Marker-Assisted Selection for Early Maturing E Loci in Soybean Yielded Prospective Breeding Lines for High Latitudes of Northern Kazakhstan
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Hybridization
2.3. DNA Extraction, PCR, Molecular Markers, and Marker-Assisted Selection (MAS)
2.4. Research Field Experiments and Field Trials
2.5. Statistical Treatment
3. Results
3.1. Selection of Parents for Crosses and Hybrid Analysis
3.2. Molecular Markers and MAS for Four Genes, E1, E3, E4, and E7, Controlling Photoperiod Sensitivity in F2–F3 Hybrids of Soybean
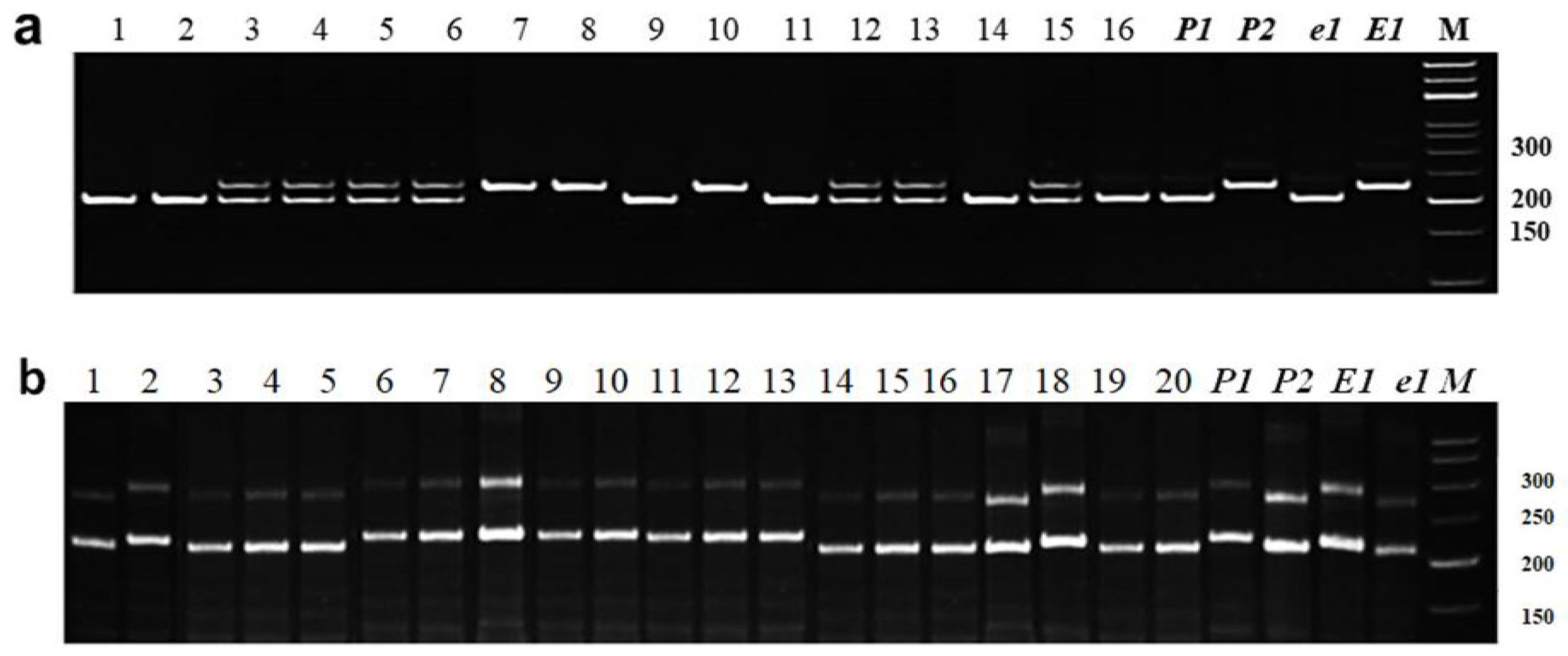
3.2.1. Identification of E1 Allelic Variation
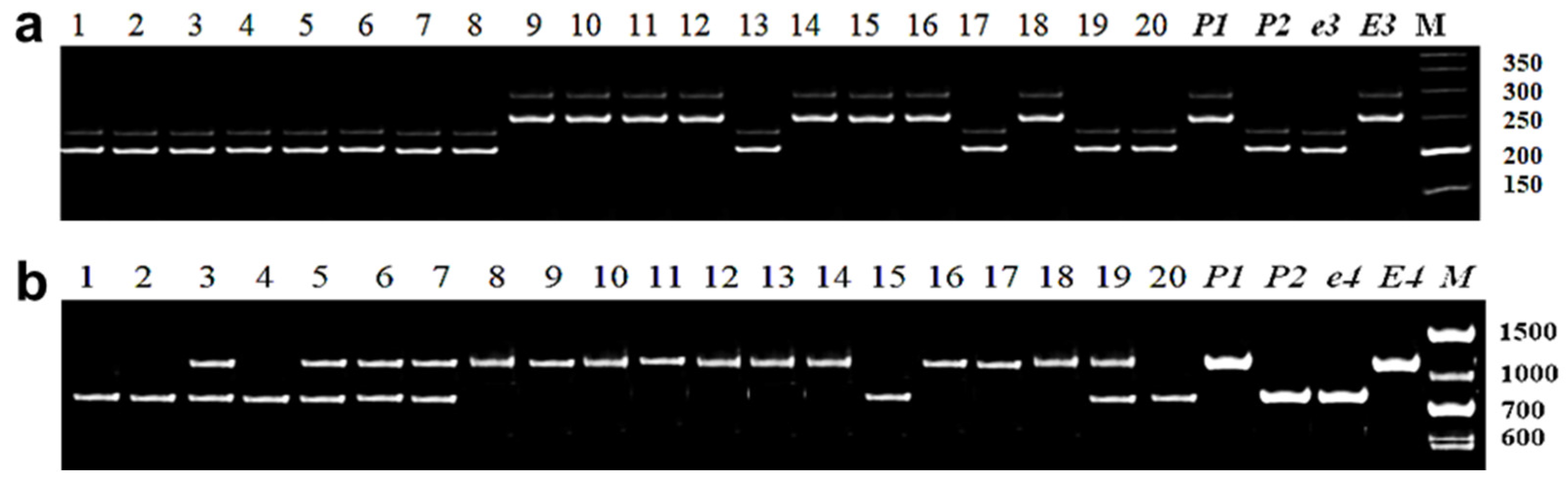
3.2.2. Identification of E3 Allelic Variation
3.2.3. Identification of E4 Allelic Variation
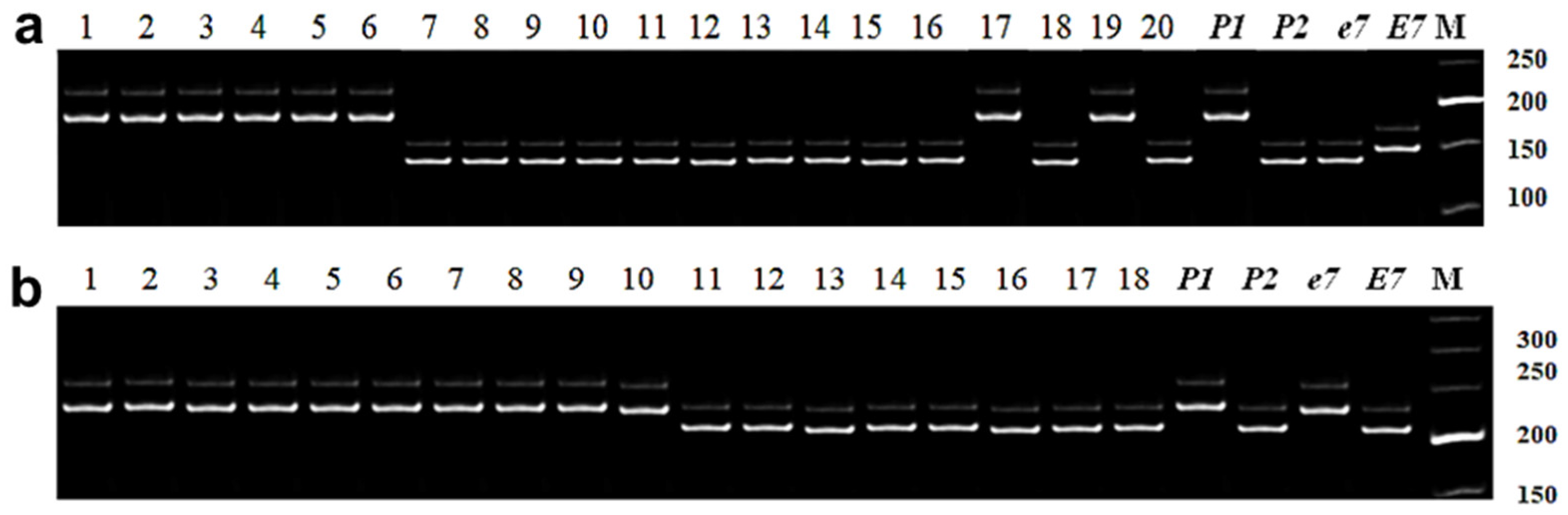
3.2.4. Identification of E7 Allelic Variation
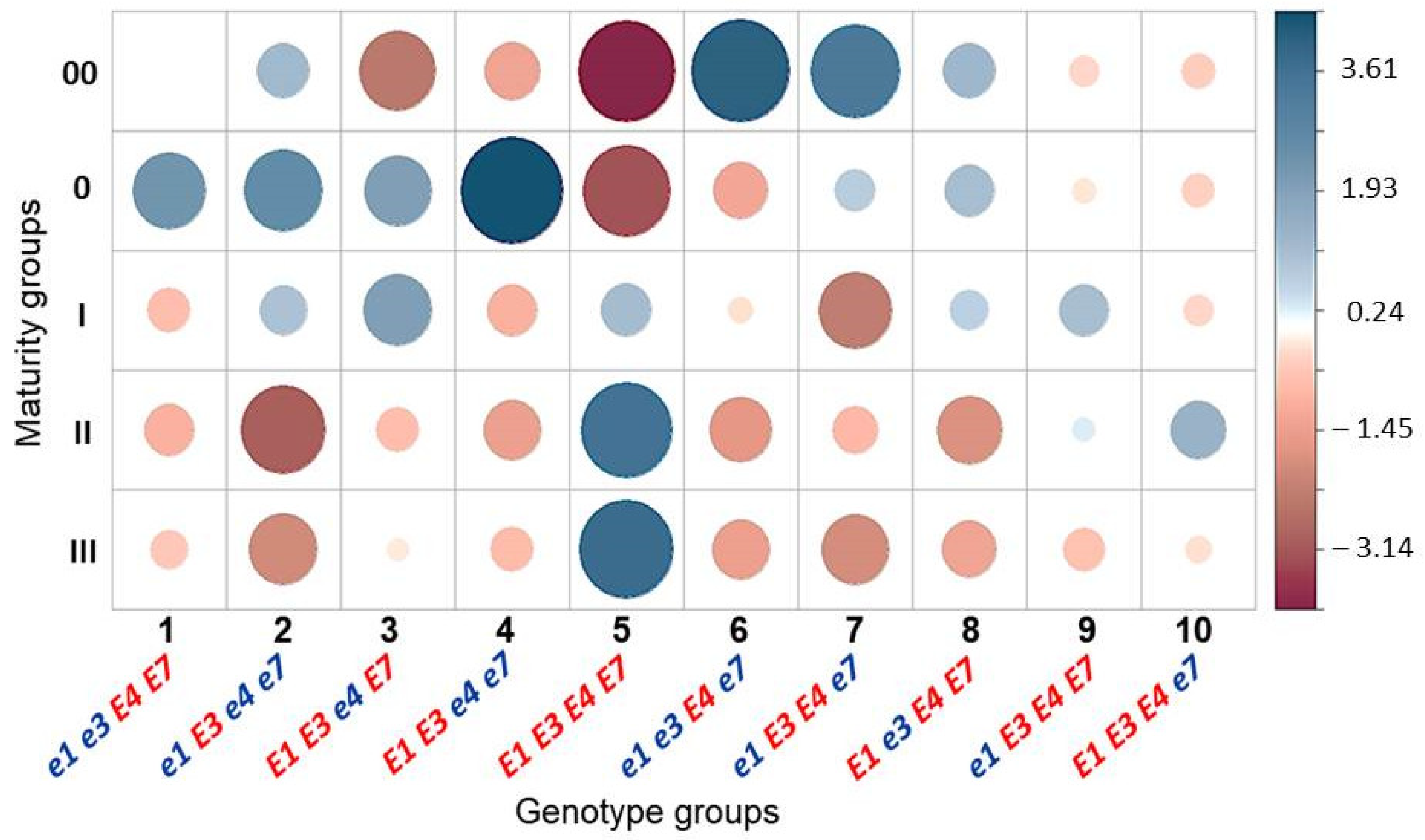
3.3. Association Analysis for Genotypes of Four E Genes and Maturity Groups in Conditions of Southern Kazakhstan
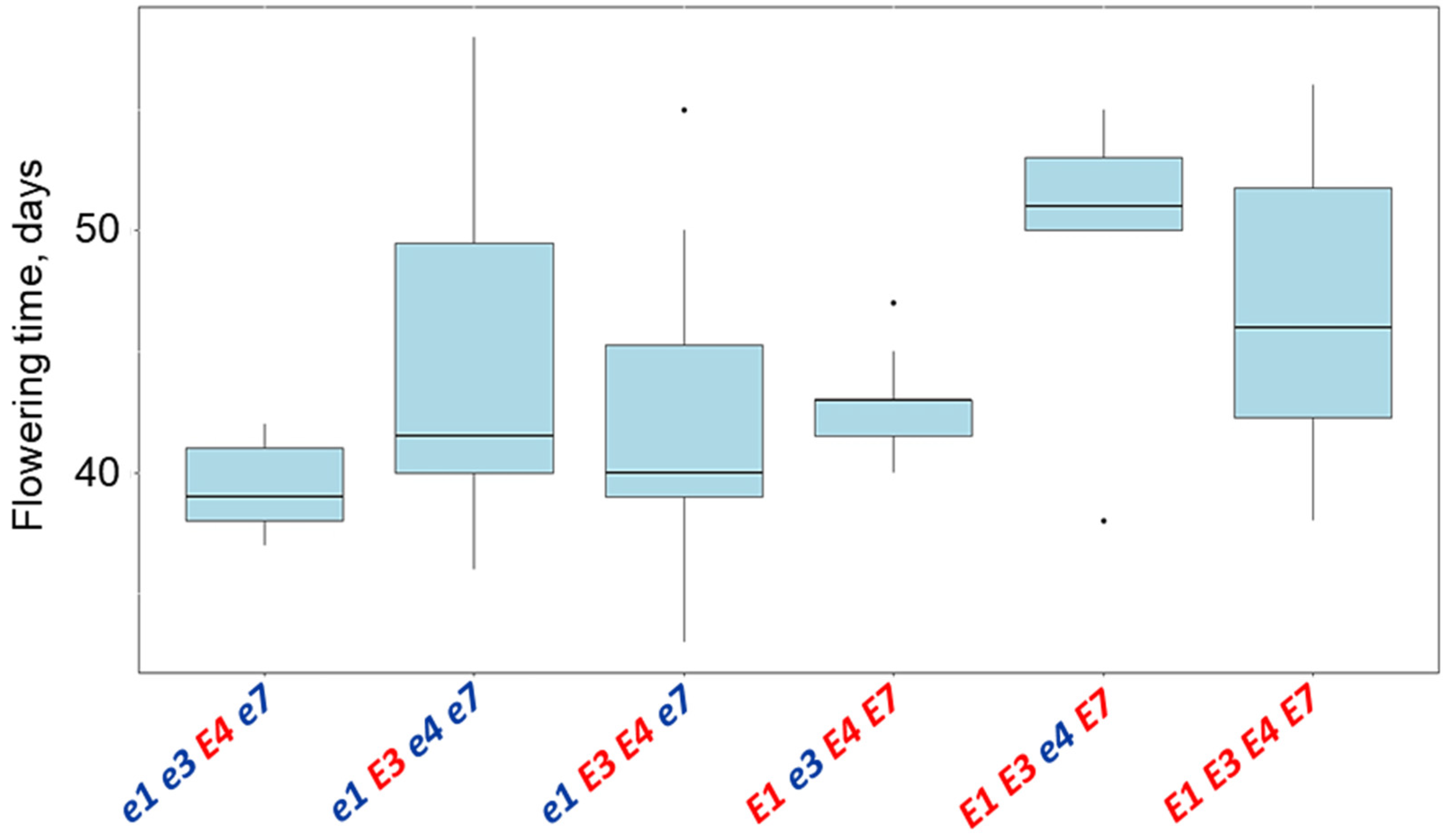
3.4. Plant Analysis and Field Trials in Southern Kazakhstan
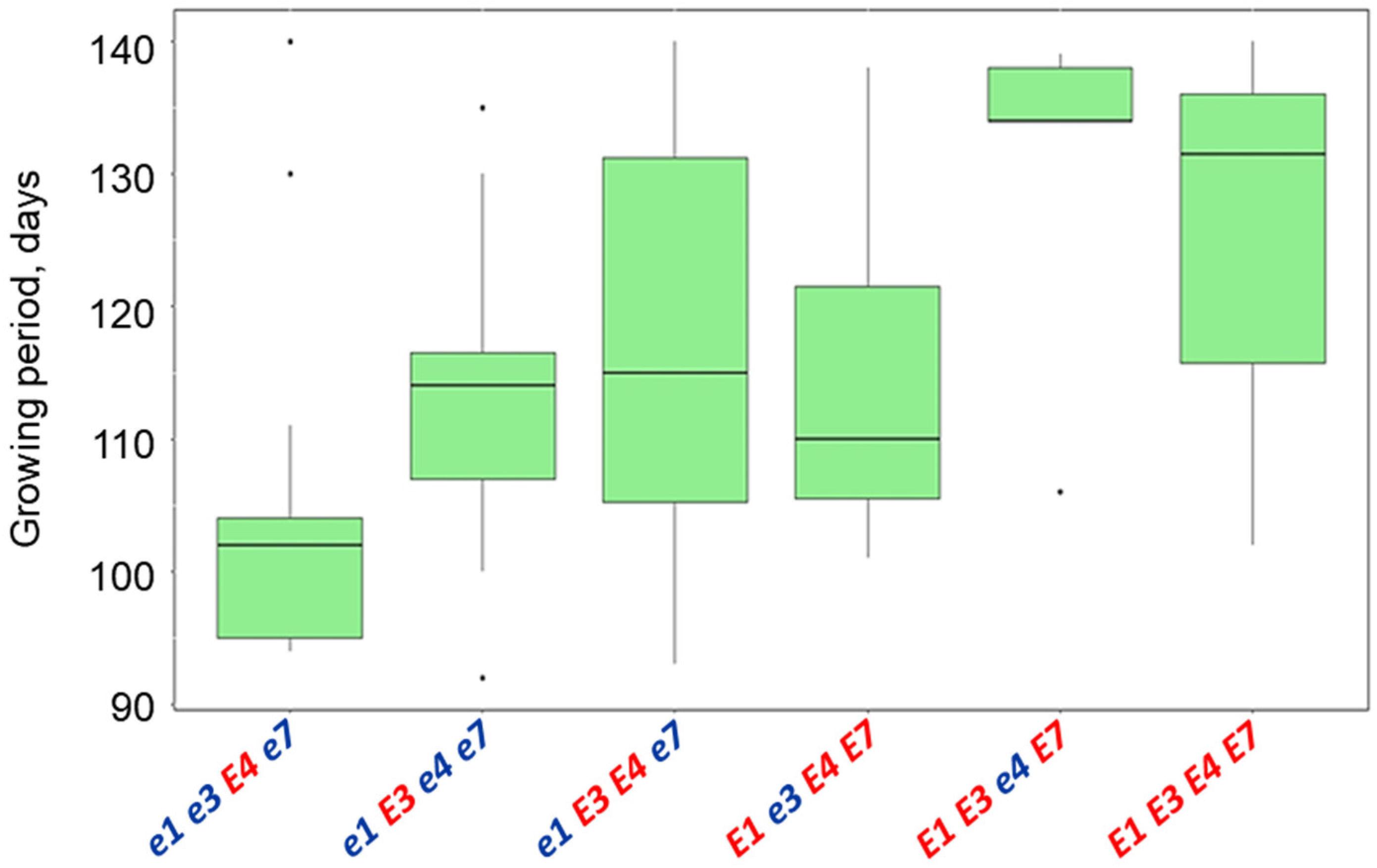
3.5. Plant Analysis and Field Trials in Northern Kazakhstan
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAS. Available online: http://www.fao.org/faostat (accessed on 24 March 2023).
- Prodöhl, I. Globalizing the Soybean. Fat, Feed, and Sometimes Food, c. 1900–1950; Routledge: London, UK, 2023; 204p. [Google Scholar] [CrossRef]
- Zhang, S.R.; Wang, H.; Wang, Z.; Ren, Y.; Niu, L.; Liu, J.; Liu, B. Photoperiodism dynamics during the domestication and improvement of soybean. Sci. China Life Sci. 2017, 60, 1416–1427. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Song, W.; Wang, L.; Sun, X.; Qi, Y.; Wu, T.; Sun, S.; Jiang, B.; Wu, C.; Hou, W.; et al. Allele combinations of maturity genes E1–E4 affect adaptation of soybean to diverse geographic regions and farming systems in China. PLoS ONE 2020, 15, e0235397. [Google Scholar] [CrossRef]
- Fedorina, J.V.; Khlestkina, E.K.; Seferova, I.V.; Vishnyakova, M.A. Genetic mechanisms underlying the expansion of soybean Glycine max (L.) Merr. cultivation to the North. Ecol. Genet. 2022, 20, 13–30. [Google Scholar] [CrossRef]
- Lin, X.; Liu, B.; Weller, L.; Abe, J.; Kong, F. Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J. Integr. Plant Biol. 2020, 63, 981–994. [Google Scholar] [CrossRef]
- Yang, W.Y.; Wu, T.T.; Zhang, X.Y.; Song, W.W.; Xu, C.L.; Sun, S.; Hou, W.S.; Jiang, B.J.; Han, T.F.; Wu, C.X. Critical photoperiod measurement of soybean varieties in different maturity groups. Crop Sci. 2019, 59, 2055–2061. [Google Scholar] [CrossRef]
- Zhang, L.X.; Liu, W.; Tsegaw, M.; Xu, X.; Qi, Y.P.; Sapey, E.; Liu, L.P.; Wu, T.T.; Sun, S.; Han, T.T. Principles and practices of the photo-thermal adaptability improvement in soybean. J. Integr. Agric. 2020, 19, 295–310. [Google Scholar] [CrossRef]
- Mourtzinis, S.; Conley, S.P. Delineating soybean maturity groups across the United States. Agron. J. 2017, 109, 1397–1403. [Google Scholar] [CrossRef]
- Jia, H.; Jiang, B.; Wu, C.; Lu, W.; Hou, W.; Sun, S.; Yan, H.; Han, T. Maturity group classification and maturity locus genotyping of early-maturing soybean varieties from high-latitude cold regions. PLoS ONE 2014, 9, e94139. [Google Scholar] [CrossRef]
- Owen, F.V. Inheritance studies in soybeans. II. Glabrousness, color of pubescence, time of maturity, and linkage relations. Genetics 1927, 12, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Kumawat, G.; Gupta, S.; Ratnaparkhe, M.B.; Maranna, S.; Satpute, G.K. QTLomics in soybean: A way forward for translational genomics and breeding. Front. Plant Sci. 2016, 7, 1852. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Takeshima, R.; Zhao, C.; Liu, B.; Jun, A.; Kong, F. Molecular mechanisms of flowering under long days and stem growth habit in soybean. J. Exp. Bot. 2017, 68, 1873–1884. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, S.; Wang, Z.; Yuan, Y.; Zhang, Z.; Liang, Q.; Yang, X.; Duan, Z.; Liu, Y.; Kong, F.; et al. Progress in soybean functional genomics over the past decade. Plant Biotechnol. J. 2022, 20, 256–282. [Google Scholar] [CrossRef]
- Bernard, R.L. Two major genes for time of flowering and maturity in soybeans. Crop Sci. 1971, 11, 242–244. [Google Scholar] [CrossRef]
- Buzzell, R.I. Inheritance of a soybean flowering response to fluorescent day length conditions. Can. J. Genet. Cytol. 1971, 13, 703–707. [Google Scholar] [CrossRef]
- Buzzell, R.I.; Voldeng, H.D. Inheritance of insensitivity to long day length. Soybean Genet. Newslet. 1980, 7, 26–29. [Google Scholar]
- McBlain, B.A.; Bernard, R.L. A new gene affecting the time of flowering maturity in soybeans. J. Hered. 1987, 178, 160–162. [Google Scholar] [CrossRef]
- Dissanayaka, A.; Rodriguez, T.O.; Di, S.; Yan, F.; Githiri, S.M.; Rodas, F.R.; Abe, J.; Takahashi, R. Quantitative trait locus mapping of soybean maturity gene E5. Breed. Sci. 2016, 66, 407–415. [Google Scholar] [CrossRef]
- Bonato, E.R.; Vello, N.A. E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet. Mol. Biol. 1999, 22, 229–232. [Google Scholar] [CrossRef]
- Cober, E.R.; Voldeng, H.D. A new soybean maturity and photoperiod-sensitivity locus linked to E1 and T. Crop Sci. 2001, 41, 698–701. [Google Scholar] [CrossRef]
- Cober, E.R.; Molnar, S.J.; Charette, M.; Voldeng, H.D. A new locus for early maturity in soybean. Crop Sci. 2010, 50, 524–527. [Google Scholar] [CrossRef]
- Kong, F.; Nan, H.; Cao, D.; Li, Y.; Wu, F.; Wang, J.; Lu, S.; Yuan, X.; Cober, E.R.; Abe, J.; et al. A new dominant gene E9 conditions early flowering and maturity in soybean. Crop Sci. 2014, 54, 2529–2535. [Google Scholar] [CrossRef]
- Zhao, C.; Takeshima, R.; Zhu, J.; Xu, M.; Sato, M.; Watanabe, S.; Kanazawa, A.; Liu, B.; Kong, F.; Yamada, T.; et al. A recessive allele for delayed flowering at the soybean maturity locus E9 is a leaky allele of FT2a, a Flowering Locus T ortholog. BMC Plant Biol. 2016, 16, 20. [Google Scholar] [CrossRef]
- Samanfar, B.; Molnar, S.J.; Charette, M.; Schoenrock, A.; Dehne, F.; Golshani, A.; Belzile, F.; Cober, E.R. Mapping and identification of a potential candidate gene for a novel maturity locus, E10, in soybean. Theor. Appl. Genet. 2017, 130, 377–390. [Google Scholar] [CrossRef]
- Wang, F.; Nan, H.; Chen, L.; Fang, C.; Zhang, H.; Su, T.; Li, S.; Cheng, Q.; Dong, L.; Liu, B.; et al. A new dominant locus, E11, controls early flowering time and maturity in soybean. Mol. Breed. 2019, 39, 70. [Google Scholar] [CrossRef]
- Ray, J.D.; Hinson, K.; Mankono, J.E.B.; Malo, M.F. Genetic control of a long-juvenile trait in soybean. Crop Sci. 1995, 35, 1001–1006. [Google Scholar] [CrossRef]
- Gupta, S.; Agrawal, N.; Tripathi, R.; Kumawat, G.; Nataraj, V.; Maranna, S.; Satpute, G.K.; Ratnaparkhe, M.B.; Rajesh, V.; Chandra, S.; et al. Long juvenility trait: A vehicle for commercial utilization of soybean (Glycine max) in lower latitudes. Plant Breed. 2021, 140, 543–560. [Google Scholar] [CrossRef]
- Lu, S.; Zhao, X.; Hu, Y.; Liu, S.; Nan, H.; Li, X.; Fang, C.; Cao, D.; Shi, X.; Kong, L.; et al. Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 2017, 49, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Liu, N.; Jiang, B.; Li, M.; Wang, H.; Jiang, Z.; Pan, H.; Xia, Q.; Ma, Q.; Han, T.; et al. A single nucleotide deletion in J encoding GmELF3 confers long juvenility and is associated with adaption of tropic soybean. Mol. Plant 2017, 10, 656–658. [Google Scholar] [CrossRef]
- Jiang, B.; Nan, H.; Gao, Y.; Tang, L.; Yue, Y.; Lu, S.; Ma, L.; Cao, D.; Sun, S.; Wang, J.; et al. Allelic combinations of soybean maturity loci E1, E2, E3 and E4 result in diversity of maturity and adaptation to different latitudes. PLoS ONE 2014, 9, e106042. [Google Scholar] [CrossRef]
- Tsubokura, Y.; Watanabe, S.; Xia, Z.; Kanamori, H.; Yamagata, H.; Kaga, A.; Katayose, Y.; Abe, J.; Ishimoto, M.; Harada, K. Natural variation in the genes responsible for maturity loci E1, E2, E3 and E4 in soybean. Ann. Bot. 2014, 113, 429–441. [Google Scholar] [CrossRef]
- Molnar, S.J.; Rai, S.; Charette, M.; Cober, E.R. Simple sequence repeat (SSR) markers linked to E1, E3, E4, and E7 maturity genes in soybean. Genome 2003, 46, 1024–1036. [Google Scholar] [CrossRef]
- Rosenzweig, V.E.; Aksyonova, E.A.; Milash, S.B.; Goloenko, D.V.; Davydenko, O.G. Prospects of exploiting of photoperiod sensitivity gene E7 in early soybean breeding and revealing of its sources with SSR-markers. Soybean Genet. Newslet. 2008, 35, 1–7. [Google Scholar]
- Shavrukov, Y.N. (Ed.) Plant Genotyping: Methods and Protocols. Methods in Molecular Biology, Vol. 2638; Springer Nature: New York, NY, USA, 2023; 469p. [Google Scholar] [CrossRef]
- Koh, H.J.; Kwon, S.Y.; Thomson, M. (Eds.) Current Technologies in Plant Molecular Breeding: A Guide Book of Plant Molecular Breeding for Researchers; Springer: Dordrecht, The Netherlands, 2015; 352p. [Google Scholar] [CrossRef]
- Rani, A.; Kumar, V. Translational genomics and breeding in soybean. In Accelerated Plant Breeding, Vol. 3, Food Legumes; Gosal, S.S., Wani, S.H., Eds.; Springer Nature: Cham, Switzerland, 2020; pp. 343–367. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef]
- Taheri, S.; Abdullah, T.L.; Yusop, M.R.; Hanafi, M.M.; Sahebi, M.; Azizi, P.; Shamshiri, R.R. Mining and development of novel SSR markers using Next generation sequencing (NGS) data in plants. Molecules 2018, 23, 399. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Shi, J.; Carlson, S.R.; Cregan, P.B.; Ward, R.W.; Diers, B.W. A low-cost, high-throughput polyacrylamide gel electrophoresis system for genotyping with microsatellite DNA markers. Crop Sci. 2003, 43, 1828–1832. [Google Scholar] [CrossRef]
- Jiang, G.L. Molecular marker-assisted breeding: A plant breeder’s review. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer International: Cham, Switzerland, 2015; pp. 431–472. [Google Scholar] [CrossRef]
- Boopathi, N.M. Genetic Mapping and Marker Assisted Selection: Basics, Practice and Benefits, 2nd ed.; Springer Nature: Singapore, 2020; 504p. [Google Scholar] [CrossRef]
- Didorenko, S.V.; Abugaliyeva, A.I.; Yerzhebayeva, R.S.; Plotnikov, V.G.; Ageyepko, A.V. Monitoring quality and yield capacity of soybean varieties during the creation of various ecotypes in Kazakhstan. Agrivita 2021, 43, 558–568. [Google Scholar] [CrossRef]
- Abugalieva, S.; Didorenko, S.; Anuarbek, S.; Volkova, L.; Gerasimova, Y.; Sidorik, I.; Turuspekov, Y. Assessment of soybean flowering and seed maturation time in different latitude regions of Kazakhstan. PLoS ONE 2016, 11, e0166894. [Google Scholar] [CrossRef]
- Didorenko, S.V.; Zakiyeva, A.A.; Sidorik, I.V.; Abugalieva, A.I.; Kudaibergenov, M.S.; Iskakov, A.R. Diversification of crop production by means of spreading soybeans to the northern regions of the republic of Kazakhstan. Biosci. Biotechol. Res. Asia 2016, 13, 23–30. [Google Scholar] [CrossRef]
- Bernard, R.L.; Weiss, M.G. Qualitative genetics. In Soybeans, Improvement, and Uses. Agronomy; Galdwell, B.E., Ed.; American Society of Agronomy: Madison, WI, USA, 1973; Volume 16, pp. 117–154. [Google Scholar]
- Walker, A.K.; Cianzio, S.R.; Bravo, J.A.; Fehr, W.R. Comparison of emasculation and nonemasculation for hybridization of soybeans. Crop Sci. 1979, 19, 285–286. [Google Scholar] [CrossRef]
- Didorenko, S.V.; Koryagin, Y.G.; Bulatova, K.M. Method of Soybean Hybridization. Patent No. 31427, 21 July 2016. Available online: https://gosreestr.kazpatent.kz/Invention/Details?docNumber=170061 (accessed on 6 January 2023). (In Kazakh and Russian).
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Liu, B.; Kanazawa, A.; Matsumura, H.; Takahashi, R.; Harada, K.; Abe, J. Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 2008, 180, 995–1007. [Google Scholar] [CrossRef] [PubMed]
- Köppen Climate Classification. Wikipedia. Available online: https://en.wikipedia.org/wiki/K%C3%B6ppen_climate_classification (accessed on 16 July 2023).
- Climate in Kazakhstan. Climate Data. Available online: https://ru.climate-data.org (accessed on 16 July 2023).
- Orf, J.H. Breeding, genetics, and production of soybeans. In Soybeans: Chemistry, Production, Processing, and Utilization; Johnson, L.A., White, P.J., Galloway, R., Eds.; AOCS Press: Urbana, IL, USA, 2008; pp. 33–65. [Google Scholar] [CrossRef]
- Fehr, W.R.; Cavines, C.E. Stages of Soybean Development; Cooperative Extension Service; Iowa State University: Ames, IA, USA, 1977; 12p, Available online: https://dr.lib.iastate.edu/handle/20.500.12876/90239 (accessed on 1 March 1977).
- Vishniyakova, M.A.; Seferova, I.V.; Buravtseva, T.V.; Burlyaeva, M.O.; Semenova, E.V.; Filipenko, G.I.; Aleksandrova, T.G.; Egorova, G.P.; Yankov, I.I.; Bulyntsev, S.V.; et al. VIR Global Collection of Grain Legume Crop Genetic Resources: Replenishment, Conservation and Studying; Ministry and Science and Higher Education: St. Petersburg, Russia, 2018; 143p. (In Russian) [Google Scholar] [CrossRef]
- Governmental Standard 12042-80; Seeds of Agricultural Crops. Methods of Determination of 1000 Seed Weight. Standardinform: Moscow, Russia, 2011; 4p. Available online: https://fsvps.gov.ru/ru/fsvps/laws/4628.html (accessed on 16 July 2023). (In Russian)
- R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org (accessed on 16 July 2023).
- Xia, Z.; Watanabe, S.; Yamada, T.; Tsubokura, Y.; Nakashima, H.; Zhai, H.; Anai, T.; Sato, S.; Yamazaki, T.; Lü, S.; et al. Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. USA 2012, 109, E2155–E2164. [Google Scholar] [CrossRef]
- Xu, M.; Xu, Z.; Liu, B.; Kong, F.; Tsubokura, Y.; Watanabe, S.; Xia, Z.; Harada, K.; Kanazawa, A.; Yamada, T.; et al. Genetic variation in four maturity genes affects photoperiod insensitivity and PHYA-regulated post-flowering responses of soybean. BMC Plant Biol. 2013, 13, 91. [Google Scholar] [CrossRef]
- Miladinović, J.; Ćeran, M.; Đorđević, V.; Balešević-Tubić, S.; Petrović, K.; Đukić, V.; Miladinović, D. Allelic variation and distribution of the major maturity genes in different soybean collections. Front. Plant Sci. 2018, 94, 1286. [Google Scholar] [CrossRef] [PubMed]
- Kurasch, A.K.; Hahn, V.; Leiser, W.L.; Vollmann, J.; Schori, A.; Bétrix, C.A.; Mayr, B.; Winkler, J.; Mechtler, K.; Aper, J.; et al. Identification of mega-environments in Europe and effect of allelic variation at maturity E loci on adaptation of European soybean. Plant Cell Environ. 2017, 40, 765–778. [Google Scholar] [CrossRef] [PubMed]
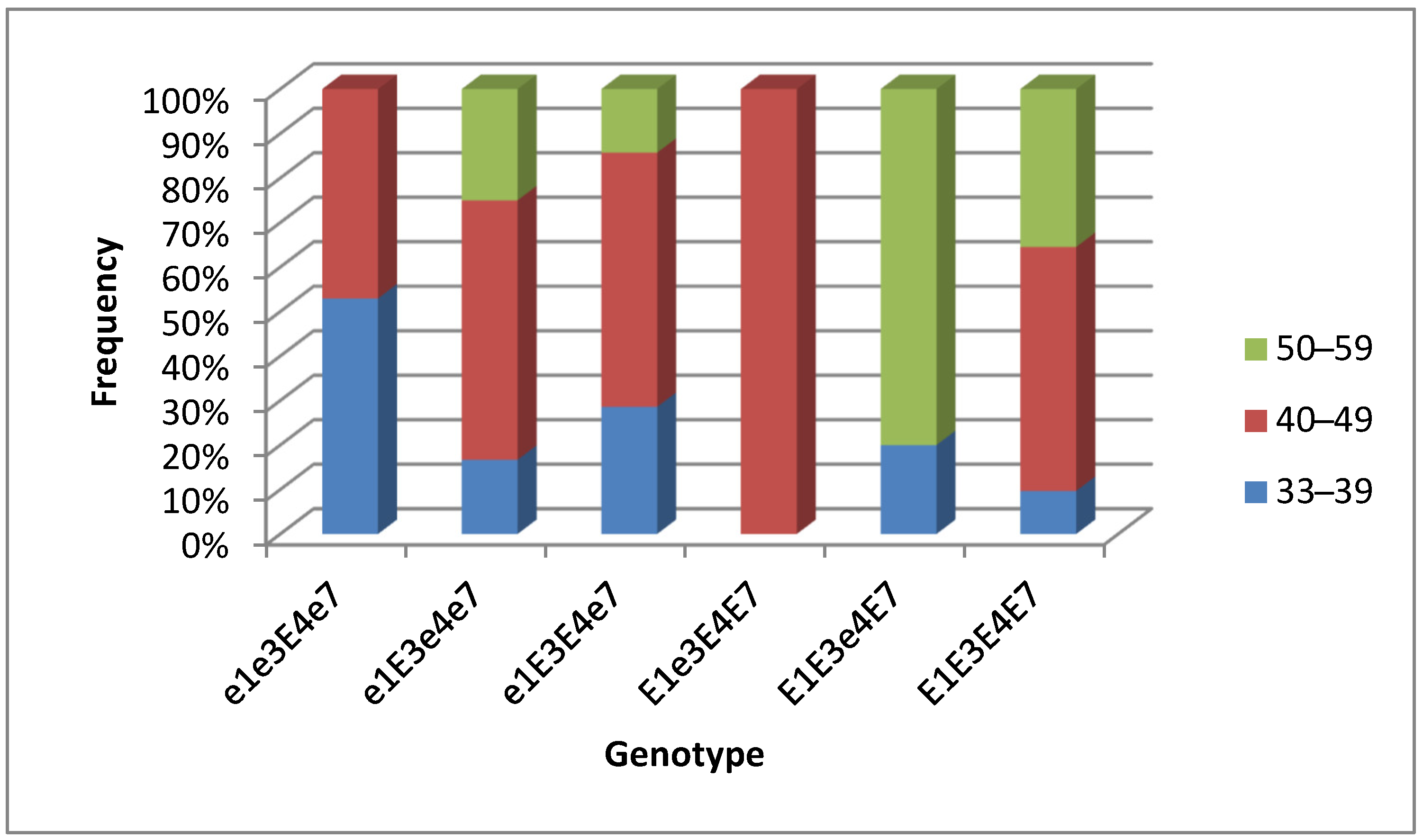
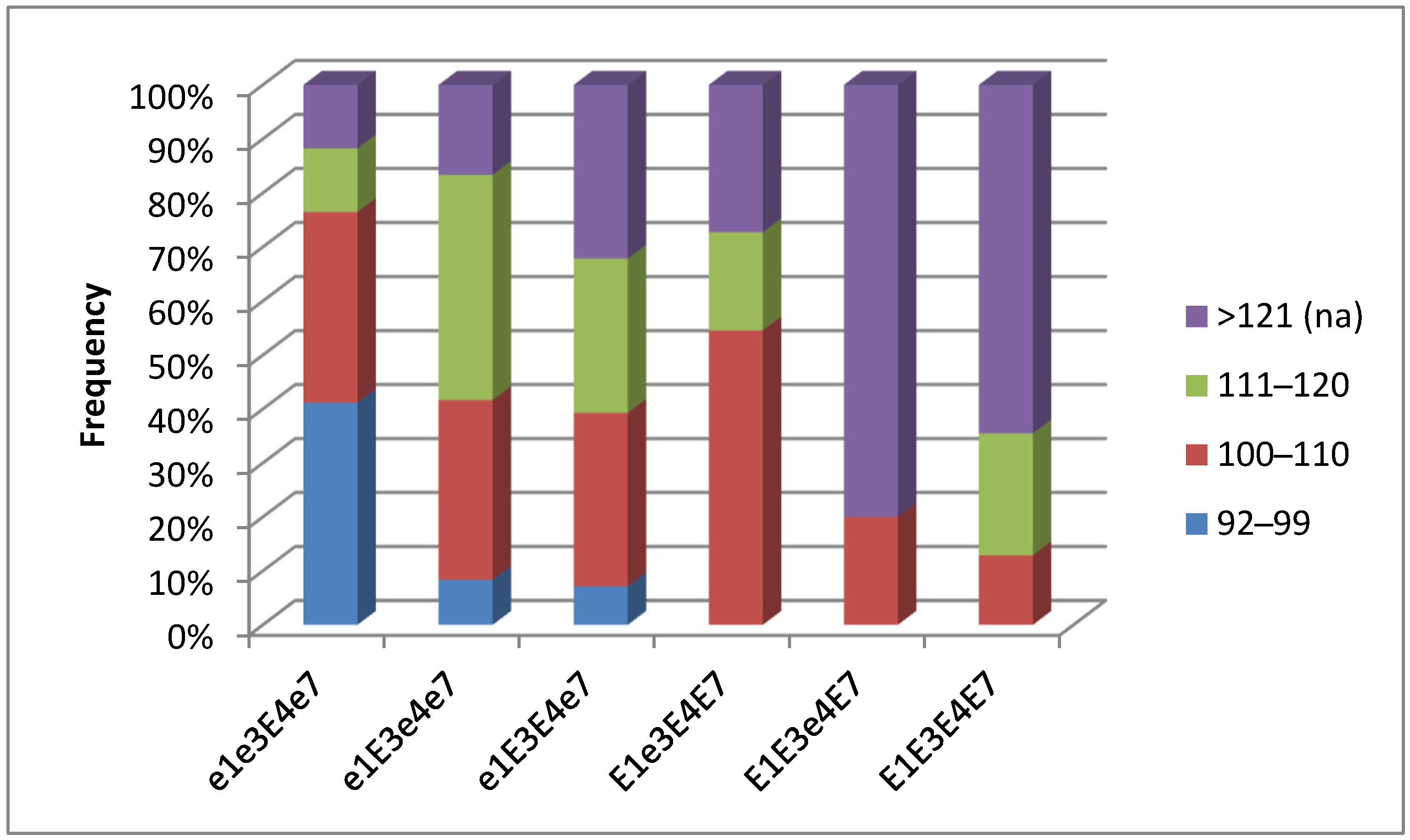

| Name of Variety | Origin | M or F | Color of Flower | Maturity Group | E1 | E3 | E4 | E7 | Growth Type | Seed Yield in Plot (g) | TSW (g) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A. Maternal parents | |||||||||||
| Birlik KV | Kazakhstan | ♀ | white | 00 | e1 | E3 | E4 | e7 | indetermin. | 178.0 a | 163.6 b |
| Zara | Kazakhstan | ♀ | white | I | E1 | E3 | E4 | E7 | determin. | 195.1 a | 167.1 b |
| B. Paternal parents crossed with cv. Birlik KV | |||||||||||
| Rana | Czech | ♂ | purple | 00 | E1 | e3 | E4 | E7 | indetermin. | 109.8 b | 145.1 b |
| Toury | Czech | ♂ | purple | 0 | e1 | E3 | E4 | e7 | semi-determ. | 170.9 a | 207.9 a |
| Pripyat | Belarus | ♂ | purple | 00 | e1 | E3 | e4 | e7 | determin. | 134.9 b | 163.5 b |
| Grignon 5 | France | ♂ | purple | I | E1 | E3 | E4 | E7 | indetermin. | 110.8 b | 153.6 b |
| Pamyat YGK | Kazakhstan | ♂ | purple | I | E1 | E3 | E4 | E7 | indetermin. | 235.3 a | 180.5 a |
| Soer 345 | Russia | ♂ | purple | 0 | E1 | E3 | E4 | E7 | determin. | 124.9 b | 152.7 b |
| Altom | Russia | ♂ | purple | 00 | e1 | e3 | e4 | e7 | indetermin. | 142.0 b | 217.0 a |
| C. Paternal parents crossed with cv. Zara | |||||||||||
| Bara | Russia | ♂ | purple | 00 | E1 | E3 | e4 | E7 | semi-determ. | 162.5 a | 182.1 a |
| Maleta | Russia | ♂ | purple | 000 | e1 | e3 | e4 | e7 | semi-determ. | 94.1 b | 173.6 ab |
| Soer 3 | Russia | ♂ | purple | 00 | e1 | E3 | E4 | E7 | determin. | 136.2 b | 162.2 b |
| Ustya | Ukraine | ♂ | purple | 00 | e1 | E3 | e4 | e7 | determin. | 134.1 b | 173.1 ab |
| Yaselda | Belarus | ♂ | purple | 00 | e1 | E3 | e4 | e7 | indetermin. | 123.2 b | 160.5 b |
| Maple Amber | Canada | ♂ | purple | 000 | e1 | E3 | E4 | e7 | determin. | 126.0 b | 203.1 a |
| Fiskeby III | Sweden | ♂ | purple | 00 | E1 | e3 | e4 | e7 | determin. | 139.3 b | 199.6 a |
| Jhony | Unknown | ♂ | purple | 00 | E1 | E3 | e4 | e7 | indetermin. | 135.7 b | 193.3 a |
| Hybrid Combination | Number of Identified Homozygote Plants with Recessive Alleles of Four E Genes | ||||
|---|---|---|---|---|---|
| Number of F2–F3 Studied Plants | e1e1 | e3e3 | e4e4 | e7e7 | |
| Birlik KV × Rana | 70 | 28 | 40 | - 1 | 28 |
| Birlik KV × Pripyat | 75 | 75 | - | 26 | 75 |
| Birlik KV × Grignon 5 | 74 | 38 | - | - | 38 |
| Birlik KV × Pamyat YGK | 37 | 16 | - | - | 16 |
| Birlik KV × Toury | 77 | 77 | - | - | 77 |
| Birlik KV × Soer 345 | 56 | 14 | - | - | 14 |
| Zara × Bara | 24 | - | - | 8 | - |
| Zara × Maleta | 146 | 48 | 17 | 22 | 48 |
| Zara × Ustya | 35 | 2 | - | 3 | 2 |
| Zara × Soer 3 | 64 | 13 | - | - | - |
| Zara × Jhony | 9 | - | - | 6 | 0 |
| Total | 667 | 311 | 57 | 65 | 298 |
| Name and Genotype | Periods from Seedling Emergence to Flowering (Days) | Periods from Seedling Emergence to Maturity (Days) |
|---|---|---|
| Hybrid breeding lines | ||
| E1 E3 e4 E7 | 49.4 a | 130.0 a |
| E1 E3 E4 E7 | 46.7 a | 127.3 a |
| e1 E3 e4 e7 | 44.3 ab | 116.7 ab |
| E1 e3 E4 E7 | 42.7 ab | 114.0 ab |
| e1 E3 E4 e7 | 42.0 ab | 113.9 ab |
| e1e3E4e7 | 39.4 b | 104.0 b |
| Isolines, and local and international standards | ||
| Harosoy OT94-47 (e1 e3 e4 e7) | 41 ab | 108 ab |
| Harosoy OT89-5 (e1 e3 e4 E7) | 43 ab | 110 ab |
| Harosoy (e1 E3 E4 E7) | 46 ab | 113 ab |
| Maleta (e1 e3 e4 E7) | 37 b | 98 b |
| Ivushka (E1 E3 E4 E7) | 39 b | 102 b |
| SIBNIIK-315 (e1 E3 e4 e7) | 39 b | 95 b |
| Maple Amber (e1 E3 E4 e7) | 38 b | 102 b |
| Fiskeby III (E1 e3 e4 e7) | 43 ab | 108 ab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yerzhebayeva, R.; Didorenko, S.; Amangeldiyeva, A.; Daniyarova, A.; Mazkirat, S.; Zinchenko, A.; Shavrukov, Y. Marker-Assisted Selection for Early Maturing E Loci in Soybean Yielded Prospective Breeding Lines for High Latitudes of Northern Kazakhstan. Biomolecules 2023, 13, 1146. https://doi.org/10.3390/biom13071146
Yerzhebayeva R, Didorenko S, Amangeldiyeva A, Daniyarova A, Mazkirat S, Zinchenko A, Shavrukov Y. Marker-Assisted Selection for Early Maturing E Loci in Soybean Yielded Prospective Breeding Lines for High Latitudes of Northern Kazakhstan. Biomolecules. 2023; 13(7):1146. https://doi.org/10.3390/biom13071146
Chicago/Turabian StyleYerzhebayeva, Raushan, Svetlana Didorenko, Aigul Amangeldiyeva, Aliya Daniyarova, Shynar Mazkirat, Alyona Zinchenko, and Yuri Shavrukov. 2023. "Marker-Assisted Selection for Early Maturing E Loci in Soybean Yielded Prospective Breeding Lines for High Latitudes of Northern Kazakhstan" Biomolecules 13, no. 7: 1146. https://doi.org/10.3390/biom13071146
APA StyleYerzhebayeva, R., Didorenko, S., Amangeldiyeva, A., Daniyarova, A., Mazkirat, S., Zinchenko, A., & Shavrukov, Y. (2023). Marker-Assisted Selection for Early Maturing E Loci in Soybean Yielded Prospective Breeding Lines for High Latitudes of Northern Kazakhstan. Biomolecules, 13(7), 1146. https://doi.org/10.3390/biom13071146







