Notes from the Underground: Heme Homeostasis in C. elegans
Abstract
1. Introduction
2. Heme Import
3. Heme Storage and Mobilization
4. Transport of Heme to Other Tissues
5. Regulation of Heme-Responsive Genes in C. elegans
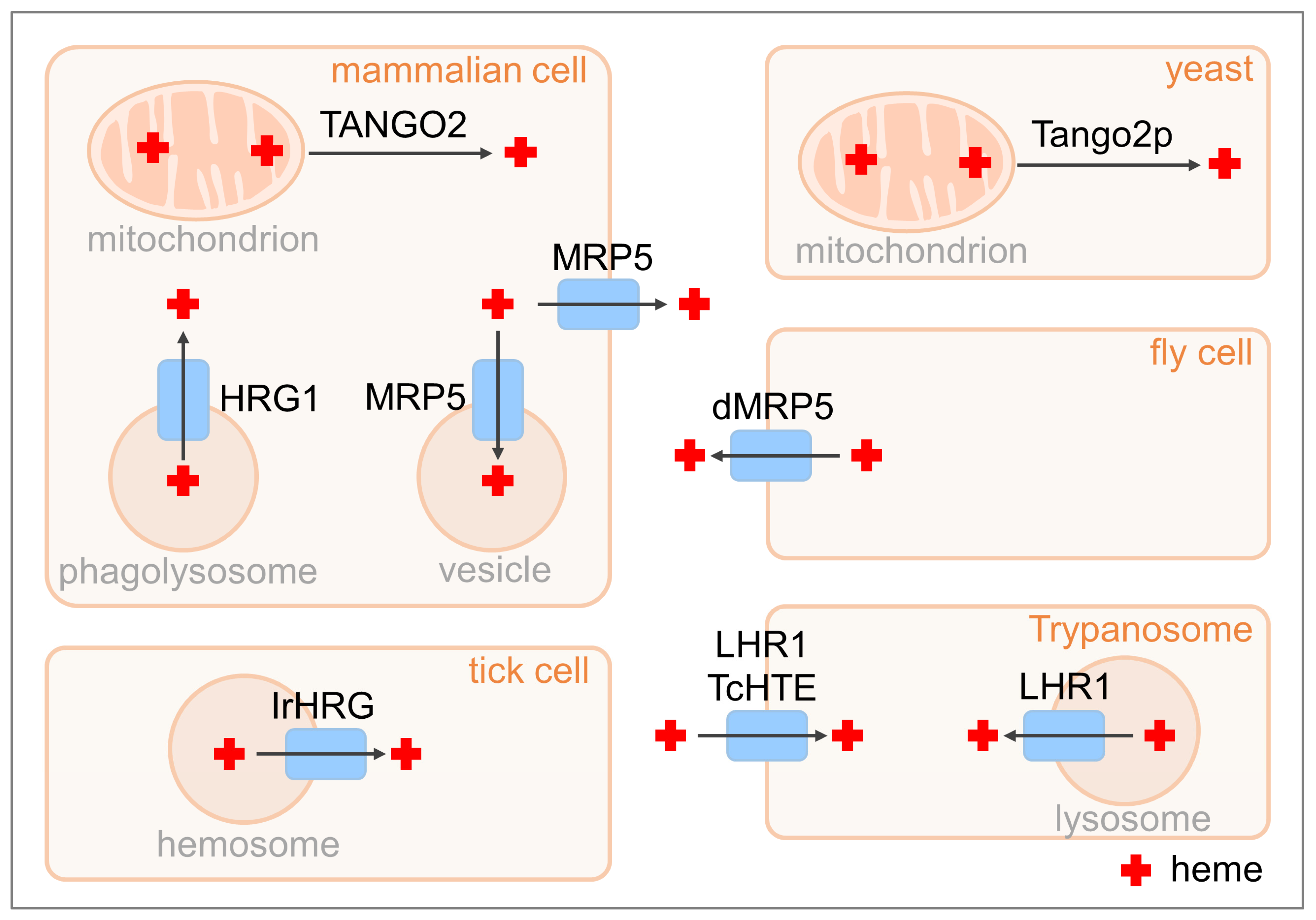
6. Homologs of C. elegans Heme-Trafficking Proteins in Other Organisms
6.1. HRG-1 Homologs
6.2. MRP-5 Homologs
6.3. HRG-9 Homologs
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Severance, S.; Hamza, I. Trafficking of heme and porphyrins in metazoa. Chem. Rev. 2009, 109, 4596–4616. [Google Scholar] [CrossRef]
- Reddi, A.R.; Hamza, I. Heme Mobilization in Animals: A Metallolipid’s Journey. Acc. Chem. Res. 2016, 49, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Chambers, I.G.; Willoughby, M.M.; Hamza, I.; Reddi, A.R. One ring to bring them all and in the darkness bind them: The trafficking of heme without deliverers. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118881. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Huang, D.; Yan, F.; Stranava, M.; Bartosova, M.; Fojtikova, V.; Martinkova, M. Gaseous O2, NO, and CO in signal transduction: Structure and function relationships of heme-based gas sensors and heme-redox sensors. Chem. Rev. 2015, 115, 6491–6533. [Google Scholar] [CrossRef] [PubMed]
- Mense, S.M.; Zhang, L. Heme: A versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res. 2006, 16, 681–692. [Google Scholar] [CrossRef]
- Ogawa, K.; Sun, J.; Taketani, S.; Nakajima, O.; Nishitani, C.; Sassa, S.; Hayashi, N.; Yamamoto, M.; Shibahara, S.; Fujita, H.; et al. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 2001, 20, 2835–2843. [Google Scholar] [CrossRef]
- Suzuki, H.; Tashiro, S.; Hira, S.; Sun, J.; Yamazaki, C.; Zenke, Y.; Ikeda-Saito, M.; Yoshida, M.; Igarashi, K. Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. EMBO J. 2004, 23, 2544–2553. [Google Scholar] [CrossRef]
- Zenke-Kawasaki, Y.; Dohi, Y.; Katoh, Y.; Ikura, T.; Ikura, M.; Asahara, T.; Tokunaga, F.; Iwai, K.; Igarashi, K. Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol. Cell. Biol. 2007, 27, 6962–6971. [Google Scholar] [CrossRef]
- Shen, J.; Sheng, X.P.; Chang, Z.N.; Wu, Q.; Wang, S.; Xuan, Z.L.; Li, D.; Wu, Y.L.; Shang, Y.J.; Kong, X.T.; et al. Iron Metabolism Regulates p53 Signaling through Direct Heme-p53 Interaction and Modulation of p53 Localization, Stability, and Function. Cell Rep. 2014, 7, 180–193. [Google Scholar] [CrossRef]
- Dioum, E.M.; Rutter, J.; Tuckerman, J.R.; Gonzalez, G.; Gilles-Gonzalez, M.A.; McKnight, S.L. NPAS2: A gas-responsive transcription factor. Science 2002, 298, 2385–2387. [Google Scholar] [CrossRef]
- Kaasik, K.; Lee, C.C. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 2004, 430, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Faller, M.; Matsunaga, M.; Yin, S.; Loo, J.A.; Guo, F. Heme Is Involved in microRNA Processing. Nat. Struct. Mol. Biol. 2007, 14, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Quick-Cleveland, J.; Jacob, J.P.; Weitz, S.H.; Shoffner, G.; Senturia, R.; Guo, F. The DGCR8 RNA-Binding Heme Domain Recognizes Primary Micrornas by Clamping the Hairpin. Cell Rep. 2014, 7, 1994–2005. [Google Scholar] [CrossRef]
- Weitz, S.H.; Gong, M.; Barr, I.; Weiss, S.; Guo, F. Processing of microRNA Primary Transcripts Requires Heme in Mammalian Cells. Proc. Natl. Acad. Sci. USA 2014, 111, 1861–1866. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Wu, N.; Curtin, J.C.; Qatanani, M.; Szwergold, N.R.; Reid, R.A.; Waitt, G.M.; Parks, D.J.; Pearce, K.H.; Wisely, G.B.; et al. Reverbalpha, a Heme Sensor That Coordinates Metabolic and Circadian Pathways. Science 2007, 318, 1786–1789. [Google Scholar] [CrossRef] [PubMed]
- Jeney, V.; Balla, J.; Yachie, A.; Varga, Z.; Vercellotti, G.M.; Eaton, J.W.; Balla, G. Pro-oxidant and cytotoxic effects of circulating heme. Blood 2002, 100, 879–887. [Google Scholar] [CrossRef]
- Kumar, S.; Bandyopadhyay, U. Free heme toxicity and its detoxification systems in human. Toxicol. Lett. 2005, 157, 175–188. [Google Scholar] [CrossRef]
- Corsi, A.K.; Wightman, B.; Chalfie, M. A Transparent Window into Biology: A Primer on Caenorhabditis elegans. Genetics 2015, 200, 387–407. [Google Scholar] [CrossRef]
- Rao, A.U.; Carta, L.K.; Lesuisse, E.; Hamza, I. Lack of heme synthesis in a free-living eukaryote. Proc. Natl. Acad. Sci. USA 2005, 102, 4270–4275. [Google Scholar] [CrossRef]
- Tilleman, L.; Germani, F.; De Henau, S.; Geuens, E.; Hoogewijs, D.; Braeckman, B.P.; Vanfleteren, J.R.; Moens, L.; Dewilde, S. Globins in Caenorhabditis elegans. IUBMB Life 2011, 63, 166–174. [Google Scholar] [CrossRef]
- Larigot, L.; Mansuy, D.; Borowski, I.; Coumoul, X.; Dairou, J. Cytochromes P450 of Caenorhabditis elegans: Implication in Biological Functions and Metabolism of Xenobiotics. Biomolecules 2022, 12, 342. [Google Scholar] [CrossRef] [PubMed]
- CES. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 1998, 282, 2012–2018. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, A.; Rao, A.U.; Amigo, J.; Tian, M.; Upadhyay, S.K.; Hall, C.; Uhm, S.; Mathew, M.K.; Fleming, M.D.; Paw, B.H.; et al. Haem homeostasis is regulated by the conserved and concerted functions of HRG-1 proteins. Nature 2008, 453, 1127–1131. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Samuel, T.K.; Sinclair, J.; Dailey, H.A.; Hamza, I. An intercellular heme-trafficking protein delivers maternal heme to the embryo during development in C. elegans. Cell 2011, 145, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Samuel, T.K.; Krause, M.; Dailey, H.A.; Hamza, I. Heme utilization in the Caenorhabditis elegans hypodermal cells is facilitated by heme-responsive gene-2. J. Biol. Chem. 2012, 287, 9601–9612. [Google Scholar] [CrossRef] [PubMed]
- Korolnek, T.; Zhang, J.; Beardsley, S.; Scheffer, G.L.; Hamza, I. Control of metazoan heme homeostasis by a conserved multidrug resistance protein. Cell Metab. 2014, 19, 1008–1019. [Google Scholar] [CrossRef]
- Sinclair, J.; Pinter, K.; Samuel, T.; Beardsley, S.; Yuan, X.; Zhang, J.; Meng, K.; Yun, S.; Krause, M.; Hamza, I. Inter-organ signalling by HRG-7 promotes systemic haem homeostasis. Nat. Cell Biol. 2017, 19, 799–807. [Google Scholar] [CrossRef]
- Sun, F.; Zhao, Z.; Willoughby, M.M.; Shen, S.; Zhou, Y.; Shao, Y.; Kang, J.; Chen, Y.; Chen, M.; Yuan, X.; et al. HRG-9 homologues regulate haem trafficking from haem-enriched compartments. Nature 2022, 610, 768–774. [Google Scholar] [CrossRef]
- Severance, S.; Rajagopal, A.; Rao, A.U.; Cerqueira, G.C.; Mitreva, M.; El-Sayed, N.M.; Krause, M.; Hamza, I. Genome-wide analysis reveals novel genes essential for heme homeostasis in Caenorhabditis elegans. PLoS Genet. 2010, 6, e1001044. [Google Scholar] [CrossRef]
- Yuan, X.; Protchenko, O.; Philpott, C.C.; Hamza, I. Topologically conserved residues direct heme transport in HRG-1-related proteins. J. Biol. Chem. 2012, 287, 4914–4924. [Google Scholar] [CrossRef]
- Zhou, J.R.; Bu, D.R.; Zhao, X.F.; Wu, F.; Chen, X.Q.; Shi, H.Z.; Yao, C.Q.; Du, A.F.; Yang, Y. Hc-hrg-2, a glutathione transferase gene, regulates heme homeostasis in the blood-feeding parasitic nematode Haemonchus contortus. Parasit. Vectors 2020, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Perally, S.; Lacourse, E.J.; Campbell, A.M.; Brophy, P.M. Heme transport and detoxification in nematodes: Subproteomics evidence of differential role of glutathione transferases. J. Proteome Res. 2008, 7, 4557–4565. [Google Scholar] [CrossRef] [PubMed]
- Zhan, B.; Perally, S.; Brophy, P.M.; Xue, J.; Goud, G.; Liu, S.; Deumic, V.; de Oliveira, L.M.; Bethony, J.; Bottazzi, M.E.; et al. Molecular cloning, biochemical characterization, and partial protective immunity of the heme-binding glutathione S-transferases from the human hookworm Necator americanus. Infect. Immun. 2010, 78, 1552–1563. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.J.; Yuan, X.; Li, J.; Dong, P.; Hamza, I.; Cheng, J.X. Label-Free Imaging of Heme Dynamics in Living Organisms by Transient Absorption Microscopy. Anal. Chem. 2018, 90, 3395–3401. [Google Scholar] [CrossRef] [PubMed]
- Mourer, T.; Normant, V.; Labbe, S. Heme Assimilation in Schizosaccharomyces pombe Requires Cell-surface-anchored Protein Shu1 and Vacuolar Transporter Abc3. J. Biol. Chem. 2017, 292, 4898–4912. [Google Scholar] [CrossRef]
- Mourer, T.; Jacques, J.F.; Brault, A.; Bisaillon, M.; Labbe, S. Shu1 is a cell-surface protein involved in iron acquisition from heme in Schizosaccharomyces pombe. J. Biol. Chem. 2015, 290, 10176–10190. [Google Scholar] [CrossRef]
- Adachi, S.; Nagano, S.; Ishimori, K.; Watanabe, Y.; Morishima, I.; Egawa, T.; Kitagawa, T.; Makino, R. Roles of proximal ligand in heme proteins: Replacement of proximal histidine of human myoglobin with cysteine and tyrosine by site-directed mutagenesis as models for P-450, chloroperoxidase, and catalase. Biochemistry 1993, 32, 241–252. [Google Scholar] [CrossRef]
- Liu, Y.; Moenne-Loccoz, P.; Hildebrand, D.P.; Wilks, A.; Loehr, T.M.; Mauk, A.G.; Ortiz de Montellano, P.R. Replacement of the proximal histidine iron ligand by a cysteine or tyrosine converts heme oxygenase to an oxidase. Biochemistry 1999, 38, 3733–3743. [Google Scholar] [CrossRef]
- Goodwin, D.C.; Rowlinson, S.W.; Marnett, L.J. Substitution of tyrosine for the proximal histidine ligand to the heme of prostaglandin endoperoxide synthase 2: Implications for the mechanism of cyclooxygenase activation and catalysis. Biochemistry 2000, 39, 5422–5432. [Google Scholar] [CrossRef]
- Luck, A.N.; Yuan, X.; Voronin, D.; Slatko, B.E.; Hamza, I.; Foster, J.M. Heme acquisition in the parasitic filarial nematode Brugia malayi. FASEB J. 2016, 30, 3501–3514. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, J.; Wu, F.; Tong, D.; Chen, X.; Jiang, S.; Duan, Y.; Yao, C.; Wang, T.; Du, A.; et al. Haem transporter HRG-1 is essential in the barber’s pole worm and an intervention target candidate. PLoS Pathog. 2023, 19, e1011129. [Google Scholar] [CrossRef] [PubMed]
- Picelli, S.; Faridani, O.R.; Bjorklund, A.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014, 9, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Rietzschel, N.; Kwon, H.; Walter Nuno, A.B.; Hanna, D.A.; Phillips, J.D.; Raven, E.L.; Reddi, A.R.; Hamza, I. Regulation of intracellular heme trafficking revealed by subcellular reporters. Proc. Natl. Acad. Sci. USA 2016, 113, E5144–E5152. [Google Scholar] [CrossRef]
- Marciano, O.; Moskovitz, Y.; Hamza, I.; Ruthstein, S. Histidine residues are important for preserving the structure and heme binding to the C. elegans HRG-3 heme-trafficking protein. J. Biol. Inorg. Chem. 2015, 20, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Raghuram, S.; Stayrook, K.R.; Huang, P.; Rogers, P.M.; Nosie, A.K.; McClure, D.B.; Burris, L.L.; Khorasanizadeh, S.; Burris, T.P.; Rastinejad, F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat. Struct. Mol. Biol. 2007, 14, 1207–1213. [Google Scholar] [CrossRef]
- Haldar, M.; Kohyama, M.; So, A.Y.; Kc, W.; Wu, X.; Briseno, C.G.; Satpathy, A.T.; Kretzer, N.M.; Arase, H.; Rajasekaran, N.S.; et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell 2014, 156, 1223–1234. [Google Scholar] [CrossRef] [PubMed]
- Mosure, S.A.; Strutzenberg, T.S.; Shang, J.; Munoz-Tello, P.; Solt, L.A.; Griffin, P.R.; Kojetin, D.J. Structural basis for heme-dependent NCoR binding to the transcriptional repressor REV-ERBbeta. Sci. Adv. 2021, 7, eabc6479. [Google Scholar] [CrossRef]
- Sinclair, J.; Hamza, I. A novel heme-responsive element mediates transcriptional regulation in Caenorhabditis elegans. J. Biol. Chem. 2010, 285, 39536–39543. [Google Scholar] [CrossRef]
- White, C.; Yuan, X.; Schmidt, P.J.; Bresciani, E.; Samuel, T.K.; Campagna, D.; Hall, C.; Bishop, K.; Calicchio, M.L.; Lapierre, A.; et al. HRG1 is essential for heme transport from the phagolysosome of macrophages during erythrophagocytosis. Cell Metab. 2013, 17, 261–270. [Google Scholar] [CrossRef]
- Zhang, J.; Chambers, I.; Yun, S.; Phillips, J.; Krause, M.; Hamza, I. Hrg1 promotes heme-iron recycling during hemolysis in the zebrafish kidney. PLoS Genet. 2018, 14, e1007665. [Google Scholar] [CrossRef]
- Pek, R.H.; Yuan, X.; Rietzschel, N.; Zhang, J.; Jackson, L.; Nishibori, E.; Ribeiro, A.; Simmons, W.; Jagadeesh, J.; Sugimoto, H.; et al. Hemozoin produced by mammals confers heme tolerance. Elife 2019, 8, e49503. [Google Scholar] [CrossRef] [PubMed]
- Perner, J.; Hatalova, T.; Cabello-Donayre, M.; Urbanova, V.; Sojka, D.; Frantova, H.; Hartmann, D.; Jirsova, D.; Perez-Victoria, J.M.; Kopacek, P. Haem-responsive gene transporter enables mobilization of host haem in ticks. Open Biol. 2021, 11, 210048. [Google Scholar] [CrossRef] [PubMed]
- Huynh, C.; Yuan, X.; Miguel, D.C.; Renberg, R.L.; Protchenko, O.; Philpott, C.C.; Hamza, I.; Andrews, N.W. Heme uptake by Leishmania amazonensis is mediated by the transmembrane protein LHR1. PLoS Pathog. 2012, 8, e1002795. [Google Scholar] [CrossRef] [PubMed]
- Miguel, D.C.; Flannery, A.R.; Mittra, B.; Andrews, N.W. Heme uptake mediated by LHR1 is essential for Leishmania amazonensis virulence. Infect. Immun. 2013, 81, 3620–3626. [Google Scholar] [CrossRef]
- Renberg, R.L.; Yuan, X.; Samuel, T.K.; Miguel, D.C.; Hamza, I.; Andrews, N.W.; Flannery, A.R. The Heme Transport Capacity of LHR1 Determines the Extent of Virulence in Leishmania amazonensis. PLoS Negl. Trop. Dis. 2015, 9, e0003804. [Google Scholar] [CrossRef]
- Andrews, N.C. Disorders of iron metabolism. N. Engl. J. Med. 1999, 341, 1986–1995. [Google Scholar] [CrossRef]
- Muckenthaler, M.U.; Rivella, S.; Hentze, M.W.; Galy, B. A Red Carpet for Iron Metabolism. Cell 2017, 168, 344–361. [Google Scholar] [CrossRef]
- Egan, T.J. Recent advances in understanding the mechanism of hemozoin (malaria pigment) formation. J. Inorg. Biochem. 2008, 102, 1288–1299. [Google Scholar] [CrossRef]
- Coronado, L.M.; Nadovich, C.T.; Spadafora, C. Malarial hemozoin: From target to tool. Biochim. Biophys. Acta 2014, 1840, 2032–2041. [Google Scholar] [CrossRef]
- Orkin, S.H.; Zon, L.I. Hematopoiesis: An evolving paradigm for stem cell biology. Cell 2008, 132, 631–644. [Google Scholar] [CrossRef]
- Perner, J.; Sobotka, R.; Sima, R.; Konvickova, J.; Sojka, D.; Oliveira, P.L.; Hajdusek, O.; Kopacek, P. Acquisition of exogenous haem is essential for tick reproduction. eLife 2016, 5, e12318. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.P.; Chang, C.S.; Sassa, S. Heme biosynthesis in bacterium-protozoon symbioses: Enzymic defects in host hemoflagellates and complemental role of their intracellular symbiotes. Proc. Natl. Acad. Sci. USA 1975, 72, 2979–2983. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Furuyama, K.; Sassa, S.; Chang, K.P. Leishmania spp.: Delta-aminolevulinate-inducible neogenesis of porphyria by genetic complementation of incomplete heme biosynthesis pathway. Exp. Parasitol. 2008, 118, 629–636. [Google Scholar] [CrossRef]
- Merli, M.L.; Pagura, L.; Hernandez, J.; Barison, M.J.; Pral, E.M.; Silber, A.M.; Cricco, J.A. The Trypanosoma cruzi Protein TcHTE Is Critical for Heme Uptake. PLoS Negl. Trop. Dis. 2016, 10, e0004359. [Google Scholar] [CrossRef]
- Pagura, L.; Tevere, E.; Merli, M.L.; Cricco, J.A. A new model for Trypanosoma cruzi heme homeostasis depends on modulation of TcHTE protein expression. J. Biol. Chem. 2020, 295, 13202–13212. [Google Scholar] [CrossRef]
- Sinclair, J.; Hamza, I. Lessons from bloodless worms: Heme homeostasis in C. elegans. Biometals 2015, 28, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zeng, P.; Zhou, B. Identification and characterization of a heme exporter from the MRP family in Drosophila melanogaster. BMC Biol. 2022, 20, 126. [Google Scholar] [CrossRef]
- Keppler, D. Multidrug resistance proteins (MRPs, ABCCs): Importance for pathophysiology and drug therapy. Handb. Exp. Pharmacol. 2011, 201, 299–323. [Google Scholar] [CrossRef]
- Chambers, I.G.; Kumar, P.; Lichtenberg, J.; Wang, P.; Yu, J.; Phillips, J.D.; Kane, M.A.; Bodine, D.; Hamza, I. MRP5 and MRP9 play a concerted role in male reproduction and mitochondrial function. Proc. Natl. Acad. Sci. USA 2022, 119, e2111617119. [Google Scholar] [CrossRef]
- Jedlitschky, G.; Burchell, B.; Keppler, D. The multidrug resistance protein 5 functions as an ATP-dependent export pump for cyclic nucleotides. J. Biol. Chem. 2000, 275, 30069–30074. [Google Scholar] [CrossRef]
- Wielinga, P.; Hooijberg, J.H.; Gunnarsdottir, S.; Kathmann, I.; Reid, G.; Zelcer, N.; van der Born, K.; de Haas, M.; van der Heijden, I.; Kaspers, G.; et al. The human multidrug resistance protein MRP5 transports folates and can mediate cellular resistance against antifolates. Cancer Res. 2005, 65, 4425–4430. [Google Scholar] [CrossRef] [PubMed]
- Schulz, T.; Schumacher, U.; Prehm, P. Hyaluronan export by the ABC transporter MRP5 and its modulation by intracellular cGMP. J. Biol. Chem. 2007, 282, 20999–21004. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.S.; Mahakena, S.; de Haas, M.; Borst, P.; van de Wetering, K. ATP-binding Cassette Subfamily C Member 5 (ABCC5) Functions as an Efflux Transporter of Glutamate Conjugates and Analogs. J. Biol. Chem. 2015, 290, 30429–30440. [Google Scholar] [CrossRef] [PubMed]
- Na, H.; Ponomarova, O.; Giese, G.E.; Walhout, A.J.M.C. elegans MRP-5 Exports Vitamin B12 from Mother to Offspring to Support Embryonic Development. Cell Rep. 2018, 22, 3126–3133. [Google Scholar] [CrossRef]
- Bard, F.; Casano, L.; Mallabiabarrena, A.; Wallace, E.; Saito, K.; Kitayama, H.; Guizzunti, G.; Hu, Y.; Wendler, F.; Dasgupta, R.; et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 2006, 439, 604–607. [Google Scholar] [CrossRef]
- Milev, M.P.; Saint-Dic, D.; Zardoui, K.; Klopstock, T.; Law, C.; Distelmaier, F.; Sacher, M. The phenotype associated with variants in TANGO2 may be explained by a dual role of the protein in ER-to-Golgi transport and at the mitochondria. J. Inherit. Metab. Dis. 2021, 44, 426–437. [Google Scholar] [CrossRef]
- Hanna, D.A.; Harvey, R.M.; Martinez-Guzman, O.; Yuan, X.; Chandrasekharan, B.; Raju, G.; Outten, F.W.; Hamza, I.; Reddi, A.R. Heme dynamics and trafficking factors revealed by genetically encoded fluorescent heme sensors. Proc. Natl. Acad. Sci. USA 2016, 113, 7539–7544. [Google Scholar] [CrossRef]
- Chiabrando, D.; Marro, S.; Mercurio, S.; Giorgi, C.; Petrillo, S.; Vinchi, F.; Fiorito, V.; Fagoonee, S.; Camporeale, A.; Turco, E.; et al. The mitochondrial heme exporter FLVCR1b mediates erythroid differentiation. J. Clin. Investig. 2012, 122, 4569–4579. [Google Scholar] [CrossRef]
- Piel, R.B., 3rd; Shiferaw, M.T.; Vashisht, A.A.; Marcero, J.R.; Praissman, J.L.; Phillips, J.D.; Wohlschlegel, J.A.; Medlock, A.E. A Novel Role for Progesterone Receptor Membrane Component 1 (PGRMC1): A Partner and Regulator of Ferrochelatase. Biochemistry 2016, 55, 5204–5217. [Google Scholar] [CrossRef]
- Galmozzi, A.; Kok, B.P.; Kim, A.S.; Montenegro-Burke, J.R.; Lee, J.Y.; Spreafico, R.; Mosure, S.; Albert, V.; Cintron-Colon, R.; Godio, C.; et al. PGRMC2 is an intracellular haem chaperone critical for adipocyte function. Nature 2019, 576, 138–142. [Google Scholar] [CrossRef]
- Kremer, L.S.; Distelmaier, F.; Alhaddad, B.; Hempel, M.; Iuso, A.; Kupper, C.; Muhlhausen, C.; Kovacs-Nagy, R.; Satanovskij, R.; Graf, E.; et al. Bi-allelic Truncating Mutations in TANGO2 Cause Infancy-Onset Recurrent Metabolic Crises with Encephalocardiomyopathy. Am. J. Hum. Genet. 2016, 98, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Lalani, S.R.; Liu, P.; Rosenfeld, J.A.; Watkin, L.B.; Chiang, T.; Leduc, M.S.; Zhu, W.; Ding, Y.; Pan, S.; Vetrini, F.; et al. Recurrent Muscle Weakness with Rhabdomyolysis, Metabolic Crises, and Cardiac Arrhythmia Due to Bi-allelic TANGO2 Mutations. Am. J. Hum. Genet. 2016, 98, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Miyake, C.Y.; Lay, E.J.; Soler-Alfonso, C.; Glinton, K.E.; Houck, K.M.; Tosur, M.; Moran, N.E.; Stephens, S.B.; Scaglia, F.; Howard, T.S.; et al. Natural history of TANGO2 deficiency disorder: Baseline assessment of 73 patients. Genet. Med. 2022, 25, 100352. [Google Scholar] [CrossRef]
- Asadi, P.; Milev, M.P.; Saint-Dic, D.; Gamberi, C.; Sacher, M. Vitamin B5, a coenzyme A precursor, rescues TANGO2 deficiency disease-associated defects in Drosophila and human cells. J. Inherit. Metab. Dis. 2023, 46, 358–368. [Google Scholar] [CrossRef]
- Jennions, E.; Hedberg-Oldfors, C.; Berglund, A.K.; Kollberg, G.; Tornhage, C.J.; Eklund, E.A.; Oldfors, A.; Verloo, P.; Vanlander, A.V.; De Meirleir, L.; et al. TANGO2 deficiency as a cause of neurodevelopmental delay with indirect effects on mitochondrial energy metabolism. J. Inherit. Metab. Dis. 2019, 42, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Mingirulli, N.; Pyle, A.; Hathazi, D.; Alston, C.L.; Kohlschmidt, N.; O’Grady, G.; Waddell, L.; Evesson, F.; Cooper, S.B.T.; Turner, C.; et al. Clinical presentation and proteomic signature of patients with TANGO2 mutations. J. Inherit. Metab. Dis. 2020, 43, 297–308. [Google Scholar] [CrossRef]
- Heiman, P.; Mohsen, A.W.; Karunanidhi, A.; St Croix, C.; Watkins, S.; Koppes, E.; Haas, R.; Vockley, J.; Ghaloul-Gonzalez, L. Mitochondrial dysfunction associated with TANGO2 deficiency. Sci. Rep. 2022, 12, 3045. [Google Scholar] [CrossRef]
- Han, S.; Guo, K.; Wang, W.; Gao, H. Bacterial TANGO2 homologs are heme-trafficking proteins that facilitate biosynthesis of cytochromes c. mBio 2023, e0132023. [Google Scholar] [CrossRef]
- Meyer, T.E.; Tsapin, A.I.; Vandenberghe, I.; de Smet, L.; Frishman, D.; Nealson, K.H.; Cusanovich, M.A.; van Beeumen, J.J. Identification of 42 possible cytochrome C genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. OMICS 2004, 8, 57–77. [Google Scholar] [CrossRef]
- Quigley, J.G.; Yang, Z.T.; Worthington, M.T.; Phillips, J.D.; Sabo, K.M.; Sabath, D.E.; Berg, C.L.; Sassa, S.; Wood, B.L.; Abkowitz, J.L. Identification of a human heme exporter that is essential for erythropoiesis. Cell 2004, 118, 757–766. [Google Scholar] [CrossRef]
- Keel, S.B.; Doty, R.T.; Yang, Z.; Quigley, J.G.; Chen, J.; Knoblaugh, S.; Kingsley, P.D.; De Domenico, I.; Vaughn, M.B.; Kaplan, J.; et al. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science 2008, 319, 825–828. [Google Scholar] [CrossRef]
- Doty, R.T.; Phelps, S.R.; Shadle, C.; Sanchez-Bonilla, M.; Keel, S.B.; Abkowitz, J.L. Coordinate expression of heme and globin is essential for effective erythropoiesis. J. Clin. Investig. 2015, 125, 4681–4691. [Google Scholar] [CrossRef]
- Doty, R.T.; Yan, X.; Lausted, C.; Munday, A.D.; Yang, Z.; Yi, D.; Jabbari, N.; Liu, L.; Keel, S.B.; Tian, Q.; et al. Single-cell analyses demonstrate that a heme-GATA1 feedback loop regulates red cell differentiation. Blood 2019, 133, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.P.; Shing, J.; Saraon, P.; Berger, L.C.; Eiden, M.V.; Wilde, A.; Tailor, C.S. The Fowler syndrome-associated protein FLVCR2 is an importer of heme. Mol. Cell. Biol. 2010, 30, 5318–5324. [Google Scholar] [CrossRef] [PubMed]
- Cabello-Donayre, M.; Orrego, L.M.; Herraez, E.; Vargas, P.; Martinez-Garcia, M.; Campos-Salinas, J.; Perez-Victoria, I.; Vicente, B.; Marin, J.J.G.; Perez-Victoria, J.M. Leishmania heme uptake involves LmFLVCRb, a novel porphyrin transporter essential for the parasite. Cell. Mol. Life Sci. 2020, 77, 1827–1845. [Google Scholar] [CrossRef] [PubMed]
- Chakravarti, R.; Aulak, K.S.; Fox, P.L.; Stuehr, D.J. GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proc. Natl. Acad. Sci. USA 2010, 107, 18004–18009. [Google Scholar] [CrossRef]
- Ghosh, A.; Stuehr, D.J. Soluble guanylyl cyclase requires heat shock protein 90 for heme insertion during maturation of the NO-active enzyme. Proc. Natl. Acad. Sci. USA 2012, 109, 12998–13003. [Google Scholar] [CrossRef]
- Ghosh, A.; Garee, G.; Sweeny, E.A.; Nakamura, Y.; Stuehr, D.J. Hsp90 chaperones hemoglobin maturation in erythroid and nonerythroid cells. Proc. Natl. Acad. Sci. USA 2018, 115, E1117–E1126. [Google Scholar] [CrossRef]
- Sweeny, E.A.; Singh, A.B.; Chakravarti, R.; Martinez-Guzman, O.; Saini, A.; Haque, M.M.; Garee, G.; Dans, P.D.; Hannibal, L.; Reddi, A.R.; et al. Glyceraldehyde-3-phosphate dehydrogenase is a chaperone that allocates labile heme in cells. J. Biol. Chem. 2018, 293, 14557–14568. [Google Scholar] [CrossRef]
- Dai, Y.; Sweeny, E.A.; Schlanger, S.; Ghosh, A.; Stuehr, D.J. GAPDH delivers heme to soluble guanylyl cyclase. J. Biol. Chem. 2020, 295, 8145–8154. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Hamza, I. Notes from the Underground: Heme Homeostasis in C. elegans. Biomolecules 2023, 13, 1149. https://doi.org/10.3390/biom13071149
Chen C, Hamza I. Notes from the Underground: Heme Homeostasis in C. elegans. Biomolecules. 2023; 13(7):1149. https://doi.org/10.3390/biom13071149
Chicago/Turabian StyleChen, Caiyong, and Iqbal Hamza. 2023. "Notes from the Underground: Heme Homeostasis in C. elegans" Biomolecules 13, no. 7: 1149. https://doi.org/10.3390/biom13071149
APA StyleChen, C., & Hamza, I. (2023). Notes from the Underground: Heme Homeostasis in C. elegans. Biomolecules, 13(7), 1149. https://doi.org/10.3390/biom13071149






