Fingerprinting Interactions between Proteins and Ligands for Facilitating Machine Learning in Drug Discovery
Abstract
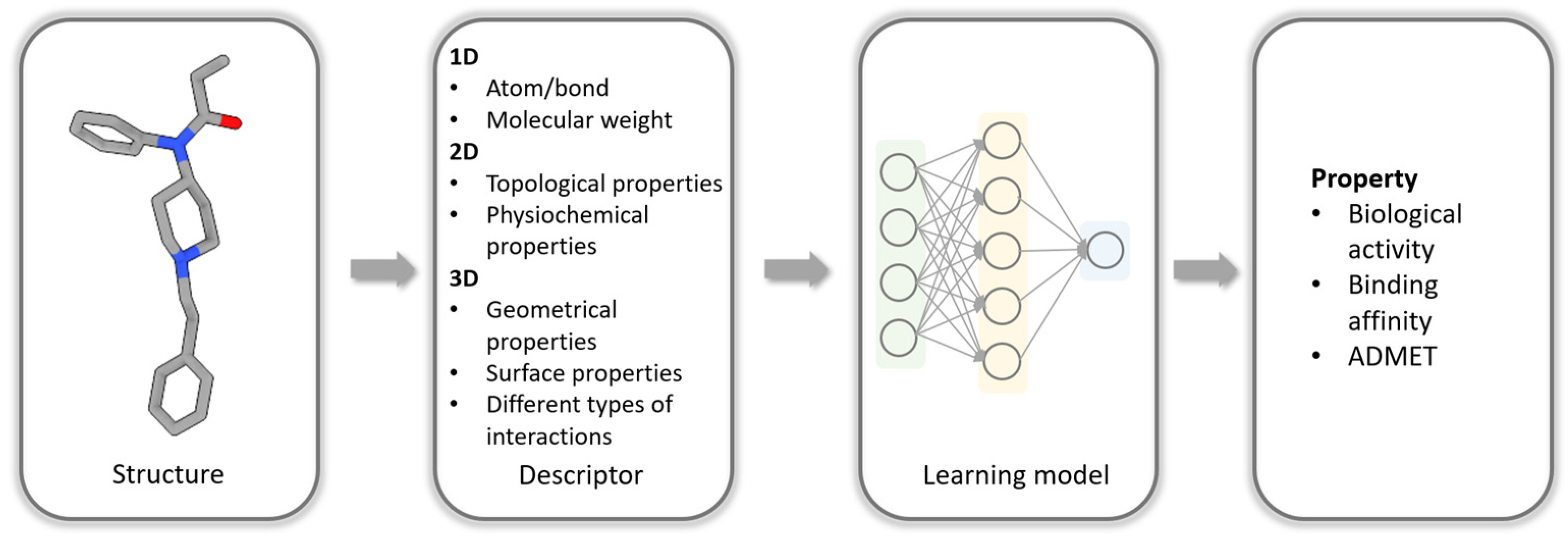
1. Introduction
2. Types of Structural Interaction Fingerprints
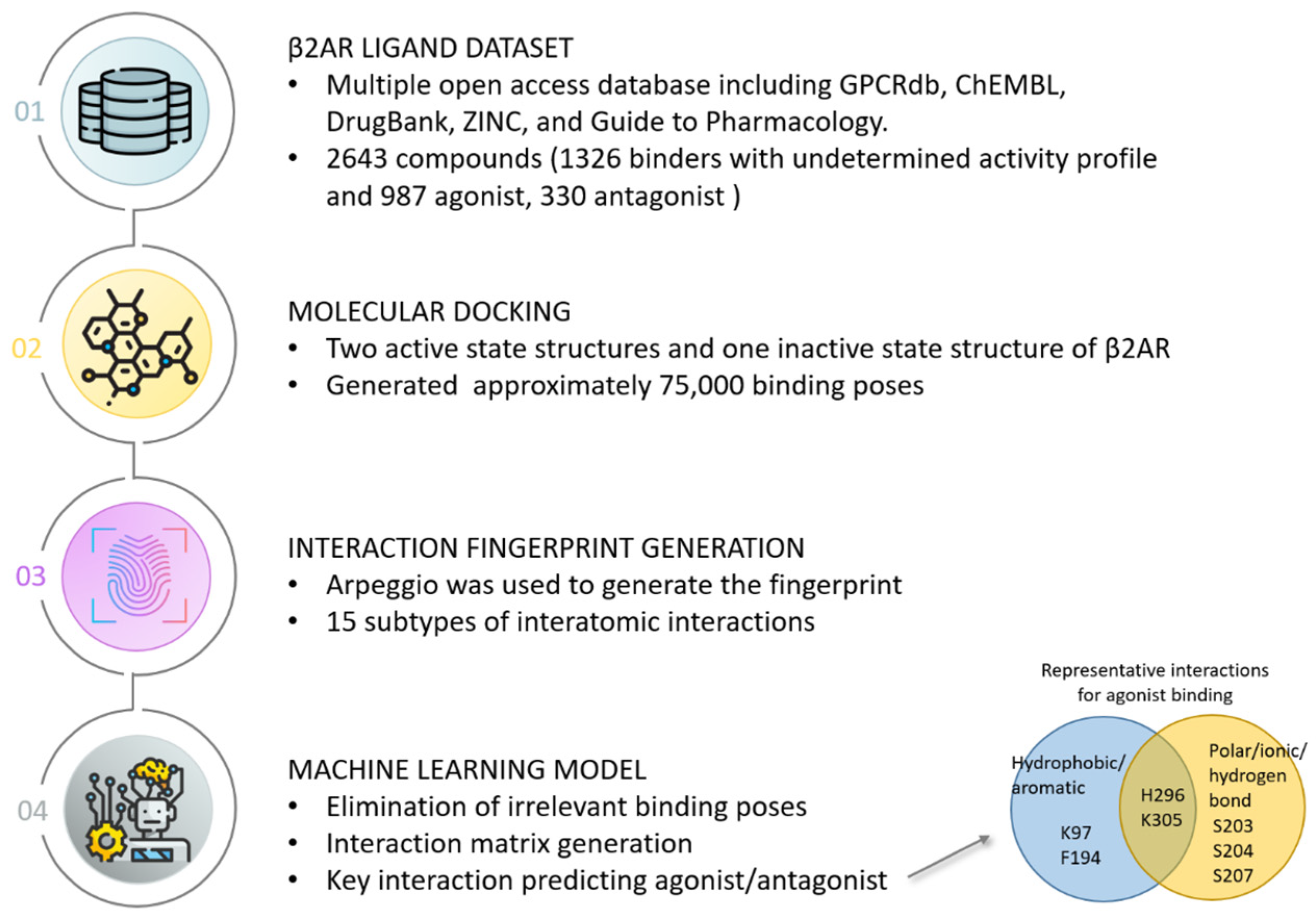
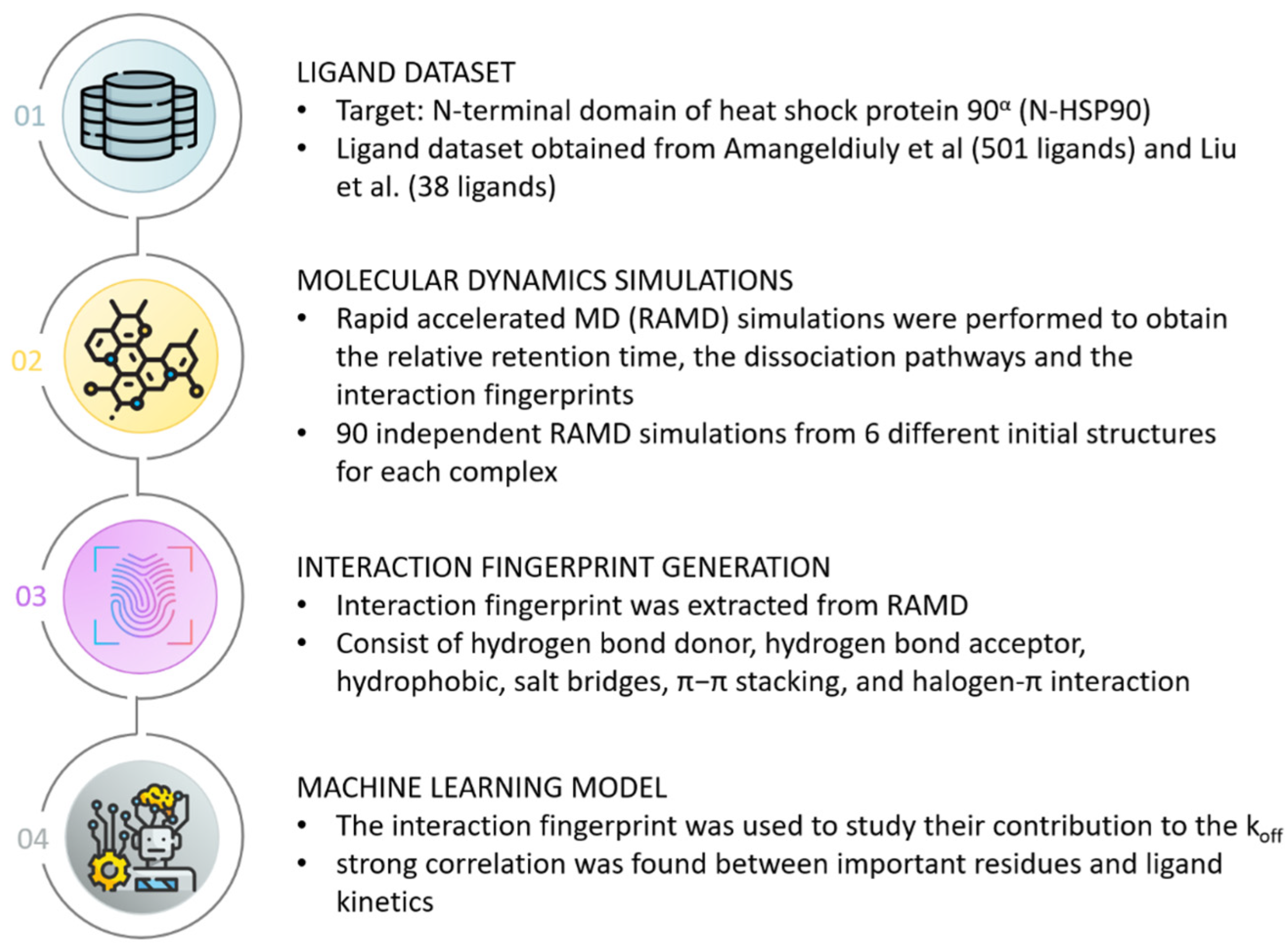
3. Case Study of Structural Interaction Fingerprint Application
4. Future Perspective
Author Contributions
Funding
Conflicts of Interest
Authors’ Disclaimer
References
- Janin, J. Protein-Protein Recognition. Prog. Biophys. Mol. Biol. 1995, 64, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, Y.; Xia, Y.-L.; Ai, S.-M.; Liang, J.; Sang, P.; Ji, X.-L.; Liu, S.-Q. Insights into Protein–Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef] [PubMed]
- Kairys, V.; Baranauskiene, L.; Kazlauskiene, M.; Matulis, D.; Kazlauskas, E. Binding Affinity in Drug Design: Experimental and Computational Techniques. Expert Opin. Drug Discov. 2019, 14, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Colwell, L.J. Statistical and Machine Learning Approaches to Predicting Protein–Ligand Interactions. Curr. Opin. Struct. Biol. 2018, 49, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Nguyen, D.D.; Sresht, V.; Mathiowetz, A.M.; Tu, M.; Wei, G.-W. Are 2D Fingerprints Still Valuable for Drug Discovery? Phys. Chem. Chem. Phys. 2020, 22, 8373–8390. [Google Scholar] [CrossRef] [PubMed]
- Hansch, C.; Maloney, P.P.; Fujita, T.; Muir, R.M. Correlation of Biological Activity of Phenoxyacetic Acids with Hammett Substituent Constants and Partition Coefficients. Nature 1962, 194, 178–180. [Google Scholar] [CrossRef]
- Hong, H.; Xie, Q.; Ge, W.; Qian, F.; Fang, H.; Shi, L.; Su, Z.; Perkins, R.; Tong, W. Mold2, Molecular Descriptors from 2D Structures for Chemoinformatics and Toxicoinformatics. J. Chem. Inf. Model. 2008, 48, 1337–1344. [Google Scholar] [CrossRef]
- Hong, H.; Liu, J.; Ge, W.; Sakkiah, S.; Guo, W.; Yavas, G.; Zhang, C.; Gong, P.; Tong, W.; Patterson, T.A. Mold2 Descriptors Facilitate Development of Machine Learning and Deep Learning Models for Predicting Toxicity of Chemicals. In Machine Learning and Deep Learning in Computational Toxicology; Computational Methods in Engineering & the Sciences; Hong, H., Ed.; Springer International Publishing: Cham, Switzerland, 2023; pp. 297–321. ISBN 978-3-031-20729-7. [Google Scholar]
- Guo, W.; Liu, J.; Dong, F.; Chen, R.; Das, J.; Ge, W.; Xu, X.; Hong, H. Deep Learning Models for Predicting Gas Adsorption Capacity of Nanomaterials. Nanomaterials 2022, 12, 3376. [Google Scholar] [CrossRef]
- Liu, J.; Guo, W.; Dong, F.; Aungst, J.; Fitzpatrick, S.; Patterson, T.A.; Hong, H. Machine Learning Models for Rat Multigeneration Reproductive Toxicity Prediction. Front. Pharmacol. 2022, 13, 1018226. [Google Scholar] [CrossRef]
- Liu, J.; Guo, W.; Sakkiah, S.; Ji, Z.; Yavas, G.; Zou, W.; Chen, M.; Tong, W.; Patterson, T.A.; Hong, H. Machine Learning Models for Predicting Liver Toxicity. In In Silico Methods for Predicting Drug Toxicity; Methods in Molecular Biology; Benfenati, E., Ed.; Springer US: New York, NY, USA, 2022; Volume 2425, pp. 393–415. ISBN 978-1-07-161959-9. [Google Scholar]
- Huang, Y.; Li, X.; Xu, S.; Zheng, H.; Zhang, L.; Chen, J.; Hong, H.; Kusko, R.; Li, R. Quantitative Structure–Activity Relationship Models for Predicting Inflammatory Potential of Metal Oxide Nanoparticles. Environ. Health Perspect. 2020, 128, 067010. [Google Scholar] [CrossRef]
- Idakwo, G.; Thangapandian, S.; Luttrell, J.; Li, Y.; Wang, N.; Zhou, Z.; Hong, H.; Yang, B.; Zhang, C.; Gong, P. Structure–Activity Relationship-Based Chemical Classification of Highly Imbalanced Tox21 Datasets. J. Cheminform. 2020, 12, 66. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, J.; Hong, H. Developing QSAR Models with Defined Applicability Domains on PPARγ Binding Affinity Using Large Data Sets and Machine Learning Algorithms. Environ. Sci. Technol. 2021, 55, 6857–6866. [Google Scholar] [CrossRef] [PubMed]
- Geppert, H.; Vogt, M.; Bajorath, J. Current Trends in Ligand-Based Virtual Screening: Molecular Representations, Data Mining Methods, New Application Areas, and Performance Evaluation. J. Chem. Inf. Model. 2010, 50, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.T.H. Predictions of the ADMET Properties of Candidate Drug Molecules Utilizing Different QSAR/QSPR Modelling Approaches. Curr. Drug Metab. 2010, 11, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.; Mitra, I. Electrotopological State Atom (E-State) Index in Drug Design, QSAR, Property Prediction and Toxicity Assessment. Curr. Comput. Aided-Drug Des. 2012, 8, 135–158. [Google Scholar] [CrossRef] [PubMed]
- Rogers, D.; Hahn, M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.-C.; Rensi, S.E.; Torng, W.; Altman, R.B. Machine Learning in Chemoinformatics and Drug Discovery. Drug Discov. Today 2018, 23, 1538–1546. [Google Scholar] [CrossRef]
- Cereto-Massagué, A.; Ojeda, M.J.; Valls, C.; Mulero, M.; Garcia-Vallvé, S.; Pujadas, G. Molecular Fingerprint Similarity Search in Virtual Screening. Methods 2015, 71, 58–63. [Google Scholar] [CrossRef]
- Hall, L.H.; Kier, L.B. Electrotopological State Indices for Atom Types: A Novel Combination of Electronic, Topological, and Valence State Information. J. Chem. Inf. Comput. Sci. 1995, 35, 1039–1045. [Google Scholar] [CrossRef]
- Durant, J.L.; Leland, B.A.; Henry, D.R.; Nourse, J.G. Reoptimization of MDL Keys for Use in Drug Discovery. J. Chem. Inf. Comput. Sci. 2002, 42, 1273–1280. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Cai, Y.; Zhao, K.; Xie, H.; Chen, X. Concepts and Applications of Chemical Fingerprint for Hit and Lead Screening. Drug Discov. Today 2022, 27, 103356. [Google Scholar] [CrossRef] [PubMed]
- Dudek, A.; Arodz, T.; Galvez, J. Computational Methods in Developing Quantitative Structure-Activity Relationships (QSAR): A Review. Comb. Chem. High Throughput Screen. 2006, 9, 213–228. [Google Scholar] [CrossRef]
- Jiménez, J.; Škalič, M.; Martínez-Rosell, G.; De Fabritiis, G. KDEEP: Protein–Ligand Absolute Binding Affinity Prediction via 3D-Convolutional Neural Networks. J. Chem. Inf. Model. 2018, 58, 287–296. [Google Scholar] [CrossRef]
- Feinberg, E.N.; Sur, D.; Wu, Z.; Husic, B.E.; Mai, H.; Li, Y.; Sun, S.; Yang, J.; Ramsundar, B.; Pande, V.S. PotentialNet for Molecular Property Prediction. ACS Cent. Sci. 2018, 4, 1520–1530. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.D.; Cang, Z.; Wu, K.; Wang, M.; Cao, Y.; Wei, G.-W. Mathematical Deep Learning for Pose and Binding Affinity Prediction and Ranking in D3R Grand Challenges. J. Comput. Aided Mol. Des. 2019, 33, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Ain, Q.U.; Aleksandrova, A.; Roessler, F.D.; Ballester, P.J. Machine-Learning Scoring Functions to Improve Structure-Based Binding Affinity Prediction and Virtual Screening: Machine-Learning SFs to Improve Structure-Based Binding Affinity Prediction and Virtual Screening. WIREs Comput. Mol. Sci. 2015, 5, 405–424. [Google Scholar] [CrossRef]
- Desaphy, J.; Raimbaud, E.; Ducrot, P.; Rognan, D. Encoding Protein–Ligand Interaction Patterns in Fingerprints and Graphs. J. Chem. Inf. Model. 2013, 53, 623–637. [Google Scholar] [CrossRef]
- Salentin, S.; Haupt, V.J.; Daminelli, S.; Schroeder, M. Polypharmacology Rescored: Protein–Ligand Interaction Profiles for Remote Binding Site Similarity Assessment. Prog. Biophys. Mol. Biol. 2014, 116, 174–186. [Google Scholar] [CrossRef]
- Crisman, T.J.; Sisay, M.T.; Bajorath, J. Ligand-Target Interaction-Based Weighting of Substructures for Virtual Screening. J. Chem. Inf. Model. 2008, 48, 1955–1964. [Google Scholar] [CrossRef]
- Deng, Z.; Chuaqui, C.; Singh, J. Structural Interaction Fingerprint (SIFt): A Novel Method for Analyzing Three-Dimensional Protein−Ligand Binding Interactions. J. Med. Chem. 2004, 47, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Mordalski, S.; Kosciolek, T.; Kristiansen, K.; Sylte, I.; Bojarski, A.J. Protein Binding Site Analysis by Means of Structural Interaction Fingerprint Patterns. Bioorganic Med. Chem. Lett. 2011, 21, 6816–6819. [Google Scholar] [CrossRef] [PubMed]
- Vass, M.; Kooistra, A.J.; Ritschel, T.; Leurs, R.; de Esch, I.J.; de Graaf, C. Molecular Interaction Fingerprint Approaches for GPCR Drug Discovery. Curr. Opin. Pharmacol. 2016, 30, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Marcou, G.; Rognan, D. Optimizing Fragment and Scaffold Docking by Use of Molecular Interaction Fingerprints. J. Chem. Inf. Model. 2007, 47, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, F.; Desaphy, J.; Rognan, D. IChem: A Versatile Toolkit for Detecting, Comparing, and Predicting Protein-Ligand Interactions. ChemMedChem 2018, 13, 507–510. [Google Scholar] [CrossRef]
- Zhao, Z.; Bourne, P.E. Harnessing Systematic Protein–Ligand Interaction Fingerprints for Drug Discovery. Drug Discov. Today 2022, 27, 103319. [Google Scholar] [CrossRef] [PubMed]
- Radifar, M.; Yuniarti, N.; Istyastono, E.P. PyPLIF: Python-Based Protein-Ligand Interaction Fingerprinting. Bioinformation 2013, 9, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Nueno, V.I.; Rabal, O.; Borrell, J.I.; Teixidó, J. APIF: A New Interaction Fingerprint Based on Atom Pairs and Its Application to Virtual Screening. J. Chem. Inf. Model. 2009, 49, 1245–1260. [Google Scholar] [CrossRef]
- Chupakhin, V.; Marcou, G.; Gaspar, H.; Varnek, A. Simple Ligand–Receptor Interaction Descriptor (SILIRID) for Alignment-Free Binding Site Comparison. Comput. Struct. Biotechnol. J. 2014, 10, 33–37. [Google Scholar] [CrossRef]
- Da, C.; Kireev, D. Structural Protein–Ligand Interaction Fingerprints (SPLIF) for Structure-Based Virtual Screening: Method and Benchmark Study. J. Chem. Inf. Model. 2014, 54, 2555–2561. [Google Scholar] [CrossRef]
- Wójcikowski, M.; Kukiełka, M.; Stepniewska-Dziubinska, M.M.; Siedlecki, P. Development of a Protein–Ligand Extended Connectivity (PLEC) Fingerprint and Its Application for Binding Affinity Predictions. Bioinformatics 2019, 35, 1334–1341. [Google Scholar] [CrossRef] [PubMed]
- Jubb, H.C.; Higueruelo, A.P.; Ochoa-Montaño, B.; Pitt, W.R.; Ascher, D.B.; Blundell, T.L. Arpeggio: A Web Server for Calculating and Visualising Interatomic Interactions in Protein Structures. J. Mol. Biol. 2017, 429, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Szulc, N.A.; Mackiewicz, Z.; Bujnicki, J.M.; Stefaniak, F. FingeRNAt—A Novel Tool for High-Throughput Analysis of Nucleic Acid-Ligand Interactions. PLoS Comput. Biol. 2022, 18, e1009783. [Google Scholar] [CrossRef] [PubMed]
- Fassio, A.V.; Shub, L.; Ponzoni, L.; McKinley, J.; O’Meara, M.J.; Ferreira, R.S.; Keiser, M.J.; de Melo Minardi, R.C. Prioritizing Virtual Screening with Interpretable Interaction Fingerprints. J. Chem. Inf. Model. 2022, 62, 4300–4318. [Google Scholar] [CrossRef] [PubMed]
- Kokh, D.B.; Doser, B.; Richter, S.; Ormersbach, F.; Cheng, X.; Wade, R.C. A Workflow for Exploring Ligand Dissociation from a Macromolecule: Efficient Random Acceleration Molecular Dynamics Simulation and Interaction Fingerprint Analysis of Ligand Trajectories. J. Chem. Phys. 2020, 153, 125102. [Google Scholar] [CrossRef] [PubMed]
- Wójcikowski, M.; Zielenkiewicz, P.; Siedlecki, P. Open Drug Discovery Toolkit (ODDT): A New Open-Source Player in the Drug Discovery Field. J. Cheminform. 2015, 7, 26. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully Automated Protein–Ligand Interaction Profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef]
- Bouysset, C.; Fiorucci, S. ProLIF: A Library to Encode Molecular Interactions as Fingerprints. J. Cheminform. 2021, 13, 72. [Google Scholar] [CrossRef]
- Sastry, M.; Lowrie, J.F.; Dixon, S.L.; Sherman, W. Large-Scale Systematic Analysis of 2D Fingerprint Methods and Parameters to Improve Virtual Screening Enrichments. J. Chem. Inf. Model. 2010, 50, 771–784. [Google Scholar] [CrossRef]
- Duan, J.; Dixon, S.L.; Lowrie, J.F.; Sherman, W. Analysis and Comparison of 2D Fingerprints: Insights into Database Screening Performance Using Eight Fingerprint Methods. J. Mol. Graph. Model. 2010, 29, 157–170. [Google Scholar] [CrossRef]
- Jiménez-Rosés, M.; Morgan, B.A.; Jimenez Sigstad, M.; Tran, T.D.Z.; Srivastava, R.; Bunsuz, A.; Borrega-Román, L.; Hompluem, P.; Cullum, S.A.; Harwood, C.R.; et al. Combined Docking and Machine Learning Identify Key Molecular Determinants of Ligand Pharmacological Activity on Β2 Adrenoceptor. Pharmacol. Res. Perspect. 2022, 10, e00994. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yin, S.; Xiao, Y.; Lin, Z.; Fu, W.; Zhang, Y.J. Structure–Kinetic Relationship for Drug Design Revealed by a PLS Model with Retrosynthesis-Based Pre-Trained Molecular Representation and Molecular Dynamics Simulation. ACS Omega 2023, 8, 18312–18322. [Google Scholar] [CrossRef] [PubMed]
- Amangeldiuly, N.; Karlov, D.; Fedorov, M.V. Baseline Model for Predicting Protein-Ligand Unbinding Kinetics through Machine Learning. J. Chem. Inf. Model. 2020, 60, 5946–5956. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Su, M.; Lin, H.-X.; Wang, R.; Li, Y. Public Data Set of Protein-Ligand Dissociation Kinetic Constants for Quantitative Structure-Kinetics Relationship Studies. ACS Omega 2022, 7, 18985–18996. [Google Scholar] [CrossRef]

| Types of Protein–Ligand Interaction Fingerprints | Characteristics and Pattern Types | Length | Reference |
|---|---|---|---|
| Structural IFP | Uses well-defined interaction types such as hydrogen bond, halogen bonds, and π-π stacking | Each residue is represented by a seven-bit long bit string | [33,34] |
| Python-based protein–ligand interaction fingerprint (PyPLIF) | Uses well-defined interaction types such as hydrogen bond, halogen bonds, and π-π stacking | Seven bits represent seven different interactions for each residue | [39] |
| Triplet IFP | Uses two interacting atoms and an interaction pseudoatom positioned at three potential locations: the geometric center of the interacting atoms, the interacting protein atom, and the interacting ligand atom to encode different interaction types (7 types) at defined distance ranges (6 ranges) | 210 integers | [30] |
| Atom-pairs-based interaction fingerprint (APIF) | Considers the relative positions of the atom pairs instead of the absolute locations of the individual interactions | 294 bits | [40] |
| Simple ligand–receptor interaction descriptor (SILIRID) | Groups interactions by residue type, the interactions included are hydrophobic, aromatic face to face, aromatic edge to face, H-bond donated by the protein, H-bond donated by the ligand, ionic bond with protein cation and protein anion, and interaction with metal ion | 168 integers (corresponds to the product of 20 amino acids and 1 co-factor and 8 interaction types per amino acid) | [41] |
| Structural protein–ligand interaction fingerprint (SPLIF) | Encodes interacting ligand and protein fragments by representing them as circular fingerprints using Extended Connectivity Fingerprints (ECFP2) and generates integer identifiers to represent each substructure fragment | Length depends on the number of interacting fragments identified | [42] |
| Protein–ligand extended connectivity fingerprint (PLECFP) | Pairs and hashes the ECFP environment from the interacting ligand and protein atoms to represent contacts and interactions between the molecules | The raw folded fingerprint consists of integers between 0 and 232 (32 bits) | [43] |
| Software/Web Server | Types of Input Complex | Input Format | MD Trajectory Analysis | Reference |
|---|---|---|---|---|
| Arpeggio | All combinations between ligand, protein, DNA and RNA molecules | PDB | N/A | [44] |
| fingeRNAt | All combinations between ligand, protein, DNA and RNA molecules | PDB and SDF | N/A | [45] |
| getContacts | All combinations between ligand, protein, DNA and RNA molecules | VMD | N/A | getcontacts.github.io (accessed on 2 November 2023) |
| Ichem | Protein ligand complex only | Mol2 | N/A | [37] |
| LUNA | Protein ligand and protein–protein complex | PDB, Mol, Mol2, and RDKit | N/A | [46] |
| MD-IFP | Ligand protein complex only | MDAnalysis | Yes | [47] |
| ODDT | Ligand protein complex only | OpenBabel and RDKit | N/A | [48] |
| PLIP | All combinations between ligand, protein, DNA and RNA molecules | PDB | N/A | [49] |
| ProLIF | All combinations between ligand, protein, DNA and RNA molecules | MDAnalysis and RDKit | Yes | [50] |
| PyPLIF HIPPOS | Ligand protein complex only | PDBQT and Mol2 | N/A | [39] |
| Schrodinger | Ligand protein complex only | SDF, PDB, and MAE | N/A | [51,52] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Huang, R.; Xia, M.; Patterson, T.A.; Hong, H. Fingerprinting Interactions between Proteins and Ligands for Facilitating Machine Learning in Drug Discovery. Biomolecules 2024, 14, 72. https://doi.org/10.3390/biom14010072
Li Z, Huang R, Xia M, Patterson TA, Hong H. Fingerprinting Interactions between Proteins and Ligands for Facilitating Machine Learning in Drug Discovery. Biomolecules. 2024; 14(1):72. https://doi.org/10.3390/biom14010072
Chicago/Turabian StyleLi, Zoe, Ruili Huang, Menghang Xia, Tucker A. Patterson, and Huixiao Hong. 2024. "Fingerprinting Interactions between Proteins and Ligands for Facilitating Machine Learning in Drug Discovery" Biomolecules 14, no. 1: 72. https://doi.org/10.3390/biom14010072
APA StyleLi, Z., Huang, R., Xia, M., Patterson, T. A., & Hong, H. (2024). Fingerprinting Interactions between Proteins and Ligands for Facilitating Machine Learning in Drug Discovery. Biomolecules, 14(1), 72. https://doi.org/10.3390/biom14010072








