxidative phosphorylation. The pink module exhibits enrichment in cytoplasmic translation and ribosome biogenesis, while the turquoise module shows enrichment in RNA splicing and ribosome biogenesis. The results of functional enrichment analysis for all gene modules are presented in the Supplementary Data.
3.3. Identification of the Hub Genes as Drug Targets
Out of the co-expression network modules, we selected only the brown and blue modules for our downstream analysis as they have both the lowest p-value and the highest correlation score with the disease state. Upon examining the intersection of the top 1% of the brown module, as determined by the module membership parameter and the upregulated DEGs, we discovered 22 common genes (Figure 4A). However, second step control from the Human Protein Atlas dataset revealed that none of these genes exhibited muscle tissue-specific expression RNA levels. Therefore, we did not select any of them as targets. On the other hand, when comparing the top 1% of the blue module with the downregulated DEGs, we identified 24 common genes. Among these 24 genes, 12 displayed muscle tissue-specific RNA expression levels (Figure 4B). As a result, we selected these genes as our targets: CHCHD10, MRPS24, TRIP10, RNF123, MRPS15, NDUFB4, COX10, FUNDC2, MDH2, RPL3L, NDUFB11, and PARVB (Table 1).
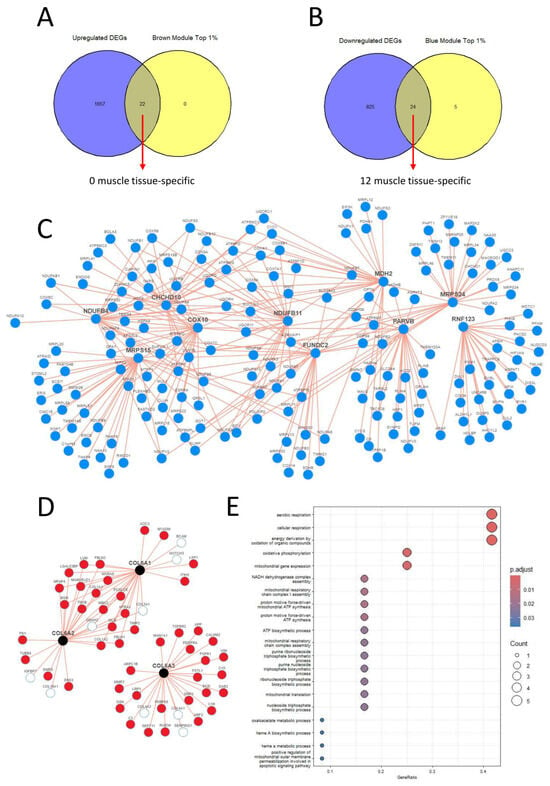
Figure 4.
(A,B) We identified 22 genes shared between the top 1% upregulated modules and upregulated DEGs, and 24 genes common to the top 1% downregulated modules and downregulated DEGs. However, only 12 genes that exclusively have specific expression patterns for muscle tissue, based on the Human Protein Atlas database, were selected as targets. (C) Using the iNetModels database, a network graph of coexpression patterns for 10 selected genes was created. Node limits were set to 25 and red lines indicate positive correlations. (D) A coexpression map for COL6A1, COL6A2, and COL6A3 was created using the iNetModels database. Red nodes show upregulated genes in our study, while white nodes are not differentially expressed. Red lines represent positive correlations between nodes. The node limit was set to 25, and the correlation parameter was set to “Both”. (E) Functional enrichment analysis results for the target genes are presented here, which were subsequently utilized in the Gene2drug tool.

Table 1.
List of downregulated target genes for COL6RD and their descriptions.
To provide a more comprehensive overview of COL6RD’s gene expression profile, we also examined COL6A1, COL6A2, and COL6A3 gene co-expression patterns using the iNetmodels database [61,62]. We observed several positive correlations between COL6A1 and COL6A2, but no correlations were observed with COL6A3. This indicated that COL6A1 and COL6A2 had a stronger correlation with each other compared to COL6A3. This variation might result from COL6A3 being regulated diversely and having additional biological functions compared to the other two alpha chains [63]. Furthermore, when examining all the connection nodes among COL6A1, COL6A2, and COL6A3, we found that 46 nodes exhibited upregulation in our dataset, whereas 9 nodes showed no differential expression in our study. Figure 4D presents this co-expression comparison map and the correlations between the nodes.
3.4. Drug Repositioning to Identify Potential Drug Candidates
We utilized gene ontology terms representing biological pathways obtained from gene set enrichment analysis of 12 target genes as input for the Gene2drug database (Figure 4E). This yielded 222 drugs (p < 0.05), among which 64 drugs were found to upregulate our target pathways (with positive enrichment scores), while 158 drugs downregulated them (with negative enrichment scores). Given that all our gene targets for COL6RD were downregulated, we aimed to upregulate them to treat the disease and restore cell status closer to the healthy state. Consequently, we focused on drugs with positive enrichment scores. Next, we selected the top 10 drugs based on the lowest p-values, excluding antineoplastic agents due to their cytotoxic effects. These drugs include apigenin, flunarizine, deferoxamine, luteolin, verteporfin, ursodeoxycholic acid, ioxaglic acid, risperidone, fipexide, and naftifine. More detailed information about these drugs can be found in Table 2.

Table 2.
Top 10 repurposed drugs for COL6RD and their enrichment scores, p-values and brief descriptions.
3.5. Comparison Between the Context-Specific Genome-Scale Metabolic Models
To comprehend the metabolic context of the disease, we constructed four GEMs for COL6RD and one model for a healthy state based on their transcriptomic profiles using tINIT. The COL6RD models include separate representations for three histology grades, denoted as COL6RD-1, COL6RD-2, and COL6RD-3. Additionally, a common COL6RD model was developed by averaging the TPM values across all muscular dystrophy patients at all three stages, providing a general view of the disease. Following this, we carried out an analytical evaluation using methodologies based on GEMs. The GEMs for COL6RD-1, COL6RD-2, COL6RD-3, and COL6RD-Common varied in their numbers of genes, reactions, and metabolites. Specifically, COL6RD-1 comprised 1877 genes, 6810 reactions, and 5621 metabolites, while COL6RD-2 included 1899 genes, 6576 reactions, and 5258 metabolites. Similarly, COL6RD-3 consisted of 1907 genes, 6697 reactions, and 5375 metabolites, and COL6RD-Common encompassed 1909 genes, 7038 reactions, and 5880 metabolites. In contrast, the healthy model consisted of 1740 genes, 6409 reactions, and 5280 metabolites.
Upon inspection of the Hamming distance between the GEMs, it was observed that the healthy and COL6RD-Common models exhibit the most distinct pattern, as expected. Furthermore, COL6RD-3 and COL6RD-2 displayed the highest similarity, with a Hamming distance score of 0.96. This was followed by the COL6RD-1 and COL6RD-Common models (Figure 5A). When looking at subsystem coverage between models, we saw upregulation in peptide metabolism in the healthy model compared to COL6RD-Common. In contrast, the healthy model had downregulation in vitamin B2 metabolism, beta-oxidation of fatty acids, bile acid metabolism, and phenylalanine metabolism compared to the disease model (Figure 5B). DPA, DHA, 12,15,18,21-tetracosatetraenoic acid and adrenic acid de novo synthesis were represented in the healthy model but were not represented in the COL6RD-Common model. On the other hand, various tasks were represented in the COL6RD-Common model and not in the healthy model such as ATP salvage from hypoxanthine, lauric acid myristic acid de novo synthesis, etc. (Figure 5C).
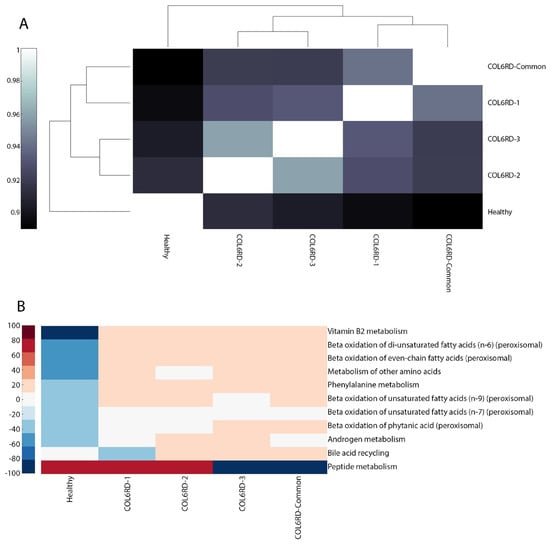

Figure 5.
(A) A cluster gram displays Hamming distances between COL6RD and healthy GEMs, based on their reaction content and gene expressions. Notably, the Healthy GEM and COL6RD-Common model, generated from the average TPM of all COL6RD patients regardless of their histological grade, exhibit the most distinct pattern compared to others. (B) Another cluster gram highlights variations in subsystem coverage, focusing on those with at least a 25% variance in one or more GEMs. Redder tones signify higher coverage, while bluer tones denote lower coverage compared to other pathways. (C) A scatter plot depicts variations in the success or failure of metabolic tasks within genome-scale models. It is worth noting that genes with expression levels below 1 TPM are considered not expressed in the model while being built with the tINIT algorithm.
3.6. Revealing the Differences by Performing Reporter Metabolite Analysis
RMA reveals important key metabolites that are associated with significant changes in gene expression levels. In this study, we identified 224 upregulated and 73 downregulated reporter metabolites for COL6RD (p < 0.05) (Figure 6A,B). Namely, the top five upregulated metabolites were nicotinate ribonucleotide, phosphatidate-LD-PC pool, phosphatidate-LD-PE pool, phosphatidate-LD-PS pool, and (7Z)-octadecenoyl-CoA, while ubiquinol, ubiquinone, ferricytochrome C, ferrocytochrome C, and 3-phospho-D-glycerate were the top five downregulated metabolites, based on ascending p-values (Table 3). We observed that these metabolites were mostly associated with mitochondrial dysfunction [74,75,76].
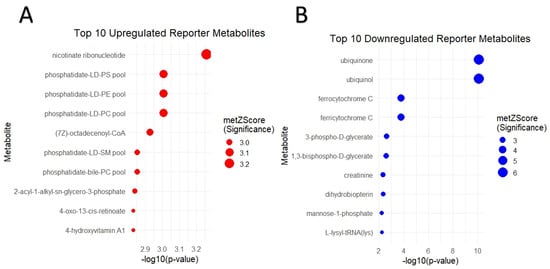
Figure 6.
(A,B) Dot plots showcase the top ten upregulated and downregulated reporter metabolites, ranked by the lowest p-value. Upregulation is denoted by red dots, while downregulation is represented by blue dots. Each dot’s size corresponds to the metabolite’s Z score, with larger sizes indicating higher Z scores.

Table 3.
Catalog of the top 10 reporter metabolites, including their regulatory status and concise explanations.
4. Discussion
To reveal the molecular mechanisms involved in COL6RD, we employed transcriptomic data from a prior cohort study. We conducted differential expression and gene ontology analysis to understand molecular patterns of the disease. Although these two analyses were conducted in the original study as well, our study provided different results due to using the latest reference genome (GRCh38.p14) and different bioinformatic tools (kallisto, DESeq2, clusterProfiler, etc.). In addition, our research has extended the original findings through further downstream analyses. For example, using gene co-expression analysis, we found gene modules and hub genes linked to COL6RD. We also identified candidate drugs suitable for therapeutic intervention by employing a pathway-based drug repurposing strategy. Also, genome-scale metabolic models for both diseased and healthy conditions were reconstructed to compare the global metabolic changes in response to COL6RD. Lastly, we identified important changes at the metabolite level by utilizing reporter metabolite analysis. To sum up, our analysis uncovers insights into the molecular pathophysiology of COL6RD, identifying potential drug candidates and reporter metabolites to advance its diagnosis, screening, and treatment strategies.
In this study, we observed downregulation in mitochondria and aerobic respiration processes in COL6RD. Similarly, previous research has shown that in mouse models and muscle cells from patients with COL6RD, mitochondria exhibit structural and functional anomalies, including enlarged size, reduced matrix density, distorted cristae, and impaired respiratory capacity [85,86,87]. While these conditions are associated with mitochondrial dysfunction and impaired oxidative phosphorylation, they lead to reduced ATP production and increased oxidative stress in muscle cells [85,88]. The combination of mitochondrial dysfunction and oxidative stress leads to muscle fiber apoptosis, which worsens muscle damage and weakness in muscular dystrophies, making them key pathogenic mechanisms.
Examination of the iNetModels database indicated that 10 out of 12 of our target genes were interconnected (Figure 4C). The proteins were mainly located in mitochondria and play roles in oxidative phosphorylation. These genes were shown as drug targets for many muscular diseases by previous researchers. For instance, COX10 encodes a key component of cytochrome c oxidase, with mutations linked to congenital muscular dystrophy and mitochondrial myopathies [89,90,91]. Another example is MDH2 (malate dehydrogenase 2), a mitochondrial enzyme identified as a promising prognostic biomarker for Duchenne muscular dystrophy [92]. Serum levels of MDH2 significantly decrease with age in Duchenne muscular dystrophy patients, correlating with disease progression [93]. Likewise, prior studies aimed to boost mitochondrial activity in COL6RD patients using cyclosporine A and its non-immunosuppressive derivatives [31,86]. The treatment improved mitochondrial function, muscle regeneration, and reduced apoptosis, supporting these compounds as potential therapies for mitochondrial dysfunction in collagen VI-related dystrophies. In this context, CMAs can also be used to activate the mitochondrial metabolism in the muscle tissue of COL6RD patients since their effect in activating mitochondria has been demonstrated previously [24,25,26,27,28,29].
Our repurposed drugs, apigenin and luteolin, are flavonoids that occur naturally in fruits and vegetables. They exhibit anti-inflammatory, antioxidant, and anticancer characteristics, offering potential advantages for diverse cancers and other oxidative-associated ailments [94,95,96]. They are recommended for muscular dystrophies and muscle atrophy due to their ability to enhance muscle growth and myogenic differentiation, and reduce oxidative stress [97,98,99,100,101]. Another medication recommended through our pathway-based drug repurposing is deferoxamine, which functions as a chelating agent utilized in the treatment of iron or aluminum toxicity [102]. Previous studies propose its potential use in treating Duchenne muscular dystrophy by preventing oxidative damage through the removal of surplus iron, potentially aiding in the reversal of muscle damage associated with the condition [103,104]. While these proposed medications have not been specifically tested for COL6RD, their potential effectiveness is anticipated based on similarities in disease mechanisms with other muscular dystrophies.
In this research, we created context-specific GEMs to represent both healthy controls and COL6RD patients in three histologic states, comparing them to identify metabolic differences. We found increased lipid metabolism and decreased energy metabolism in COL6RD patients. Later, reporter metabolite analysis showed elevated lipid-associated metabolites, e.g., phosphatidates, and reduced energy-related metabolites such as ubiquinol and ferricytochrome C. These findings align with existing literature knowledge. For instance, it is known that in many muscular dystrophies, adipose tissue takes over muscle tissue, a process known as fatty infiltration [105,106,107]. Likewise, previous research indicates that COL6RD is linked to obesity and causes a rise in overall adipose tissue, emphasizing the need to consider this in patient management [108]. Additionally, several prior studies have observed a decrease in energy metabolism in COL6RD patients [85,86,88]. Since reporter metabolites can be detected in plasma and urine and hold significance as biomarkers [75], we hypothesized that phosphatidate, ubiquinol, ferricytochrome C, and related metabolites might be candidate biomarkers for COL6RD. Nonetheless, further metabolomic and clinical studies are necessary to confirm these results.
Our study faces three main limitations. First, the small sample size (n = 36) restricts the generalizability of our findings, despite the rarity of the disease. Second, the cohort consists only of cross-sectional samples, which prevents us from observing changes over time. Lastly, our study is based solely on computational analysis without biochemical or molecular validation. To address these issues, we plan to collect more samples from COL6RD patients and validate our findings using Western blot experiments and metabolomic analysis. Although our study makes modest contributions to the understanding of COL6RD, it provides valuable insights and lays the groundwork for further research.
5. Conclusions
In conclusion, our study on COL6RD using systems biology has offered important insights into the disease’s pathophysiology. By integrating differential expression analysis with WGCNA, we pinpointed hub genes and identified 12 potential drug targets. Then, we applied the pathway-based drug repositioning strategy to discover pharmacotherapy candidates aimed at modulating these genes. Additionally, we explored metabolic differences between healthy individuals and COL6RD patients by reconstructing and comparing context-specific genome-scale metabolic models. This was followed by reporter metabolite analysis, which helped identify key metabolites that may be valuable for early detection and monitoring. Briefly, our study identified hub genes, drug targets, key metabolites and pathways linked to COL6RD, and promising drug candidates for improving patient care. It provides a foundation for future metabolomic and clinical research to validate these findings.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biom14111376/s1, Supplementry data: The metadata of gene modules.
Author Contributions
A.M. and S.G.T. conceptualized the study. A.M. managed the project and secured funding. A.B.C. organized the data and performed computational analyses. C.Z. and A.K. assisted with developing the computational methodology. A.M., O.A., S.G.T., H.T. and C.Z. provided supervision. A.B.C. wrote, A.M., S.G.T., O.A. and C.Z. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The article processing charges were covered by KTH-Royal Institute of Technology. A.C.’s PhD education and bench fees were funded by the Turkish Ministry of National Education. This study received financial support from Trustlife Therapeutics and the Knut and Alice Wallenberg Foundation (Grant number: 72254).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The RNA-seq data utilized in our research were obtained from publicly available data sources. They can be accessed through the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE103608, accessed on 3 June 2024.
Acknowledgments
ABC is sponsored by the Ministry of National Education (Turkey) for his postgraduate education and research to be recruited at Bursa Uludag University.
Conflicts of Interest
A.M. and H.T. are the founders and shareholders of Trustlife Therapeutics, while the remaining authors report no conflicts of interest.
References
- Muscular Dystrophy. NHS. Available online: https://www.nhs.uk/conditions/muscular-dystrophy/ (accessed on 3 August 2024).
- Mercuri, E.; Bönnemann, C.G.; Muntoni, F. Muscular dystrophies. Lancet 2019, 394, 2025–2038. [Google Scholar] [CrossRef] [PubMed]
- Topaloğlu, H.; Poorshiri, B. The congenital muscular dystrophies. Ann. Child Neurol. Soc. 2024, 2, 27–39. [Google Scholar] [CrossRef]
- Bönnemann, C.G. The collagen VI-related myopathies: Muscle meets its matrix. Nat. Rev. Neurol. 2011, 7, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, A.; Cescon, M.; D’Agostino, C.; Schilardi, F.; Sabatelli, P.; Merlini, L.; Faldini, C. Collagen VI in the Musculoskeletal System. Int. J. Mol. Sci. 2023, 24, 5095. [Google Scholar] [CrossRef] [PubMed]
- Voit, E.O.; Shah, A.M.; Olivença, D.; Vodovotz, Y. What’s next for computational systems biology? Front. Syst. Biol. 2023, 3, 1250228. [Google Scholar] [CrossRef]
- Larsson, I.; Uhlén, M.; Zhang, C.; Mardinoglu, A. Genome-Scale Metabolic Modeling of Glioblastoma Reveals Promising Targets for Drug Development. Front. Genet. 2020, 11, 381. [Google Scholar] [CrossRef]
- Agren, R.; Mardinoglu, A.; Asplund, A.; Kampf, C.; Uhlen, M.; Nielsen, J. Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol. Syst. Biol. 2014, 10, 721. [Google Scholar] [CrossRef]
- Doran, S.; Arif, M.; Lam, S.; Bayraktar, A.; Turkez, H.; Uhlen, M.; Boren, J.; Mardinoglu, A. Multi-omics approaches for revealing the complexity of cardiovascular disease. Brief. Bioinform. 2021, 22, bbab061. [Google Scholar] [CrossRef]
- Lam, S.; Hartmann, N.; Benfeitas, R.; Zhang, C.; Arif, M.; Turkez, H.; Uhlén, M.; Englert, C.; Knight, R.; Mardinoglu, A. Systems analysis reveals ageing-related perturbations in retinoids and sex hormones in alzheimer’s and parkinson’s diseases. Biomedicines 2021, 9, 1310. [Google Scholar] [CrossRef]
- Yang, H.; Arif, M.; Yuan, M.; Li, X.; Shong, K.; Türkez, H.; Nielsen, J.; Uhlén, M.; Borén, J.; Zhang, C.; et al. A network-based approach reveals the dysregulated transcriptional regulation in non-alcoholic fatty liver disease. iScience 2021, 24, 103222. [Google Scholar] [CrossRef]
- Li, X.; Kim, W.; Juszczak, K.; Arif, M.; Sato, Y.; Kume, H.; Ogawa, S.; Turkez, H.; Boren, J.; Nielsen, J.; et al. Stratification of patients with clear cell renal cell carcinoma to facilitate drug repositioning. iScience 2021, 24, 102722. [Google Scholar] [CrossRef] [PubMed]
- Ceyhan, A.B.; Ozcan, M.; Kim, W.; Li, X.; Altay, O.; Zhang, C.; Mardinoglu, A. Novel drug targets and molecular mechanisms for sarcopenia based on systems biology. Biomed. Pharmacother. 2024, 176, 116920. [Google Scholar] [CrossRef] [PubMed]
- Veenstra, T.D. Omics in Systems Biology: Current Progress and Future Outlook. Proteomics 2020, 21, 2000235. [Google Scholar] [CrossRef] [PubMed]
- Systems Biology as Defined by NIH|NIH Intramural Research Program. Available online: https://irp.nih.gov/catalyst/19/6/systems-biology-as-defined-by-nih (accessed on 31 July 2024).
- Liu, M.; Wang, Y.; Shi, W.; Yang, C.; Wang, Q.; Chen, J.; Li, J.; Chen, B.; Sun, G. PCDH7 as the key gene related to the co-occurrence of sarcopenia and osteoporosis. Front. Genet. 2023, 14, 1163162. [Google Scholar] [CrossRef]
- Bidkhori, G.; Benfeitas, R.; Elmas, E.; Kararoudi, M.N.; Arif, M.; Uhlen, M.; Nielsen, J.; Mardinoglu, A. Metabolic Network-Based Identification and Prioritization of Anticancer Targets Based on Expression Data in Hepatocellular Carcinoma. Front. Physiol. 2018, 9, 916. [Google Scholar] [CrossRef]
- Gu, C.; Kim, G.B.; Kim, W.J.; Kim, H.U.; Lee, S.Y. Current status and applications of genome-scale metabolic models. Genome Biol. 2019, 20, 121. [Google Scholar] [CrossRef]
- Zhang, C.; Hua, Q. Applications of Genome-Scale Metabolic Models in Biotechnology and Systems Medicine. Front. Physiol. 2016, 6, 413. [Google Scholar] [CrossRef]
- Mardinoglu, A.; Palsson, B. Genome-scale models in human metabologenomics. Nat. Rev. Genet. 2024. Online ahead of print. [Google Scholar] [CrossRef]
- Jourdan, J.-P.; Bureau, R.; Rochais, C.; Dallemagne, P. Drug repositioning: A brief overview. J. Pharm. Pharmacol. 2020, 72, 1145–1151. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Drug Repurposing: Approaches and Methods, and Considerations|Elsevier. Available online: https://www.elsevier.com/en-gb/industry/drug-repurposing (accessed on 22 April 2024).
- Yulug, B.; Altay, O.; Li, X.; Hanoglu, L.; Cankaya, S.; Lam, S.; Velioglu, H.A.; Yang, H.; Coskun, E.; Idil, E.; et al. Combined metabolic activators improve cognitive functions in Alzheimer’s disease patients: A randomised, double-blinded, placebo-controlled phase-II trial. Transl. Neurodegener. 2023, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, H.; Jin, H.; Turkez, H.; Ozturk, G.; Doganay, H.L.; Zhang, C.; Nielsen, J.; Uhlén, M.; Borén, J.; et al. The acute effect of different NAD+ precursors included in the combined metabolic activators. Free. Radic. Biol. Med. 2023, 205, 77–89. [Google Scholar] [CrossRef]
- Yang, H.; Mayneris-Perxachs, J.; Boqué, N.; del Bas, J.M.; Arola, L.; Yuan, M.; Türkez, H.; Uhlén, M.; Borén, J.; Zhang, C.; et al. Combined Metabolic Activators Decrease Liver Steatosis by Activating Mitochondrial Metabolism in Hamsters Fed with a High-Fat Diet. Biomedicines 2021, 9, 1440. [Google Scholar] [CrossRef]
- Altay, O.; Arif, M.; Li, X.; Yang, H.; Aydın, M.; Alkurt, G.; Kim, W.; Akyol, D.; Zhang, C.; Dinler-Doganay, G.; et al. Combined Metabolic Activators Accelerates Recovery in Mild-to-Moderate COVID-19. Adv. Sci. 2021, 8, 2101222. [Google Scholar] [CrossRef] [PubMed]
- Altay, O.; Yang, H.; Yildirim, S.; Bayram, C.; Bolat, I.; Oner, S.; Tozlu, O.O.; Arslan, M.E.; Hacimuftuoglu, A.; Shoaie, S.; et al. Combined Metabolic Activators with Different NAD+ Precursors Improve Metabolic Functions in the Animal Models of Neurodegenerative Diseases. Biomedicines 2024, 12, 927. [Google Scholar] [CrossRef] [PubMed]
- Zeybel, M.; Altay, O.; Arif, M.; Li, X.; Yang, H.; Fredolini, C.; Akyildiz, M.; Saglam, B.; Gonenli, M.G.; Ural, D.; et al. Combined metabolic activators therapy ameliorates liver fat in nonalcoholic fatty liver disease patients. Mol. Syst. Biol. 2021, 17, e10459. [Google Scholar] [CrossRef]
- Mohassel, P.; Hearn, H.; Zou, Y.; Rooney, J.; Bönnemann, C. P425 Inhibition of TGFβ signaling pathway as a therapeutic approach in collagen VI-related muscular dystrophy. Neuromuscul. Disord. 2023, 33, S158–S159. [Google Scholar] [CrossRef]
- Merlini, L.; Sabatelli, P.; Armaroli, A.; Gnudi, S.; Angelin, A.; Grumati, P.; Michelini, M.E.; Franchella, A.; Gualandi, F.; Bertini, E.; et al. Cyclosporine A in Ullrich Congenital Muscular Dystrophy: Long-Term Results. Oxidative Med. Cell. Longev. 2011, 2011, 139194. [Google Scholar] [CrossRef]
- Merlini, L.; Angelin, A.; Tiepolo, T.; Braghetta, P.; Sabatelli, P.; Zamparelli, A.; Ferlini, A.; Maraldi, N.M.; Bonaldo, P.; Bernardi, P. Cyclosporin A corrects mitochondrial dysfunction and muscle apoptosis in patients with collagen VI myopathies. Proc. Natl. Acad. Sci. USA 2008, 105, 5225–5229. [Google Scholar] [CrossRef]
- Guadagnin, E.; Mohassel, P.; Johnson, K.R.; Yang, L.; Santi, M.; Uapinyoying, P.; Dastgir, J.; Hu, Y.; Dillmann, A.; Cookson, M.R.; et al. Transcriptome analysis of collagen VI-related muscular dystrophy muscle biopsies. Ann. Clin. Transl. Neurol. 2021, 8, 2184–2198. [Google Scholar] [CrossRef]
- SRA-Toolkit. Available online: https://hpc.nih.gov/apps/sratoolkit.html (accessed on 29 September 2023).
- Harrison, P.W.; Amode, M.R.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2024. Nucleic Acids Res. 2024, 52, D891–D899. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- BioMart. Available online: https://www.ensembl.org/biomart/martview/f35bab39a3527e4733aa39bd8c7e953d (accessed on 31 July 2024).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 27 March 2024)ISBN 978-3-319-24277-4.
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Carlson, M.; Falcon, S.; Pages, H.; Li, N. org. Hs. eg. db: Genome wide annotation for Human. R Package Version 2019, 3, 3. [Google Scholar]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 1–13. [Google Scholar] [CrossRef]
- Mishra, P.; Singh, U.; Pandey, C.M.; Mishra, P.; Pandey, G. Application of student’s t-test, analysis of variance, and covariance. Ann. Card. Anaesth. 2019, 22, 407–411. [Google Scholar] [CrossRef]
- Sjöstedt, E.; Zhong, W.; Fagerberg, L.; Karlsson, M.; Mitsios, N.; Adori, C.; Oksvold, P.; Edfors, F.; Limiszewska, A.; Hikmet, F.; et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 2020, 367, eaay5947. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef]
- Napolitano, F.; Carrella, D.; Mandriani, B.; Pisonero-Vaquero, S.; Sirci, F.; Medina, D.L.; Brunetti-Pierri, N.; Di Bernardo, D. gene2drug: A computational tool for pathway-based rational drug repositioning. Bioinformatics 2018, 34, 1498–1505. [Google Scholar] [CrossRef]
- Agren, R.; Bordel, S.; Mardinoglu, A.; Pornputtapong, N.; Nookaew, I.; Nielsen, J. Reconstruction of Genome-Scale Active Metabolic Networks for 69 Human Cell Types and 16 Cancer Types Using INIT. PLoS Comput. Biol. 2012, 8, e1002518. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Marcišauskas, S.; Sánchez, B.J.; Domenzain, I.; Hermansson, D.; Agren, R.; Nielsen, J.; Kerkhoven, E.J. RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PLoS Comput. Biol. 2018, 14, e1006541. [Google Scholar] [CrossRef] [PubMed]
- CHCHD10 Coiled-Coil-Helix-Coiled-Coil-Helix Domain Containing 10 [Homo Sapiens (Human)]–Gene. NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/400916 (accessed on 10 October 2024).
- MRPS24 Mitochondrial Ribosomal Protein S24 [Homo Sapiens (Human)]–Gene. NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/64951 (accessed on 10 October 2024).
- TRIP10 Thyroid Hormone Receptor Interactor 10 [Homo Sapiens (Human)]–Gene. NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/9322 (accessed on 10 October 2024).
- RNF123 Ring Finger Protein 123 [Homo Sapiens (Human)]–Gene. NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/63891 (accessed on 10 October 2024).
- MRPS15 Mitochondrial Ribosomal Protein S15 [Homo Sapiens (Human)]–Gene. NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/64960 (accessed on 10 October 2024).
- NDUFB4 NADH:Ubiquinone Oxidoreductase Subunit B4 [Homo Sapiens (Human)]–Gene. NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/4710 (accessed on 10 October 2024).
- COX10 Cytochrome c Oxidase Assembly Factor Heme A: Farnesyltransferase COX10 [Homo Sapiens (human)]–Gene. NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/1352 (accessed on 10 October 2024).
- FUNDC2 FUN14 Domain Containing 2 [Homo Sapiens (Human)]–Gene. NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/65991 (accessed on 10 October 2024).
- MDH2 Malate Dehydrogenase 2 [Homo Sapiens (Human)]–Gene. NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/4191 (accessed on 10 October 2024).
- RPL3L Ribosomal Protein L3 Like [Homo Sapiens (Human)]–Gene. NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/6123 (accessed on 10 October 2024).
- NDUFB11 NADH:ubiquinone Oxidoreductase Subunit B11 [Homo Sapiens (Human)]–Gene. NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/54539 (accessed on 10 October 2024).
- PARVB Parvin Beta [Homo Sapiens (Human)]–Gene. NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/29780 (accessed on 10 October 2024).
- Lee, S.; Zhang, C.; Arif, M.; Liu, Z.; Benfeitas, R.; Bidkhori, G.; Deshmukh, S.; Al Shobky, M.; Lovric, A.; Boren, J.; et al. TCSBN: A database of tissue and cancer specific biological networks. Nucleic Acids Res. 2018, 46, D595–D600. [Google Scholar] [CrossRef] [PubMed]
- Arif, M.; Zhang, C.; Li, X.; Güngör, C.; Çakmak, B.; Arslantürk, M.; Tebani, A.; Özcan, B.; Subaş, O.; Zhou, W.; et al. iNetModels 2.0: An interactive visualization and database of multi-omics data. Nucleic Acids Res. 2021, 49, W271–W276. [Google Scholar] [CrossRef] [PubMed]
- Gara, S.K.; Grumati, P.; Urciuolo, A.; Bonaldo, P.; Kobbe, B.; Koch, M.; Paulsson, M.; Wagener, R. Three Novel Collagen VI Chains with High Homology to the α3 Chain. J. Biol. Chem. 2008, 283, 10658–10670. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Venditti, A.; Sharifi-Rad, M.; Kręgiel, D.; Sharifi-Rad, J.; Durazzo, A.; Lucarini, M.; Santini, A.; Souto, E.B.; Novellino, E.; et al. The Therapeutic Potential of Apigenin. Int. J. Mol. Sci. 2019, 20, 1305. [Google Scholar] [CrossRef]
- Flunarizine. Wikipedia. Available online: https://en.wikipedia.org/wiki/Flunarizine (accessed on 10 October 2024).
- Deferoxamine. Wikipedia. Available online: https://en.wikipedia.org/wiki/Deferoxamine (accessed on 10 October 2024).
- Luo, Y.; Shang, P.; Li, D. Luteolin: A Flavonoid that Has Multiple Cardio-Protective Effects and Its Molecular Mechanisms. Front. Pharmacol. 2017, 8, 692. [Google Scholar] [CrossRef]
- Verteporfin. Wikipedia. Available online: https://en.wikipedia.org/wiki/Verteporfin (accessed on 10 October 2024).
- Ursodeoxycholic Acid: Uses, Interactions, Mechanism of Action|DrugBank Online. Available online: https://go.drugbank.com/drugs/DB01586 (accessed on 10 October 2024).
- Ioxaglic Acid. Wikipedia. Available online: https://en.wikipedia.org/wiki/Ioxaglic_acid (accessed on 10 October 2024).
- Risperidone. Wikipedia. Available online: https://en.wikipedia.org/wiki/Risperidone (accessed on 10 October 2024).
- Fipexide. Wikipedia. Available online: https://en.wikipedia.org/wiki/Fipexide (accessed on 10 October 2024).
- Naftifine. Wikipedia. Available online: https://en.wikipedia.org/wiki/Naftifine (accessed on 10 October 2024).
- Pradhan, N.; Singh, C.; Singh, A. Coenzyme Q10 a mitochondrial restorer for various brain disorders. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2021, 394, 2197–2222. [Google Scholar] [CrossRef]
- Cytochrome c. Wikipedia. Available online: https://en.wikipedia.org/wiki/Cytochrome_c (accessed on 10 October 2024).
- Glycerol-3-Phosphate Dehydrogenase. Wikipedia. Available online: https://en.wikipedia.org/wiki/Glycerol-3-phosphate_dehydrogenase (accessed on 11 October 2024).
- MetaCyc Beta-Nicotinate D-Ribonucleotide. Available online: https://biocyc.org/compound?id=NICOTINATE_NUCLEOTIDE&orgid=META#showAll (accessed on 10 October 2024).
- Phosphatidate-LD-PC Pool, Metabolite in Human-GEM. Available online: https://metabolicatlas.org/explore/Human-GEM/gem-browser/metabolite/MAM02728c (accessed on 10 October 2024).
- Phosphatidate-LD-PE Pool, Metabolite in Human-GEM. Available online: https://metabolicatlas.org/explore/Human-GEM/gem-browser/metabolite/MAM02729c (accessed on 10 October 2024).
- Phosphatidate-LD-PS Pool, Metabolite in Rat-GEM. Available online: https://metabolicatlas.org/explore/Rat-GEM/gem-browser/metabolite/MAM02731c (accessed on 10 October 2024).
- (7Z)-Octadecenoyl-CoA, Metabolite in Human-GEM. Available online: https://metabolicatlas.org/explore/Human-GEM/gem-browser/metabolite/MAM00116r (accessed on 10 October 2024).
- Ubiquinol. Wikipedia. Available online: https://en.wikipedia.org/wiki/Ubiquinol#:~:text=A%20ubiquinol%20is%20an%20electron,IUPAC%20name (accessed on 10 October 2024).
- Coenzyme Q10. Wikipedia. Available online: https://en.wikipedia.org/wiki/Coenzyme_Q10 (accessed on 10 October 2024).
- Human Metabolome Database: Showing Metabocard for 3-Phosphoglyceric Acid (HMDB0000807). Available online: https://hmdb.ca/metabolites/HMDB0000807 (accessed on 10 October 2024).
- Zulian, A.; Tagliavini, F.; Rizzo, E.; Pellegrini, C.; Sardone, F.; Zini, N.; Maraldi, N.M.; Santi, S.; Faldini, C.; Merlini, L.; et al. Melanocytes from Patients Affected by Ullrich Congenital Muscular Dystrophy and Bethlem Myopathy have Dysfunctional Mitochondria That Can be Rescued with Cyclophilin Inhibitors. Front. Aging Neurosci. 2014, 6, 324. [Google Scholar] [CrossRef][Green Version]
- Bernardi, P.; Bonaldo, P. Mitochondrial Dysfunction and Defective Autophagy in the Pathogenesis of Collagen VI Muscular Dystrophies. Cold Spring Harb. Perspect. Biol. 2013, 5, a011387. [Google Scholar] [CrossRef]
- Lamandé, S.R. Collagen VI muscle disorders: Mutation types, pathogenic mechanisms and approaches to therapy. Prog. Heritable Soft Connect. Tissue Dis. 2021, 1348, 311–323. [Google Scholar]
- Irwin, W.A.; Bergamin, N.; Sabatelli, P.; Reggiani, C.; Megighian, A.; Merlini, L.; Braghetta, P.; Columbaro, M.; Volpin, D.; Bressan, G.M.; et al. Mitochondrial dysfunction and apoptosis in myopathic mice with collagen VI deficiency. Nat. Genet. 2003, 35, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Mitsuhashi, S.; Ohkuma, A.; Talim, B.; Karahashi, M.; Koumura, T.; Aoyama, C.; Kurihara, M.; Quinlivan, R.; Sewry, C.; Mitsuhashi, H.; et al. A Congenital Muscular Dystrophy with Mitochondrial Structural Abnormalities Caused by Defective De Novo Phosphatidylcholine Biosynthesis. Am. J. Hum. Genet. 2011, 88, 845–851. [Google Scholar] [CrossRef] [PubMed]
- Brischigliaro, M.; Zeviani, M. Cytochrome c oxidase deficiency. Biochim. Et Biophys. Acta (BBA)-Bioenerg. 2021, 1862, 148335. [Google Scholar] [CrossRef]
- Di Leo, V.; Bernardino Gomes, T.M.; Vincent, A.E. Interactions of mitochondrial and skeletal muscle biology in mitochondrial myopathy. Biochem. J. 2023, 480, 1767–1789. [Google Scholar] [CrossRef]
- Koutsoulidou, A.; Phylactou, L.A. Circulating Biomarkers in Muscular Dystrophies: Disease and Therapy Monitoring. Mol. Ther. Methods Clin. Dev. 2020, 18, 230–239. [Google Scholar] [CrossRef]
- Signorelli, M.; Ayoglu, B.; Johansson, C.; Lochmüller, H.; Straub, V.; Muntoni, F.; Niks, E.; Tsonaka, R.; Persson, A.; Aartsma-Rus, A.; et al. Longitudinal serum biomarker screening identifies malate dehydrogenase 2 as candidate prognostic biomarker for Duchenne muscular dystrophy. J. Cachexia Sarcopenia Muscle 2019, 11, 505–517. [Google Scholar] [CrossRef]
- Ntalouka, F.; Tsirivakou, A. Luteolin: A promising natural agent in management of pain in chronic conditions. Front. Pain Res. 2023, 4, 1114428. [Google Scholar] [CrossRef]
- Tian, C.; Liu, X.; Chang, Y.; Wang, R.; Lv, T.; Cui, C.; Liu, M. Investigation of the anti-inflammatory and antioxidant activities of luteolin, kaempferol, apigenin and quercetin. S. Afr. J. Bot. 2020, 137, 257–264. [Google Scholar] [CrossRef]
- Özcan, Ö.; Aldemir, O.; Karabulut, B. Flavones (apigenin, luteolin, chrysin) and their importance for health. Mellifera 2020, 20, 16–27. [Google Scholar]
- Jang, Y.J.; Son, H.J.; Choi, Y.M.; Ahn, J.; Jung, C.H.; Ha, T.Y. Apigenin enhances skeletal muscle hypertrophy and myoblast differentiation by regulating Prmt7. Oncotarget 2017, 8, 78300–78311. [Google Scholar] [CrossRef] [PubMed]
- Mosca, N.; Petrillo, S.; Bortolani, S.; Monforte, M.; Ricci, E.; Piemonte, F.; Tasca, G. Redox Homeostasis in Muscular Dystrophies. Cells 2021, 10, 1364. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Li, B.; Xu, Y.; Meng, S.; Wang, Y.; Jiang, Y. Luteolin reduces cancer-induced skeletal and cardiac muscle atrophy in a Lewis lung cancer mouse model. Oncol. Rep. 2018, 40, 1129–1137. [Google Scholar] [CrossRef]
- Hawila, N.; Hedya, S.; Abd Elmaaboud, M.; Abdin, A. Luteolin Attenuates Dexamethasone-Induced Skeletal Muscle Atrophy in Male Albino Rats. Med. J. Cairo Univ. 2019, 87, 3365–3374. [Google Scholar]
- Sitzia, C.; Meregalli, M.; Belicchi, M.; Farini, A.; Arosio, M.; Bestetti, D.; Villa, C.; Valenti, L.; Brambilla, P.; Torrente, Y. Preliminary Evidences of Safety and Efficacy of Flavonoids- and Omega 3-Based Compound for Muscular Dystrophies Treatment: A Randomized Double-Blind Placebo Controlled Pilot Clinical Trial. Front. Neurol. 2019, 10, 755. [Google Scholar] [CrossRef] [PubMed]
- Deferoxamine: Uses, Interactions, Mechanism of Action|DrugBank Online. Available online: https://go.drugbank.com/drugs/DB00746 (accessed on 3 May 2024).
- Alves, F.M.; Kysenius, K.; Caldow, M.K.; Hardee, J.P.; Chung, J.D.; Trieu, J.; Hare, D.J.; Crouch, P.J.; Ayton, S.; Bush, A.I.; et al. Iron overload and impaired iron handling contribute to the dystrophic pathology in models of Duchenne muscular dystrophy. J. Cachex Sarcopenia Muscle 2022, 13, 1541–1553. [Google Scholar] [CrossRef]
- Andrysiak, K.; Machaj, G.; Priesmann, D.; Woźnicka, O.; Martyniak, A.; Ylla, G.; Krüger, M.; Pyza, E.; Potulska-Chromik, A.; Kostera-Pruszczyk, A.; et al. Dysregulated iron homeostasis in dystrophin-deficient cardiomyocytes: Correction by gene editing and pharmacological treatment. Cardiovasc. Res. 2024, 120, 69–81. [Google Scholar] [CrossRef]
- Heskamp, L.; Ogier, A.; Bendahan, D.; Heerschap, A. Whole-muscle fat analysis identifies distal muscle end as disease initiation site in facioscapulohumeral muscular dystrophy. Commun. Med. 2022, 2, 155. [Google Scholar] [CrossRef]
- Díaz-Manera, J.; Llauger, J.; Gallardo, E.; Illa, I. Muscle MRI in muscular dystrophies. Acta Myol. 2015, 34, 95–108. [Google Scholar]
- Li, W.; Zheng, Y.; Zhang, W.; Wang, Z.; Xiao, J.; Yuan, Y. Progression and variation of fatty infiltration of the thigh muscles in Duchenne muscular dystrophy, a muscle magnetic resonance imaging study. Neuromuscul. Disord. 2015, 25, 375–380. [Google Scholar] [CrossRef]
- Rodríguez, M.A.; Barquero, L.M.D.R.; Ortez, C.I.; Jou, C.; Vigo, M.; Medina, J.; Febrer, A.; Ramon-Krauel, M.; Diaz-Manera, J.; Olive, M.; et al. Differences in Adipose Tissue and Lean Mass Distribution in Patients with Collagen VI Related Myopathies Are Associated with Disease Severity and Physical Ability. Front. Aging Neurosci. 2017, 9, 268. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).