Biochemical Characterization of the Copper Nitrite Reductase from Neisseria gonorrhoeae
Abstract
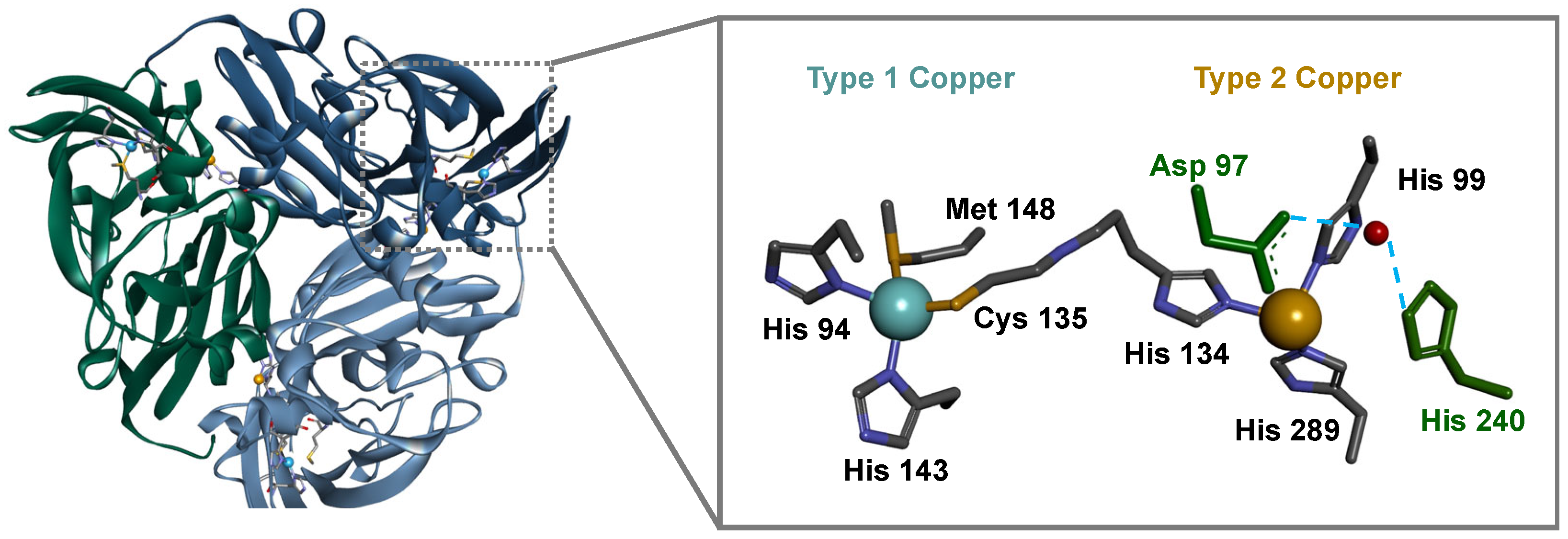
1. Introduction
2. Results and Discussion
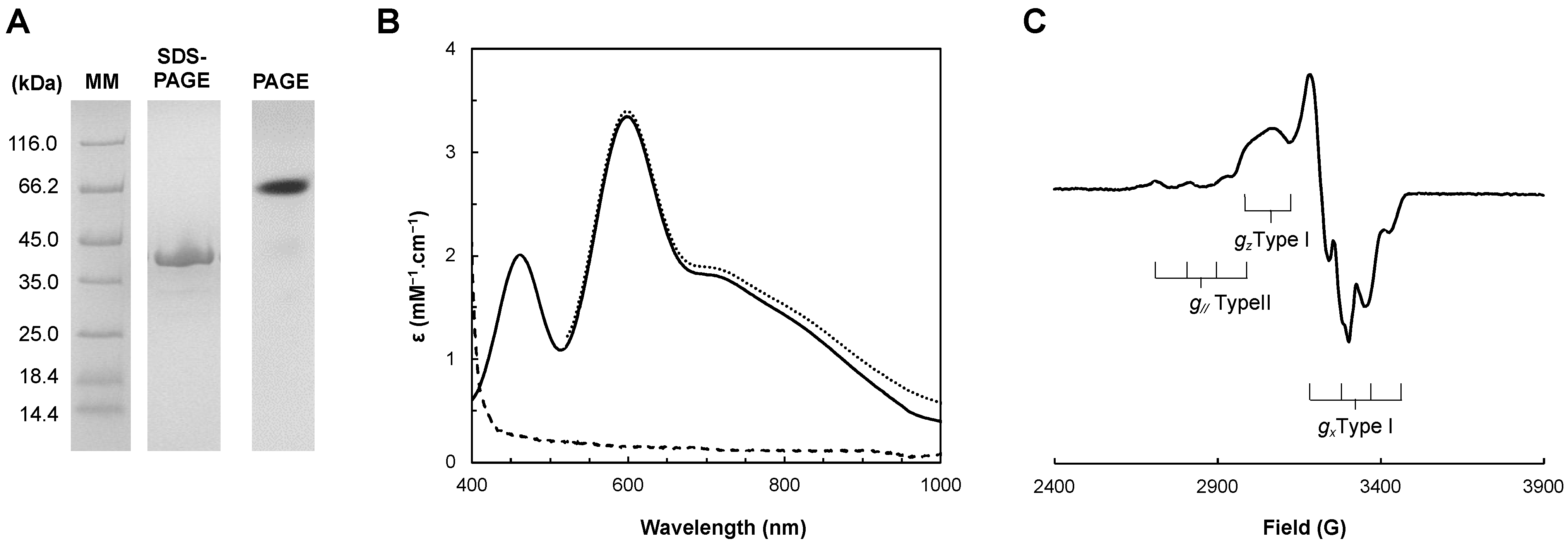
2.1. Protein Production
2.2. Spectroscopic Characterization
2.2.1. UV–Visible Spectroscopy
2.2.2. Electron Paramagnetic Resonance Spectroscopy
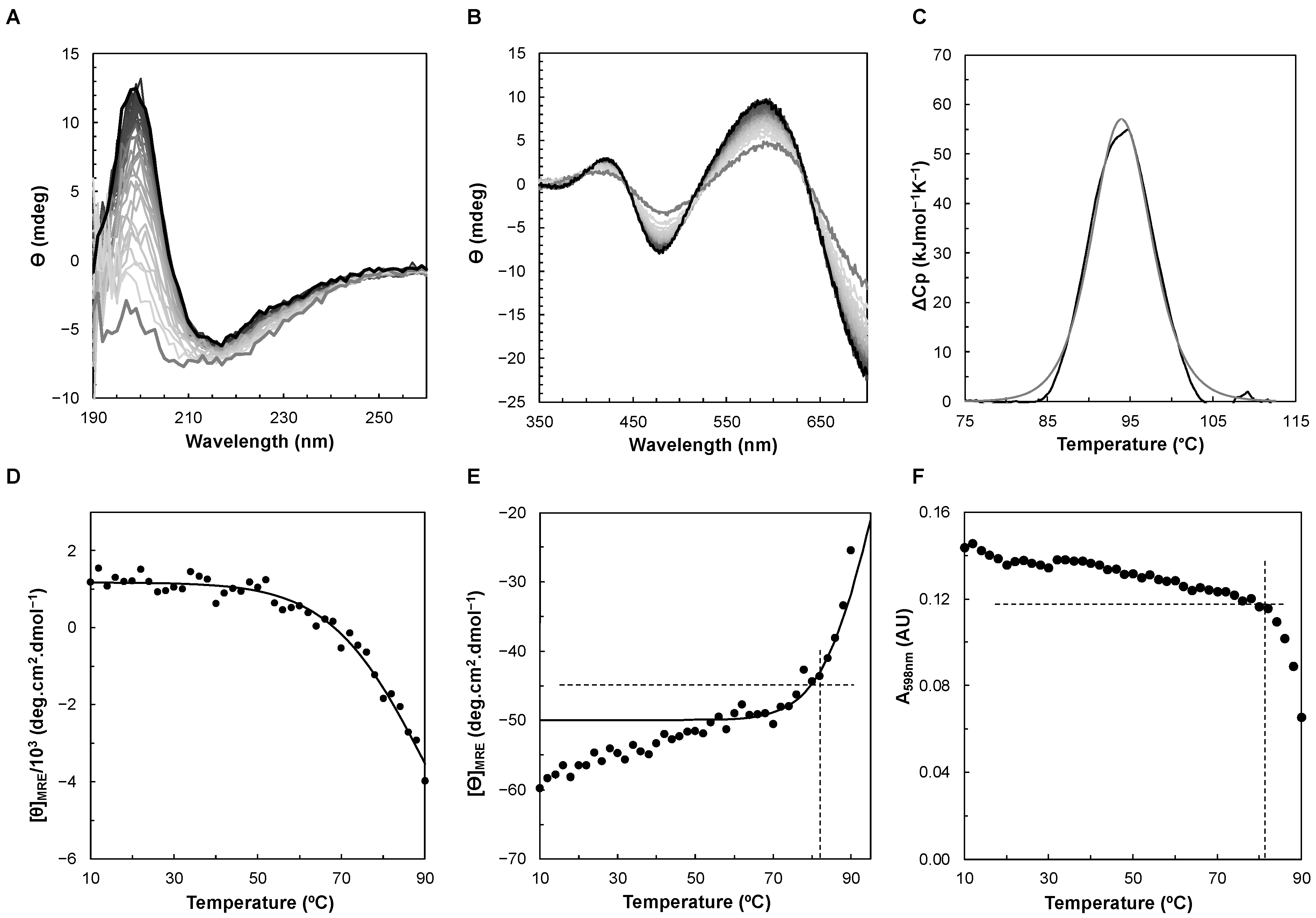
2.2.3. Circular Dichroism
2.3. Thermostability
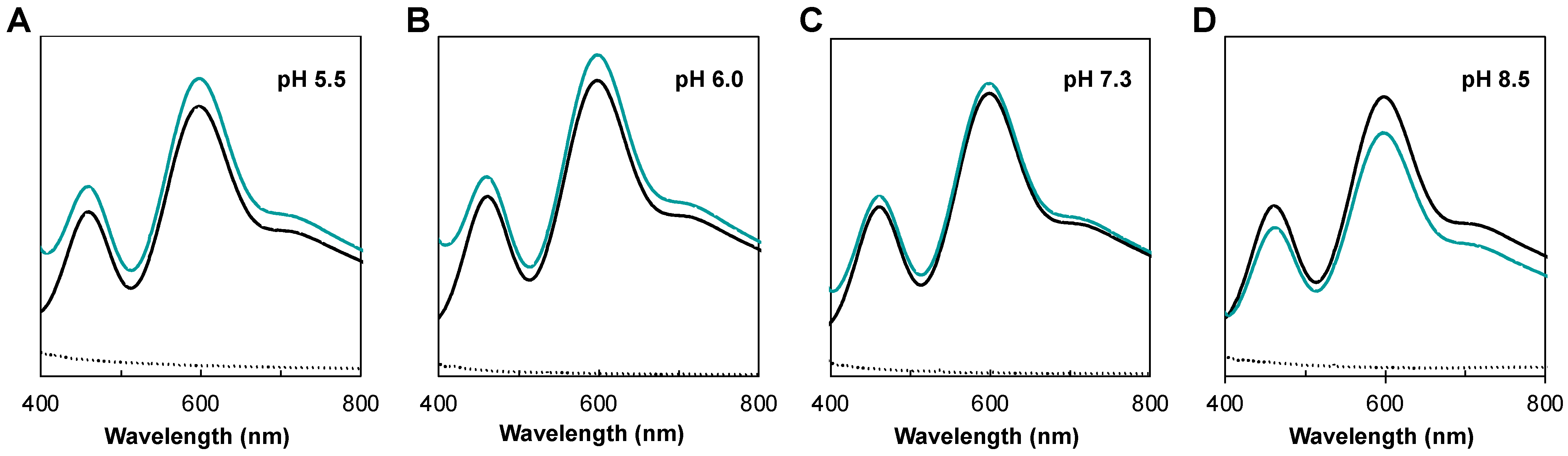
2.4. Reoxidation of NgCuNiR in the Presence of Nitrite
3. Materials and Methods
3.1. Chemicals and Solutions
3.2. Heterologous Production of NgCuNiR
3.3. Purification of NgCuNiR
3.4. Apparent Molecular Weight
3.5. Spectroscopic and Thermal Characterization
3.6. Reoxidation of NgCuNiR in the Presence of Nitrite
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zumft, W.G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 1997, 61, 533–616. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.W.; Barker, P.D.; Daltrop, O.; Stevens, J.M.; Tomlinson, E.J.; Sinha, N.; Sambongi, Y.; Ferguson, S.J. Why isn’t ‘standard’ heme good enough for c-type and d1-type cytochromes? Dalton Trans. 2005, 21, 3410–3418. [Google Scholar] [CrossRef]
- Cutruzzolà, F.; Arese, M.; Ranghino, G.; van Pouderoyen, G.; Canters, G.; Brunori, M. Pseudomonas aeruginosa cytochrome c551: Probing the role of the hydrophobic patch in electron transfer. J. Inorg. Biochem. 2002, 88, 353–361. [Google Scholar] [CrossRef]
- Pearson, I.V.; Page, M.D.; van Spanning, R.J.; Ferguson, S.J. A mutant of Paracoccus denitrificans with disrupted genes coding for cytochrome c550 and pseudoazurin establishes these two proteins as the in vivo electron donors to cytochrome cd1 nitrite reductase. J. Bacteriol. 2003, 185, 6308–6315. [Google Scholar] [CrossRef]
- Fukuda, Y.; Tse, K.M.; Nakane, T.; Nakatsu, T.; Suzuki, M.; Sugahara, M.; Inoue, S.; Masuda, T.; Yumoto, F.; Matsugaki, N.; et al. Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography. Proc. Natl. Acad. Sci. USA 2016, 113, 2928–2933. [Google Scholar] [CrossRef]
- Leferink, N.G.; Han, C.; Antonyuk, S.V.; Heyes, D.J.; Rigby, S.E.; Hough, M.A.; Eady, R.R.; Scrutton, N.S.; Hasnain, S.S. Proton-coupled electron transfer in the catalytic cycle of Alcaligenes xylosoxidans copper-dependent nitrite reductase. Biochemistry 2011, 50, 4121–4131. [Google Scholar] [CrossRef] [PubMed]
- Libby, E.; Averill, B.A. Evidence that the Type 2 copper centers are the site of nitrite reduction by Achromobacter cycloclastes nitrite reductase. Biochem. Biophys. Res. Commun. 1992, 187, 1529–1535. [Google Scholar] [CrossRef]
- Murphy, L.M.; Dodd, F.E.; Yousafzai, F.K.; Eady, R.R.; Hasnain, S.S. Electron donation between copper containing nitrite reductases and cupredoxins: The nature of protein-protein interaction in complex formation. J. Mol. Biol. 2002, 315, 859–871. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Kohzuma, T.; Deligeer; Yamaguchi, K.; Nakamura, N.; Shidara, S.; Kobayashi, K.; Tagawa, S. Pulse radiolysis studies on nitrite rreductase from Achromobacter cycloclastes IAM 1013: Evidence for intramolecular electron transfer from Type 1 Cu to Type 2 Cu. J. Am. Chem. Soc. 2002, 116, 11145–11146. [Google Scholar] [CrossRef]
- Eady, R.R.; Samar Hasnain, S. New horizons in structure-function studies of copper nitrite reductase. Coord. Chem. Rev. 2022, 460, 214463. [Google Scholar] [CrossRef]
- Solomon, E.I.; Szilagyi, R.K.; DeBeer George, S.; Basumallick, L. Electronic structures of metal sites in proteins and models: Contributions to function in blue copper proteins. Chem. Rev. 2004, 104, 419–458. [Google Scholar] [CrossRef]
- Horrell, S.; Kekilli, D.; Strange, R.W.; Hough, M.A. Recent structural insights into the function of copper nitrite reductases. Metallomics 2017, 9, 1470–1482. [Google Scholar] [CrossRef]
- Boulanger, M.J.; Kukimoto, M.; Nishiyama, M.; Horinouchi, S.; Murphy, M.E. Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase. J. Biol. Chem. 2000, 275, 23957–23964. [Google Scholar] [CrossRef]
- Kataoka, K.; Furusawa, H.; Takagi, K.; Yamaguchi, K.; Suzuki, S. Functional analysis of conserved aspartate and histidine residues located around the type 2 copper site of copper-containing nitrite reductase. J. Biochem. 2000, 127, 345–350. [Google Scholar] [CrossRef]
- Boulanger, M.J.; Murphy, M.E. Crystal structure of the soluble domain of the major anaerobically induced outer membrane protein (AniA) from pathogenic Neisseria: A new class of copper-containing nitrite reductases. J. Mol. Biol. 2002, 315, 1111–1127. [Google Scholar] [CrossRef]
- Dodd, F.E.; Van Beeumen, J.; Eady, R.R.; Hasnain, S.S. X-ray structure of a blue-copper nitrite reductase in two crystal forms. The nature of the copper sites, mode of substrate binding and recognition by redox partner. J. Mol. Biol. 1998, 282, 369–382. [Google Scholar] [CrossRef]
- Strange, R.W.; Murphy, L.M.; Dodd, F.E.; Abraham, Z.H.; Eady, R.R.; Smith, B.E.; Hasnain, S.S. Structural and kinetic evidence for an ordered mechanism of copper nitrite reductase. J. Mol. Biol. 1999, 287, 1001–1009. [Google Scholar] [CrossRef]
- Rose, S.L.; Antonyuk, S.V.; Sasaki, D.; Yamashita, K.; Hirata, K.; Ueno, G.; Ago, H.; Eady, R.R.; Tosha, T.; Yamamoto, M.; et al. An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures. Sci. Adv. 2021, 7, eabd8523. [Google Scholar] [CrossRef] [PubMed]
- Solomon, E.I. Spectroscopic methods in bioinorganic chemistry: Blue to green to red copper sites. Inorg. Chem. 2006, 45, 8012–8025. [Google Scholar] [CrossRef] [PubMed]
- Hadt, R.G.; Xie, X.; Pauleta, S.R.; Moura, I.; Solomon, E.I. Analysis of resonance Raman data on the blue copper site in pseudoazurin: Excited state pi and sigma charge transfer distortions and their relation to ground state reorganization energy. J. Inorg. Biochem. 2012, 115, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Nojiri, M. Structure and Function of Copper Nitrite Reductase. In Metalloenzymes in Denitrification: Applications and Environmental Impacts; Moura, I., Moura, J.J.G., Pauleta, S.R., Maia, L.B., Eds.; The Royal Society of Chemistry: London, UK, 2016; pp. 91–113. [Google Scholar]
- Nobrega, C.S.; Carvalho, A.L.; Romao, M.J.; Pauleta, S.R. Structural characterization of Neisseria gonorrhoeae bacterial peroxidase-insights into the catalytic cycle of Bacterial Peroxidases. Int. J. Mol. Sci. 2023, 24, 6246. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, C.S.; Raposo, M.; Van Driessche, G.; Devreese, B.; Pauleta, S.R. Biochemical characterization of the bacterial peroxidase from the human pathogen Neisseria gonorrhoeae. J. Inorg. Biochem. 2017, 171, 108–119. [Google Scholar] [CrossRef]
- Barreiro, D.S.; Oliveira, R.N.S.; Pauleta, S.R. Bacterial peroxidases–Multivalent enzymes that enable the use of hydrogen peroxide for microaerobic and anaerobic proliferation. Coord. Chem. Rev. 2023, 485, 215114. [Google Scholar] [CrossRef]
- Castiglione, N.; Rinaldo, S.; Giardina, G.; Stelitano, V.; Cutruzzola, F. Nitrite and nitrite reductases: From molecular mechanisms to significance in human health and disease. Antioxid. Redox Signal. 2012, 17, 684–716. [Google Scholar] [CrossRef]
- Sharma, J.N.; Al-Omran, A.; Parvathy, S.S. Role of nitric oxide in inflammatory diseases. Inflammopharmacology 2007, 15, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Anjum, M.F.; Stevanin, T.M.; Read, R.C.; Moir, J.W. Nitric oxide metabolism in Neisseria meningitidis. J. Bacteriol. 2002, 184, 2987–2993. [Google Scholar] [CrossRef]
- Muenzner, P.; Hauck, C.R. Neisseria gonorrhoeae blocks epithelial exfoliation by nitric-oxide-mediated metabolic cross talk to promote colonization in mice. Cell Host Microbe 2020, 27, 793–808.e795. [Google Scholar] [CrossRef]
- Mix, A.K.; Goob, G.; Sontowski, E.; Hauck, C.R. Microscale communication between bacterial pathogens and the host epithelium. Genes. Immun. 2021, 22, 247–254. [Google Scholar] [CrossRef]
- Hopper, A.C.; Li, Y.; Cole, J.A. A critical role for the cccA gene product, cytochrome c2, in diverting electrons from aerobic respiration to denitrification in Neisseria gonorrhoeae. J. Bacteriol. 2013, 195, 2518–2529. [Google Scholar] [CrossRef]
- Jen, F.E.; Djoko, K.Y.; Bent, S.J.; Day, C.J.; McEwan, A.G.; Jennings, M.P. A genetic screen reveals a periplasmic copper chaperone required for nitrite reductase activity in pathogenic Neisseria. FASEB J. 2015, 29, 3828–3838. [Google Scholar] [CrossRef]
- Li, X.; Parker, S.; Deeudom, M.; Moir, J.W. Tied down: Tethering redox proteins to the outer membrane in Neisseria and other genera. Biochem. Soc. Trans. 2011, 39, 1895–1899. [Google Scholar] [CrossRef] [PubMed]
- Shewell, L.K.; Ku, S.C.; Schulz, B.L.; Jen, F.E.; Mubaiwa, T.D.; Ketterer, M.R.; Apicella, M.A.; Jennings, M.P. Recombinant truncated AniA of pathogenic Neisseria elicits a non-native immune response and functional blocking antibodies. Biochem. Biophys. Res. Commun. 2013, 431, 215–220. [Google Scholar] [CrossRef]
- Ku, S.C.; Schulz, B.L.; Power, P.M.; Jennings, M.P. The pilin O-glycosylation pathway of pathogenic Neisseria is a general system that glycosylates AniA, an outer membrane nitrite reductase. Biochem. Biophys. Res. Commun. 2009, 378, 84–89. [Google Scholar] [CrossRef]
- Clark, V.L.; Knapp, J.S.; Thompson, S.; Klimpel, K.W. Presence of antibodies to the major anaerobically induced gonococcal outer membrane protein in sera from patients with gonococcal infections. Microb. Pathog. 1988, 5, 381–390. [Google Scholar] [CrossRef]
- Baarda, B.I.; Zielke, R.A.; Jerse, A.E.; Sikora, A.E. Lipid-modified azurin of Neisseria gonorrhoeae is not surface exposed and does not interact with the nitrite reductase AniA. Front. Microbiol. 2018, 9, 2915. [Google Scholar] [CrossRef]
- Nobrega, C.S.; Pauleta, S.R. Interaction between Neisseria gonorrhoeae bacterial peroxidase and its electron donor, the lipid-modified azurin. FEBS Lett. 2018, 592, 1473–1483. [Google Scholar] [CrossRef]
- Nobrega, C.S.; Saraiva, I.H.; Carreira, C.; Devreese, B.; Matzapetakis, M.; Pauleta, S.R. The solution structure of the soluble form of the lipid-modified azurin from Neisseria gonorrhoeae, the electron donor of cytochrome c peroxidase. Biochim. Biophys. Acta 2016, 1857, 169–176. [Google Scholar] [CrossRef]
- Cristaldi, J.C.; Ferroni, F.M.; Dure, A.B.; Ramirez, C.S.; Dalosto, S.D.; Rizzi, A.C.; Gonzalez, P.J.; Rivas, M.G.; Brondino, C.D. Heterologous production and functional characterization of Bradyrhizobium japonicum copper-containing nitrite reductase and its physiological redox partner cytochrome c550. Metallomics 2020, 12, 2084–2097. [Google Scholar] [CrossRef]
- Olesen, K.; Veselov, A.; Zhao, Y.; Wang, Y.; Danner, B.; Scholes, C.P.; Shapleigh, J.P. Spectroscopic, kinetic, and electrochemical characterization of heterologously expressed wild-type and mutant forms of copper-containing nitrite reductase from Rhodobacter sphaeroides 2.4.3. Biochemistry 1998, 37, 6086–6094. [Google Scholar] [CrossRef] [PubMed]
- Denariaz, G.; Payne, W.J.; LeGall, J. The denitrifying nitrite reductase of Bacillus halodenitrificans. Biochim. Biophys. Acta 1991, 1056, 225–232. [Google Scholar] [CrossRef]
- Xie, X.; Hadt, R.G.; Pauleta, S.R.; Gonzalez, P.J.; Un, S.; Moura, I.; Solomon, E.I. A variable temperature spectroscopic study on Paracoccus pantotrophus pseudoazurin: Protein constraints on the blue Cu site. J. Inorg. Biochem. 2009, 103, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, H.; Noji, S.; Shidara, S. Achromobacter cycloclastes nitrite reductase. The function of copper, amino acid composition, and ESR spectra. J. Biochem. 1975, 78, 355–361. [Google Scholar] [CrossRef]
- Basumallick, L.; Szilagyi, R.K.; Zhao, Y.; Shapleigh, J.P.; Scholes, C.P.; Solomon, E.I. Spectroscopic studies of the Met182Thr mutant of nitrite reductase: role of the axial ligand in the geometric and electronic structure of blue and green copper sites. J. Am. Chem. Soc. 2003, 125, 14784–14792. [Google Scholar] [CrossRef]
- Pinho, D.; Besson, S.; Brondino, C.D.; de Castro, B.; Moura, I. Copper-containing nitrite reductase from Pseudomonas chlororaphis DSM 50135. Eur. J. Biochem. 2004, 271, 2361–2369. [Google Scholar] [CrossRef]
- Gewirth, A.A.; Cohen, S.L.; Schugar, H.J.; Solomon, E.I. Spectroscopic and theoretical studies of the unusual EPR parameters of distorted tetrahedral cupric sites: Correlations to X-ray spectral features of core levels. Inorg. Chem. 1987, 26, 1133–1146. [Google Scholar] [CrossRef]
- LaCroix, L.B.; Randall, D.W.; Nersissian, A.M.; Hoitink, C.W.G.; Canters, G.W.; Valentine, J.S.; Solomon, E.I. Spectroscopic and geometric variations in perturbed blue copper centers: electronic structures of stellacyanin and cucumber basic protein. J. Am. Chem. Soc. 1998, 120, 9621–9631. [Google Scholar] [CrossRef]
- Stirpe, A.; Guzzi, R.; Wijma, H.; Verbeet, M.P.; Canters, G.W.; Sportelli, L. Calorimetric and spectroscopic investigations of the thermal denaturation of wild type nitrite reductase. Biochim. Biophys. Acta 2005, 1752, 47–55. [Google Scholar] [CrossRef]
- Abraham, Z.H.; Lowe, D.J.; Smith, B.E. Purification and characterization of the dissimilatory nitrite reductase from Alcaligenes xylosoxidans subsp. xylosoxidans (N.C.I.M.B. 11015): Evidence for the presence of both type 1 and type 2 copper centres. Biochem. J. 1993, 295 Pt 2, 587–593. [Google Scholar] [CrossRef]
- Greenfield, N.J. Analysis of circular dichroism data. In Meth Enzymol; Academic Press: Cambridge, MA, USA, 2004; Volume 383, pp. 282–317. [Google Scholar]
- Visser, N.V.; Hink, M.A.; Borst, J.W.; van der Krogt, G.N.M.; Visser, A.J.W.G. Circular dichroism spectroscopy of fluorescent proteins. FEBS Lett. 2002, 521, 31–35. [Google Scholar] [CrossRef]
- Sreerama, N.; Venyaminov, S.Y.; Woody, R.W. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999, 8, 370–380. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Structural composition of betaI- and betaII-proteins. Protein Sci. 2003, 12, 384–388. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.H.; Goto, Y.; Refregiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Bulyaki, E.; Kun, J.; Moussong, E.; Lee, Y.H.; Goto, Y.; Refregiers, M.; Kardos, J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef]
- Miles, A.J.; Ramalli, S.G.; Wallace, B.A. DichroWeb, a website for calculating protein secondary structure from circular dichroism spectroscopic data. Protein Sci. 2022, 31, 37–46. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
- LaCroix, L.B.; Shadle, S.E.; Wang, Y.; Averill, B.A.; Hedman, B.; Hodgson, K.O.; Solomon, E.I. Electronic structure of the perturbed blue copper site in nitrite reductase: spectroscopic properties, bonding, and implications for the entatic/Rack state. J. Am. Chem. Soc. 1996, 118, 7755–7768. [Google Scholar] [CrossRef]
- Suzuki, S.; Yamaguchi, K.; Kataoka, K.; Kobayashi, K.; Tagawa, S.; Kohzuma, T.; Shidara, S.; Iwasaki, H.; Deligeer. Spectroscopic characterization and intramolecular electron transfer processes of native and type 2 Cu-depleted nitrite reductases. J. Biol. Inorg. Chem. 1997, 2, 265–274. [Google Scholar] [CrossRef]
- Najmudin, S.; Pauleta, S.R.; Moura, I.; Romao, M.J. The 1.4 A resolution structure of Paracoccus pantotrophus pseudoazurin. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 627–635. [Google Scholar] [CrossRef]
- Halsted, T.P.; Yamashita, K.; Gopalasingam, C.C.; Shenoy, R.T.; Hirata, K.; Ago, H.; Ueno, G.; Blakeley, M.P.; Eady, R.R.; Antonyuk, S.V.; et al. Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography. IUCrJ 2019, 6, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, C.S.; Tolmie, C.; Opperman, D.J.; Gonzalez, P.J.; Rivas, M.G.; Brondino, C.D.; Ferroni, F.M. Copper nitrite reductase from Sinorhizobium meliloti 2011: Crystal structure and interaction with the physiological versus a nonmetabolically related cupredoxin-like mediator. Protein Sci. 2021, 30, 2310–2323. [Google Scholar] [CrossRef] [PubMed]
- Pauleta, S.R.; Guerlesquin, F.; Goodhew, C.F.; Devreese, B.; Van Beeumen, J.; Pereira, A.S.; Moura, I.; Pettigrew, G.W. Paracoccus pantotrophus pseudoazurin is an electron donor to cytochrome c peroxidase. Biochemistry 2004, 43, 11214–11225. [Google Scholar] [CrossRef] [PubMed]
- Anthis, N.J.; Clore, G.M. Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm. Protein Sci. 2013, 22, 851–858. [Google Scholar] [CrossRef]
- Almeida, A.V.; Jacinto, J.P.; Guerra, J.P.L.; Vieira, B.J.C.; Waerenborgh, J.C.; Jones, N.C.; Hoffmann, S.V.; Pereira, A.S.; Tavares, P. Structural features and stability of apo- and holo-forms of a simple iron-sulfur protein. Eur. Biophys. J. 2021, 50, 561–570. [Google Scholar] [CrossRef]
- Householder, T.C.; Belli, W.A.; Lissenden, S.; Cole, J.A.; Clark, V.L. cis- and trans-acting elements involved in regulation of aniA, the gene encoding the major anaerobically induced outer membrane protein in Neisseria gonorrhoeae. J. Bacteriol. 1999, 181, 541–551. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barreiro, D.S.; Oliveira, R.N.S.; Pauleta, S.R. Biochemical Characterization of the Copper Nitrite Reductase from Neisseria gonorrhoeae. Biomolecules 2023, 13, 1215. https://doi.org/10.3390/biom13081215
Barreiro DS, Oliveira RNS, Pauleta SR. Biochemical Characterization of the Copper Nitrite Reductase from Neisseria gonorrhoeae. Biomolecules. 2023; 13(8):1215. https://doi.org/10.3390/biom13081215
Chicago/Turabian StyleBarreiro, Daniela S., Ricardo N. S. Oliveira, and Sofia R. Pauleta. 2023. "Biochemical Characterization of the Copper Nitrite Reductase from Neisseria gonorrhoeae" Biomolecules 13, no. 8: 1215. https://doi.org/10.3390/biom13081215
APA StyleBarreiro, D. S., Oliveira, R. N. S., & Pauleta, S. R. (2023). Biochemical Characterization of the Copper Nitrite Reductase from Neisseria gonorrhoeae. Biomolecules, 13(8), 1215. https://doi.org/10.3390/biom13081215







