Yeast Models of Amyotrophic Lateral Sclerosis Type 8 Mimic Phenotypes Seen in Mammalian Cells Expressing Mutant VAPBP56S
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains, Growth Conditions, and Reagents
2.2. Plasmids
2.3. Plate-Based Growth Assays
2.4. Kinetic Growth Assays
2.5. Fluorescence Microscopy and Image Processing
2.6. Quantification of ER Inclusion Size
2.7. Comparison of Human ALS-Linked Proteins with Putative Yeast Homologs
2.8. Statistical Analysis and Generation of SuperPlots
3. Results
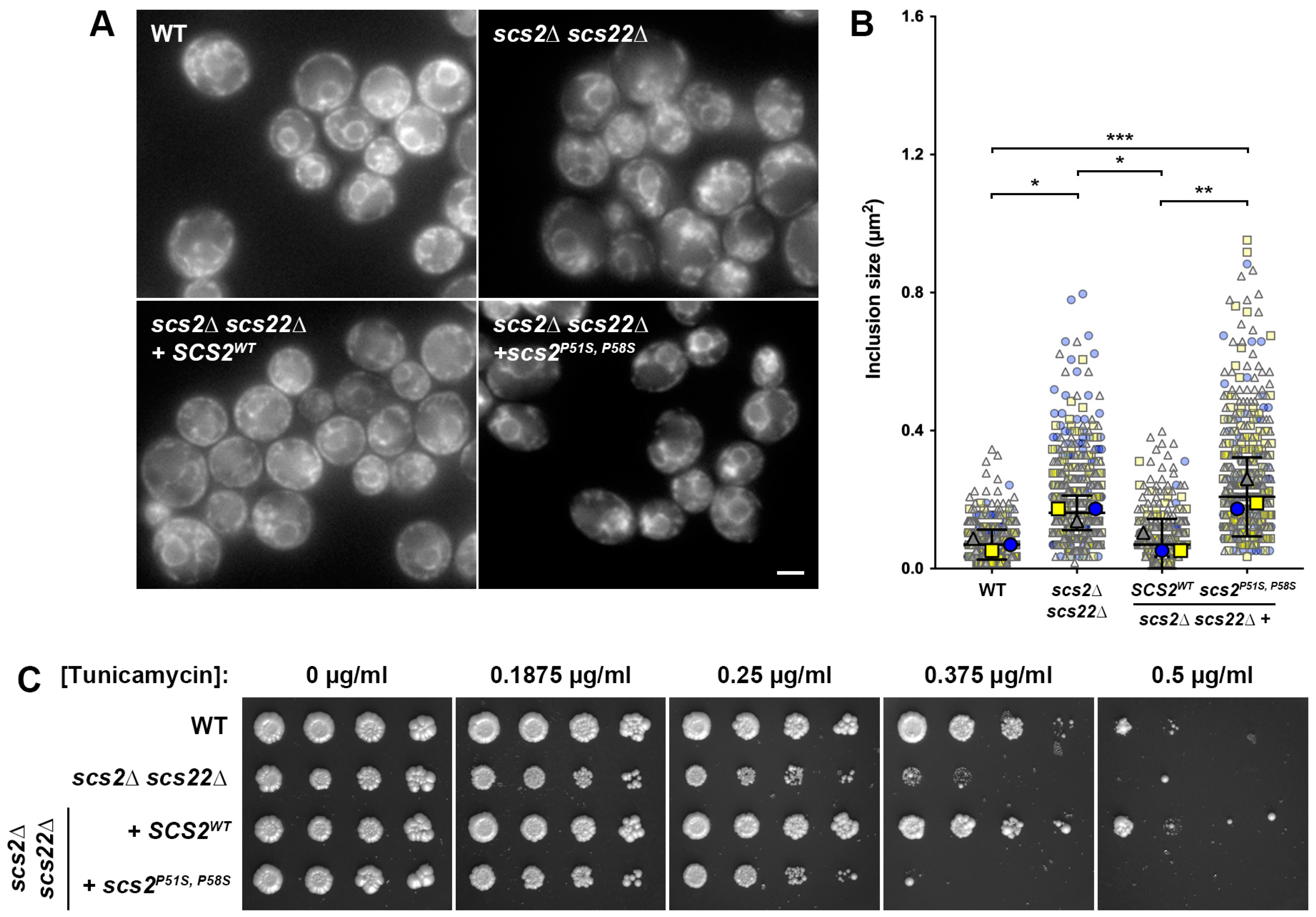
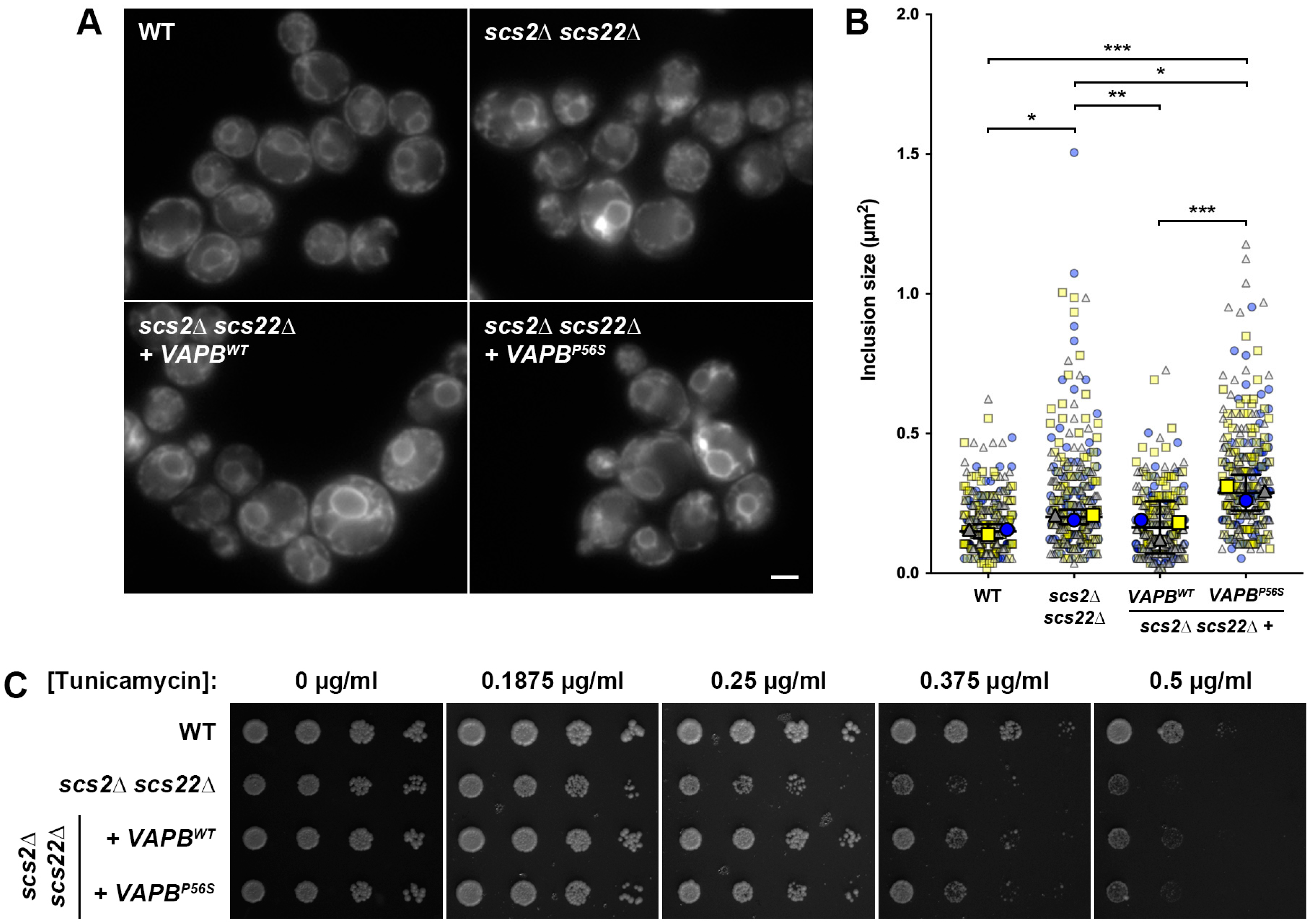
3.1. Loss of SCS2 and SCS22 Causes ER Collapse and Sensitivity to ER Stress
3.2. Expression of SCS2, but Not scs2P51S, P58S, Corrects ER Morphology and Stress Sensitivity Defects in scs2∆ scs22∆ Cells
3.3. Heterologous Expression of VAPBWT, but Not VAPBP56S, Complements ER Morphology and Stress Senstitivity in scs2∆ scs22∆ Cells
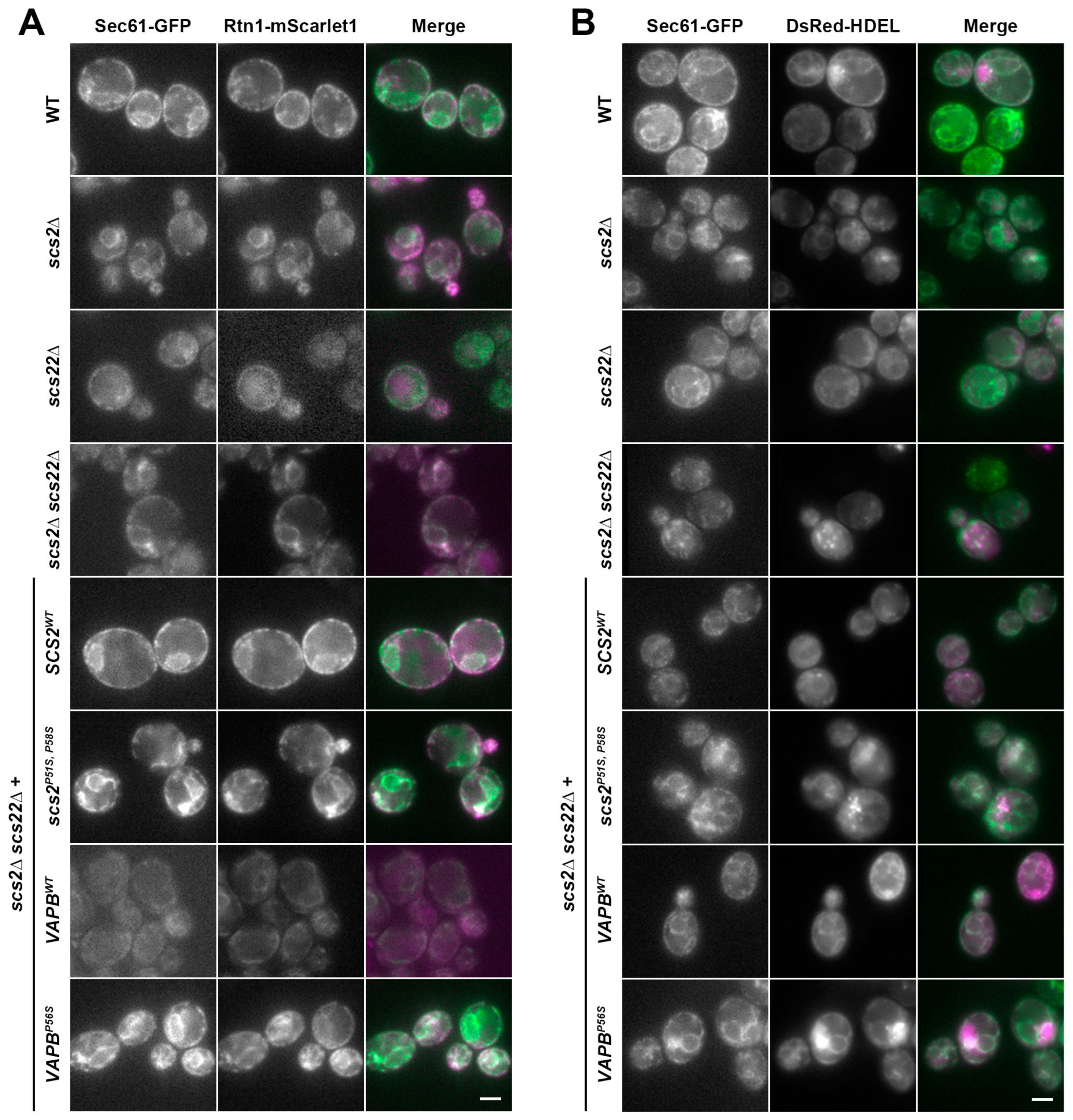
3.4. Multiple ER-Resident Proteins Localize to Inclusion-like Structures in SCS-Deficient Cells
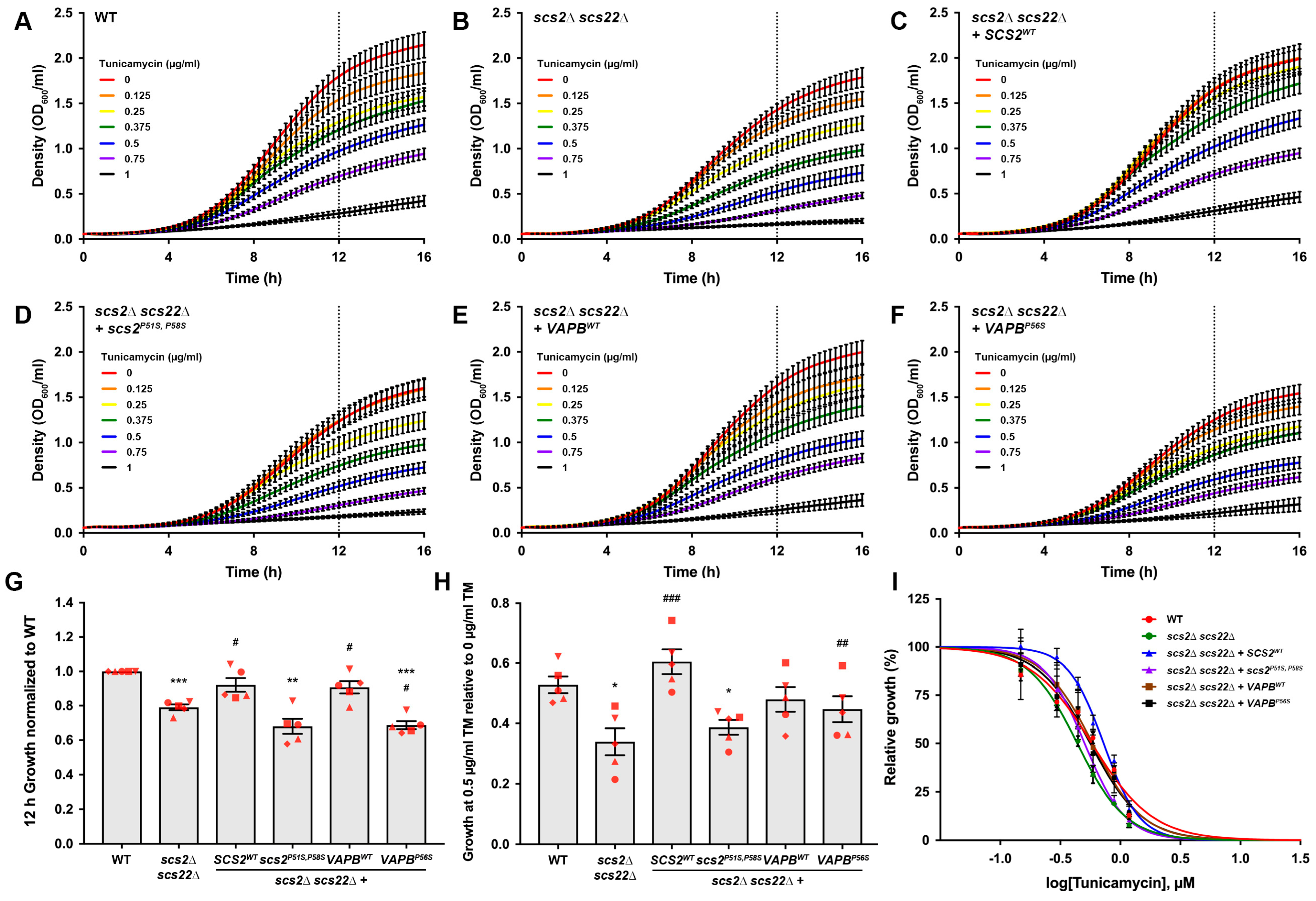
3.5. Kinetic Growth Assays Reveal Differences in Tunicamycin Sensitivity in Yeast ALS8 Model Cells
3.6. Potential Use of Yeast as a Model for Study of Other Types of ALS
4. Discussion
4.1. Yeast as a Model System for Study of Neurodegenerative Diseases
4.2. Yeast ALS8 Models May Facilitate Studies of VAPBP56S-Related Cellular Pathology
4.3. Similarities and Differences between ALS8 Models
4.4. Yeast May Provide Opportunities and Approaches for Study of Other ALS Subtypes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bruijn, L.I.; Miller, T.M.; Cleveland, D.W. Unraveling the Mechanisms Involved in Motor Neuron Degeneration in ALS. Annu. Rev. Neurosci. 2004, 27, 723–749. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Malik, Z.; Singh, M.; Rachana, R.; Mani, S.; Ponnusamy, K.; Haider, S. Amyotrophic Lateral Sclerosis Risk Genes and Suppressor. Curr. Gene Ther. 2023, 23, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Kirola, L.; Mukherjee, A.; Mutsuddi, M. Recent Updates on the Genetics of Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Mol. Neurobiol. 2022, 59, 5673–5694. [Google Scholar] [CrossRef] [PubMed]
- Deerlin, V.M.V.; Leverenz, J.B.; Bekris, L.M.; Bird, T.D.; Yuan, W.; Elman, L.B.; Clay, D.; Wood, E.M.; Chen-Plotkin, A.S.; Martinez-Lage, M.; et al. TARDBP Mutations in Amyotrophic Lateral Sclerosis with TDP-43 Neuropathology: A Genetic and Histopathological Analysis. Lancet Neurol. 2008, 7, 409–416. [Google Scholar] [CrossRef]
- Vance, C.; Rogelj, B.; Hortobágyi, T.; Vos, K.J.D.; Nishimura, A.L.; Sreedharan, J.; Hu, X.; Smith, B.; Ruddy, D.; Wright, P.; et al. Mutations in FUS, an RNA Processing Protein, Cause Familial Amyotrophic Lateral Sclerosis Type 6. Science 2009, 323, 1208–1211. [Google Scholar] [CrossRef]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef]
- Moller, A.; Bauer, C.S.; Cohen, R.N.; Webster, C.P.; Vos, K.J.D. Amyotrophic Lateral Sclerosis-Associated Mutant SOD1 Inhibits Anterograde Axonal Transport of Mitochondria by Reducing Miro1 Levels. Hum. Mol. Genet. 2017, 26, 4668–4679. [Google Scholar] [CrossRef]
- Watanabe, S.; Ilieva, H.; Tamada, H.; Nomura, H.; Komine, O.; Endo, F.; Jin, S.; Mancias, P.; Kiyama, H.; Yamanaka, K. Mitochondria-Associated Membrane Collapse Is a Common Pathomechanism in SIGMAR1- and SOD1-Linked ALS. EMBO Mol. Med. 2016, 8, 1421–1437. [Google Scholar] [CrossRef]
- Lopez-Gonzalez, R.; Lu, Y.; Gendron, T.F.; Karydas, A.; Tran, H.; Yang, D.; Petrucelli, L.; Miller, B.L.; Almeida, S.; Gao, F.-B. Poly(GR) in C9ORF72-Related ALS/FTD Compromises Mitochondrial Function and Increases Oxidative Stress and DNA Damage in IPSC-Derived Motor Neurons. Neuron 2016, 92, 383–391. [Google Scholar] [CrossRef]
- Tran, D.; Chalhoub, A.; Schooley, A.; Zhang, W.; Ngsee, J.K. A Mutation in VAPB That Causes Amyotrophic Lateral Sclerosis Also Causes a Nuclear Envelope Defect. J. Cell Sci. 2012, 125, 2831–2836. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, J.; Henderson, M.J.; Yang, P.; Hagen, B.M.; Siddique, T.; Vogel, B.E.; Deng, H.-X.; Fang, S. Nuclear Export of Misfolded SOD1 Mediated by a Normally Buried NES-like Sequence Reduces Proteotoxicity in the Nucleus. eLife 2017, 6, e23759. [Google Scholar] [CrossRef] [PubMed]
- Gkogkas, C.; Middleton, S.; Kremer, A.M.; Wardrope, C.; Hannah, M.; Gillingwater, T.H.; Skehel, P. VAPB Interacts with and Modulates the Activity of ATF6. Hum. Mol. Genet. 2008, 17, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Kanekura, K.; Levine, T.P.; Kohno, K.; Olkkonen, V.M.; Aiso, S.; Matsuoka, M. ALS-Linked P56S-VAPB, an Aggregated Loss-of-Function Mutant of VAPB, Predisposes Motor Neurons to ER Stress-Related Death by Inducing Aggregation of Co-Expressed Wild-Type VAPB. J. Neurochem. 2009, 108, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Kanekura, K.; Suzuki, H.; Aiso, S.; Matsuoka, M. ER Stress and Unfolded Protein Response in Amyotrophic Lateral Sclerosis. Mol. Neurobiol. 2009, 39, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, S.; Koyama, Y.; Katayama, T.; Taniguchi, M.; Hitomi, J.; Kato, M.; Aoki, M.; Itoyama, Y.; Kato, S.; Tohyama, M. An in Vitro Model for Lewy Body-like Hyaline Inclusion/Astrocytic Hyaline Inclusion: Induction by ER Stress with an ALS-Linked SOD1 Mutation. PLoS ONE 2007, 2, e1030. [Google Scholar] [CrossRef] [PubMed]
- Dreser, A.; Vollrath, J.T.; Sechi, A.; Johann, S.; Roos, A.; Yamoah, A.; Katona, I.; Bohlega, S.; Wiemuth, D.; Tian, Y.; et al. The ALS-Linked E102Q Mutation in Sigma Receptor-1 Leads to ER Stress-Mediated Defects in Protein Homeostasis and Dysregulation of RNA-Binding Proteins. Cell Death Differ. 2017, 24, 1655–1671. [Google Scholar] [CrossRef]
- Ilieva, E.V.; Ayala, V.; Jové, M.; Dalfó, E.; Cacabelos, D.; Povedano, M.; Bellmunt, M.J.; Ferrer, I.; Pamplona, R.; Portero-Otín, M. Oxidative and Endoplasmic Reticulum Stress Interplay in Sporadic Amyotrophic Lateral Sclerosis. Brain 2007, 130, 3111–3123. [Google Scholar] [CrossRef]
- Prosser, D.C.; Tran, D.; Gougeon, P.Y.; Verly, C.; Ngsee, J.K. FFAT Rescues VAPA-Mediated Inhibition of ER-to-Golgi Transport and VAPB-Mediated ER Aggregation. J. Cell Sci. 2008, 121, 3052–3061. [Google Scholar] [CrossRef]
- Skehel, P.A.; Martin, K.C.; Kandel, E.R.; Bartsch, D. A VAMP-Binding Protein from Aplysia Required for Neurotransmitter Release. Science 1995, 269, 1580–1583. [Google Scholar] [CrossRef]
- Nishimura, Y.; Hayashi, M.; Inada, H.; Tanaka, T. Molecular Cloning and Characterization of Mammalian Homologues of Vesicle-Associated Membrane Protein-Associated (VAMP-Associated) Proteins. Biochem. Biophys. Res. Commun. 1999, 254, 21–26. [Google Scholar] [CrossRef]
- Raychaudhuri, S.; Im, Y.J.; Hurley, J.H.; Prinz, W.A. Nonvesicular Sterol Movement from Plasma Membrane to ER Requires Oxysterol-Binding Protein–Related Proteins and Phosphoinositides. J. Cell Biol. 2006, 173, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Loewen, C.J.R.; Levine, T.P. A Highly Conserved Binding Site in Vesicle-Associated Membrane Protein-Associated Protein (VAP) for the FFAT Motif of Lipid-Binding Proteins. J. Biol. Chem. 2005, 280, 14097–14104. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.J.; Ridgway, N.D. Oxysterol-Binding Protein and Vesicle-Associated Membrane Protein-Associated Protein are Required for Sterol-Dependent Activation of the Ceramide Transport Protein. Mol. Biol. Cell 2006, 17, 2604–2616. [Google Scholar] [CrossRef] [PubMed]
- Manford, A.G.; Stefan, C.J.; Yuan, H.L.; MacGurn, J.A.; Emr, S.D. ER-to-Plasma Membrane Tethering Proteins Regulate Cell Signaling and ER Morphology. Dev. Cell 2012, 23, 1129–1140. [Google Scholar] [CrossRef]
- Loewen, C.J.R.; Young, B.P.; Tavassoli, S.; Levine, T.P. Inheritance of Cortical ER in Yeast Is Required for Normal Septin Organization. J. Cell Biol. 2007, 179, 467–483. [Google Scholar] [CrossRef]
- Rocha, N.; Kuijl, C.; van der Kant, R.; Janssen, L.; Houben, D.; Janssen, H.; Zwart, W.; Neefjes, J. Cholesterol Sensor ORP1L Contacts the ER Protein VAP to Control Rab7–RILP–P150Glued and Late Endosome Positioning. J. Cell Biol. 2009, 185, 1209–1225. [Google Scholar] [CrossRef]
- Murphy, S.E.; Levine, T.P. VAP, a Versatile Access Point for the Endoplasmic Reticulum: Review and Analysis of FFAT-like Motifs in the VAPome. Biochim. Biophys. Acta 2016, 1861, 952–961. [Google Scholar] [CrossRef]
- Nishimura, A.L.; Mitne-Neto, M.; Silva, H.C.A.; Richieri-Costa, A.; Middleton, S.; Cascio, D.; Kok, F.; Oliveira, J.R.M.; Gillingwater, T.; Webb, J.; et al. A Mutation in the Vesicle-Trafficking Protein VAPB Causes Late-Onset Spinal Muscular Atrophy and Amyotrophic Lateral Sclerosis. Am. J. Hum. Genet. 2004, 75, 822–831. [Google Scholar] [CrossRef]
- Mitne-Neto, M.; Ramos, C.R.R.; Pimenta, D.C.; Luz, J.S.; Nishimura, A.L.; Gonzales, F.A.; Oliveira, C.C.; Zatz, M. A Mutation in Human VAP-B-MSP Domain, Present in ALS Patients, Affects the Interaction with Other Cellular Proteins. Protein Expr. Purif. 2007, 55, 139–146. [Google Scholar] [CrossRef]
- Kaiser, S.E.; Brickner, J.H.; Reilein, A.R.; Fenn, T.D.; Walter, P.; Brunger, A.T. Structural Basis of FFAT Motif-Mediated ER Targeting. Structure 2005, 13, 1035–1045. [Google Scholar] [CrossRef]
- Teuling, E.; Ahmed, S.; Haasdijk, E.; Demmers, J.; Steinmetz, M.O.; Akhmanova, A.; Jaarsma, D.; Hoogenraad, C.C. Motor Neuron Disease-Associated Mutant Vesicle-Associated Membrane Protein-Associated Protein (VAP) B Recruits Wild-Type VAPs into Endoplasmic Reticulum-Derived Tubular Aggregates. J. Neurosci. 2007, 27, 9801–9815. [Google Scholar] [CrossRef] [PubMed]
- Ratnaparkhi, A.; Lawless, G.M.; Schweizer, F.E.; Golshani, P.; Jackson, G.R. A Drosophila Model of ALS: Human ALS-Associated Mutation in VAP33A Suggests a Dominant Negative Mechanism. PLoS ONE 2008, 3, e2334. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, H.; Han, S.M.; Yang, Y.; Tong, C.; Lin, Y.Q.; Mohan, K.; Haueter, C.; Zoghbi, A.; Harati, Y.; Kwan, J.; et al. The Amyotrophic Lateral Sclerosis 8 Protein VAPB Is Cleaved, Secreted, and Acts as a Ligand for Eph Receptors. Cell 2008, 133, 963–977. [Google Scholar] [CrossRef] [PubMed]
- Brewer, J.W.; Diehl, J.A. PERK Mediates Cell-Cycle Exit during the Mammalian Unfolded Protein Response. Proc. Natl. Acad. Sci. USA 2000, 97, 12625–12630. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.P.; Novoa, I.; Zhang, Y.; Zeng, H.; Wek, R.; Schapira, M.; Ron, D. Regulated Translation Initiation Controls Stress-Induced Gene Expression in Mammalian Cells. Mol. Cell. 2000, 6, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. XBP1 MRNA Is Induced by ATF6 and Spliced by IRE1 in Response to ER Stress to Produce a Highly Active Transcription Factor. Cell 2001, 107, 881–891. [Google Scholar] [CrossRef]
- Haze, K.; Yoshida, H.; Yanagi, H.; Yura, T.; Mori, K. Mammalian Transcription Factor ATF6 Is Synthesized as a Transmembrane Protein and Activated by Proteolysis in Response to Endoplasmic Reticulum Stress. Mol. Biol. Cell 1999, 10, 3787–3799. [Google Scholar] [CrossRef]
- Nakamichi, S.; Yamanaka, K.; Suzuki, M.; Watanabe, T.; Kagiwada, S. Human VAPA and the Yeast VAP Scs2p with an Altered Proline Distribution Can Phenocopy Amyotrophic Lateral Sclerosis-Associated VAPB(P56S). Biochem. Biophys. Res. Commun. 2011, 404, 605–609. [Google Scholar] [CrossRef]
- Longtine, M.S.; Mckenzie, A., III; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional Modules for Versatile and Economical PCR-based Gene Deletion and Modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Goldstein, A.L.; McCusker, J.H. Three New Dominant Drug Resistance Cassettes for Gene Disruption in Saccharomyces cerevisiae. Yeast 1999, 15, 1541–1553. [Google Scholar] [CrossRef]
- Schiestl, R.H.; Gietz, R.D. High Efficiency Transformation of Intact Yeast Cells Using Single Stranded Nucleic Acids as a Carrier. Curr. Genet. 1989, 16, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Sikorski, R.S.; Hieter, P. A System of Shuttle Vectors and Yeast Host Strains Designed for Efficient Manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Audhya, A.; Emr, S.D. Regulation of PI4,5P2 Synthesis by Nuclear-Cytoplasmic Shuttling of the Mss4 Lipid Kinase. EMBO J. 2003, 22, 4223–4236. [Google Scholar] [CrossRef] [PubMed]
- Prosser, D.C.; Wrasman, K.; Woodard, T.K.; O’Donnell, A.F.; Wendland, B. Applications of PHluorin for Quantitative, Kinetic and High-Throughput Analysis of Endocytosis in Budding Yeast. J. Vis. Exp. 2016, 116, e54587. [Google Scholar] [CrossRef]
- Deng, H.-X.; Hentati, A.; Tainer, J.A.; Iqbal, Z.; Cayabyab, A.; Hung, W.-Y.; Getzoff, E.D.; Hu, P.; Herzfeldt, B.; Roos, R.P.; et al. Amyotrophic Lateral Sclerosis and Structural Defects in Cu,Zn Superoxide Dismutase. Science 1993, 261, 1047–1051. [Google Scholar] [CrossRef]
- Rosen, D.R.; Bowling, A.C.; Patterson, D.; Usdin, T.B.; Sapp, P.; Mezey, E.; McKenna-Yasek, D.; O’Regan, J.; Rahmani, Z.; Ferrante, R.J.; et al. A Frequent Ala 4 to Val Superoxide Dismutase-1 Mutation Is Associated with a Rapidly Progressive Familial Amyotrophic Lateral Sclerosis. Hum. Mol. Genet. 1994, 3, 981–987. [Google Scholar] [CrossRef]
- Ogasawara, M.; Matsubara, Y.; Narisawa, K.; Aoki, M.; Nakamura, S.; Itoyama, Y.; Abe, K. Mild ALS in Japan Associated with Novel SOD Mutation. Nat. Genet. 1993, 5, 323–324. [Google Scholar] [CrossRef]
- Kawamata, J.; Hasegawa, H.; Shimohama, S.; Kimura, J.; Tanaka, S.; Ueda, K. Leu106-to-Val (CTC-to-GTC) Mutation of Superoxide Dismutase-1 Gene in Patient with Familial Amyotrophic Lateral Sclerosis in Japan. Lancet 1994, 343, 1501. [Google Scholar] [CrossRef]
- Andersen, P.M.; Forsgren, L.; Binzer, M.; Nilsson, P.; Ala-Hurula, V.; Keranen, M.-L.; Bergmark, L.; Saarinen, A.; Haltia, T.; Tarvainen, I.; et al. Autosomal Recessive Adult-Onset Amyotrophic Lateral Sclerosis Associated with Homozygosity for Asp90Ala CuZn-Superoxide Dismutase Mutation: A Clinical and Genealogical Study of 36 Patients. Brain 1996, 119, 1153–1172. [Google Scholar] [CrossRef]
- Ikeda, M.; Abe, K.; Aoki, M.; Sahara, M.; Watanabe, M.; Shoji, M.; George-Hyslop, P.H.S.; Hirai, S.; Itoyama, Y. Variable Clinical Symptoms in Familial Amyotrophic Lateral Sclerosis with a Novel Point Mutation in the Cu/Zn Superoxide Dismutase Gene. Neurology 1995, 45, 2038–2042. [Google Scholar] [CrossRef]
- Sapp, P.C.; Rosen, D.R.; Hosler, B.A.; Esteban, J.; McKenna-Yasek, D.; O’regan, J.P.; Horvitz, H.R.; Brown, R.H. Identification of Three Novel Mutations in the Gene for CuZn Superoxide Dismutase in Patients with Familial Amyotrophic Lateral Sclerosis. Neuromuscul. Disord. 1995, 5, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Aoki, M.; Abe, K.; Hasegawa, T.; Sakuma, R.; Onodera, Y.; Ichikawa, N.; Nishizawa, M.; Itoyama, Y. A Novel Two-Base Mutation in the CuZn Superoxide Dismutase Gene Associated with Familial Amyotrophic Lateral Sclerosis in Japan. Neurosci. Lett. 1996, 205, 79–82. [Google Scholar] [CrossRef]
- Kostrzewa, M.; Damian, M.S.; Müller, U. Superoxide Dismutase 1: Identification of a Novel Mutation in a Case of Familial Amyotrophic Lateral Sclerosis. Hum. Genet. 1996, 98, 48–50. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.T.; Swingler, R.J.; Brock, D.J. Identification of a Novel SOD1 Mutation in an Apparently Sporadic Amyotrophic Lateral Sclerosis Patient and the Detection of Ile113Thr in Three Others. Hum. Mol. Genet. 1994, 3, 649–650. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Aoki, M.; Abe, K.; Shoji, M.; Iizuka, T.; Ikeda, Y.; Hirai, S.; Kurokawa, K.; Kato, T.; Sasaki, H.; et al. A Novel Missense Point Mutation (S134N) of the Cu/Zn Superoxide Dismutase Gene in a Patient with Familial Motor Neuron Disease. Hum. Mutat. 1997, 9, 69–71. [Google Scholar] [CrossRef]
- Aoki, M.; Abe, K.; Houi, K.; Ogasawara, M.; Matsubara, Y.; Kobayashi, T.; Mochio, S.; Narisawa, K.; Itoyama, Y. Variance of Age at Onset in a Japanese Family with Amyotrophic Lateral Sclerosis Associated with a Novel Cu/Zn Superoxide Dismutase Mutation. Ann. Neurol. 1995, 37, 676–679. [Google Scholar] [CrossRef]
- Kawamata, J.; Shimohama, S.; Takano, S.; Harada, K.; Ueda, K.; Kimura, J. Novel G16S (GGC-AGC) Mutation in the SOD-1 Gene in a Patient with Apparently Sporadic Young-onset Amyotrophic Lateral Sclerosis. Hum. Mutat. 1997, 9, 356–358. [Google Scholar] [CrossRef]
- Zu, J.S.; Deng, H.-X.; Lo, T.P.; Mitsumoto, H.; Ahmed, M.S.; Hung, W.-Y.; Cai, Z.-J.; Tainer, J.A.; Siddique, T. Exon 5 Encoded Domain Is Not Required for the Toxic Function of Mutant SOD1 but Essential for the Dismutase Activity: Identification and Characterization of Two New SOD1 Mutations Associated with Familial Amyotrophic Lateral Sclerosis. Neurogenetics 1997, 1, 65–71. [Google Scholar] [CrossRef]
- Orrell, R.W.; Marklund, S.L.; de Belleroche, J.S. Familial ALS is Associated with Mutations in All Exons of SOD1: A Novel Mutation in Exon 3 (Gly72Ser). J. Neurol. Sci. 1997, 153, 46–49. [Google Scholar] [CrossRef]
- Penco, S.; Schenone, A.; Bordo, D.; Bolognesi, M.; Abbruzzese, M.; Bugiani, O.; Ajmar, F.; Garrè, C. A SOD1 Gene Mutation in a Patient with Slowly Progressing Familial ALS. Neurology 1999, 53, 404–406. [Google Scholar] [CrossRef]
- Gellera, C.; Castellotti, B.; Riggio, M.C.; Silani, V.; Morandi, L.; Testa, D.; Casali, C.; Taroni, F.; Donato, S.D.; Zeviani, M.; et al. Superoxide Dismutase Gene Mutations in Italian Patients with Familial and Sporadic Amyotrophic Lateral Sclerosis: Identification of Three Novel Missense Mutations. Neuromuscul. Disord. 2001, 11, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.D.; Traynor, B.J.; Miller, N.; Corr, B.; Frost, E.; McQuaid, S.; Brett, F.M.; Green, A.; Hardiman, O. “True” Sporadic ALS Associated with a Novel SOD-1 Mutation. Ann. Neurol. 2002, 52, 680–683. [Google Scholar] [CrossRef] [PubMed]
- Khoris, J.; Moulard, B.; Briolotti, V.; Hayer, M.; Durieux, A.; Clavelou, P.; Malafosse, A.; Rouleau, G.A.; Camu, W. Coexistence of Dominant and Recessive Familial Amyotrophic Lateral Sclerosis with the D90A Cu,Zn Superoxide Dismutase Mutation within the Same Country. Eur. J. Neurol. 2000, 7, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Elshafey, A.; Lanyon, W.G.; Connor, J.M. Identification of a New Missense Point Mutation in Exon 4 of the Cu/Zn Superoxide Dismutase (SOD-1) Gene in a Family with Amyotrophic Lateral Sclerosis. Hum. Mol. Genet. 1994, 3, 363–364. [Google Scholar] [CrossRef] [PubMed]
- Ezer, S.; Daana, M.; Park, J.H.; Yanovsky-Dagan, S.; Nordström, U.; Basal, A.; Edvardson, S.; Saada, A.; Otto, M.; Meiner, V.; et al. Infantile SOD1 Deficiency Syndrome Caused by a Homozygous SOD1 Variant with Absence of Enzyme Activity. Brain 2021, 145, 872–878. [Google Scholar] [CrossRef]
- Moreira, M.-C.; Klur, S.; Watanabe, M.; Németh, A.H.; Ber, I.L.; Moniz, J.-C.; Tranchant, C.; Aubourg, P.; Tazir, M.; Schöls, L.; et al. Senataxin, the Ortholog of a Yeast RNA Helicase, is Mutant in Ataxia-Ocular Apraxia 2. Nat. Genet. 2004, 36, 225–227. [Google Scholar] [CrossRef]
- Duquette, A.; Roddier, K.; McNabb-Baltar, J.; Gosselin, I.; St-Denis, A.; Dicaire, M.; Loisel, L.; Labuda, D.; Marchand, L.; Mathieu, J.; et al. Mutations in Senataxin Responsible for Quebec Cluster of Ataxia with Neuropathy. Ann. Neurol. 2005, 57, 408–414. [Google Scholar] [CrossRef]
- Bassuk, A.G.; Chen, Y.Z.; Batish, S.D.; Nagan, N.; Opal, P.; Chance, P.F.; Bennett, C.L. In Cis Autosomal Dominant Mutation of Senataxin Associated with Tremor/Ataxia Syndrome. Neurogenetics 2007, 8, 45–49. [Google Scholar] [CrossRef]
- Asaka, T.; Yokoji, H.; Ito, J.; Yamaguchi, K.; Matsushima, A. Autosomal Recessive Ataxia with Peripheral Neuropathy and Elevated AFP: Novel Mutations in SETX. Neurology 2006, 66, 1580–1581. [Google Scholar] [CrossRef]
- Fogel, B.L.; Perlman, S. Novel Mutations in the Senataxin DNA/RNA Helicase Domain in Ataxia with Oculomotor Apraxia 2. Neurology 2006, 67, 2083–2084. [Google Scholar] [CrossRef]
- Anheim, M.; Fleury, M.-C.; Franques, J.; Moreira, M.-C.; Delaunoy, J.-P.; Stoppa-Lyonnet, D.; Koenig, M.; Tranchant, C. Clinical and Molecular Findings of Ataxia with Oculomotor Apraxia Type 2 in 4 Families. Arch. Neurol. 2008, 65, 958–962. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-J.; Anagnostou, G.; Chai, A.; Withers, J.; Morris, A.; Adhikaree, J.; Pennetta, G.; de Belleroche, J.S. Characterization of the Properties of a Novel Mutation in VAPB in Familial Amyotrophic Lateral Sclerosis. J. Biol. Chem. 2010, 285, 40266–40281. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.Y.; Zhang, Y.; Dowling, J.J.; Jin, N.; Adamska, M.; Shiga, K.; Szigeti, K.; Shy, M.E.; Li, J.; Zhang, X.; et al. Mutation of FIG4 Causes Neurodegeneration in the Pale Tremor Mouse and Patients with CMT4J. Nature 2007, 448, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.Y.; Landers, J.E.; Bergren, S.K.; Sapp, P.C.; Grant, A.E.; Jones, J.M.; Everett, L.; Lenk, G.M.; McKenna-Yasek, D.M.; Weisman, L.S.; et al. Deleterious Variants of FIG4, a Phosphoinositide Phosphatase, in Patients with ALS. Am. J. Hum. Genet. 2009, 84, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Corona-Rivera, J.R.; Romo-Huerta, C.O.; López-Marure, E.; Ramos, F.J.; Estrada-Padilla, S.A.; Zepeda-Romero, L.C. New Ocular Findings in Two Sisters with Yunis–Varón Syndrome and Literature Review. Eur. J. Med. Genet. 2011, 54, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Campeau, P.M.; Lenk, G.M.; Lu, J.T.; Bae, Y.; Burrage, L.; Turnpenny, P.; Corona-Rivera, J.R.; Morandi, L.; Mora, M.; Reutter, H.; et al. Yunis-Varón Syndrome is Caused by Mutations in FIG4, Encoding a Phosphoinositide Phosphatase. Am. J. Hum. Genet. 2013, 92, 781–791. [Google Scholar] [CrossRef]
- Nicholson, G.; Lenk, G.M.; Reddel, S.W.; Grant, A.E.; Towne, C.F.; Ferguson, C.J.; Simpson, E.; Scheuerle, A.; Yasick, M.; Hoffman, S.; et al. Distinctive Genetic and Clinical Features of CMT4J: A Severe Neuropathy Caused by Mutations in the PI(3,5)P₂ Phosphatase FIG4. Brain J. Neurol. 2011, 134, 1959–1971. [Google Scholar] [CrossRef]
- Baulac, S.; Lenk, G.M.; Dufresnois, B.; Bencheikh, B.O.A.; Couarch, P.; Renard, J.; Larson, P.A.; Ferguson, C.J.; Noé, E.; Poirier, K.; et al. Role of the Phosphoinositide Phosphatase FIG4 Gene in Familial Epilepsy with Polymicrogyria. Neurology 2014, 82, 1068–1075. [Google Scholar] [CrossRef]
- Pulst, S.M.; Nechiporuk, A.; Nechiporuk, T.; Gispert, S.; Chen, X.N.; Lopes-Cendes, I.; Pearlman, S.; Starkman, S.; Orozco-Diaz, G.; Lunkes, A.; et al. Moderate Expansion of a Normally Biallelic Trinucleotide Repeat in Spinocerebellar Ataxia Type 2. Nat. Genet. 1996, 14, 269–276. [Google Scholar] [CrossRef]
- Watts, G.D.J.; Wymer, J.; Kovach, M.J.; Mehta, S.G.; Mumm, S.; Darvish, D.; Pestronk, A.; Whyte, M.P.; Kimonis, V.E. Inclusion Body Myopathy Associated with Paget Disease of Bone and Frontotemporal Dementia is Caused by Mutant Valosin-Containing Protein. Nat. Genet. 2004, 36, 377–381. [Google Scholar] [CrossRef]
- Haubenberger, D.; Bittner, R.E.; Rauch-Shorny, S.; Zimprich, F.; Mannhalter, C.; Wagner, L.; Mineva, I.; Vass, K.; Auff, E.; Zimprich, A. Inclusion Body Myopathy and Paget Disease Is Linked to a Novel Mutation in the VCP Gene. Neurology 2005, 65, 1304–1305. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.O.; Mandrioli, J.; Benatar, M.; Abramzon, Y.; Deerlin, V.M.V.; Trojanowski, J.Q.; Gibbs, J.R.; Brunetti, M.; Gronka, S.; Wuu, J.; et al. Exome Sequencing Reveals VCP Mutations as a Cause of Familial ALS. Neuron 2010, 68, 857–864. [Google Scholar] [CrossRef]
- Gonzalez, M.A.; Feely, S.M.; Speziani, F.; Strickland, A.V.; Danzi, M.; Bacon, C.; Lee, Y.; Chou, T.-F.; Blanton, S.H.; Weihl, C.C.; et al. A Novel Mutation in VCP Causes Charcot–Marie–Tooth Type 2 Disease. Brain 2014, 137, 2897–2902. [Google Scholar] [CrossRef] [PubMed]
- Jerath, N.U.; Crockett, C.D.; Moore, S.A.; Shy, M.E.; Weihl, C.C.; Chou, T.-F.; Grider, T.; Gonzalez, M.A.; Zuchner, S.; Swenson, A. Rare Manifestation of a c.290 C>T, p.Gly97Glu VCP Mutation. Case Rep. Genet. 2015, 2015, 239167. [Google Scholar] [CrossRef] [PubMed]
- Abrahao, A.; Neto, O.A.; Kok, F.; Zanoteli, E.; Santos, B.; de Rezende Pinto, W.B.V.; Barsottini, O.G.P.; Oliveira, A.S.B.; Pedroso, J.L. One Family, One Gene and Three Phenotypes: A Novel VCP (Valosin-Containing Protein) Mutation Associated with Myopathy with Rimmed Vacuoles, Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. J. Neurol. Sci. 2016, 368, 352–358. [Google Scholar] [CrossRef]
- Darwich, N.F.; Phan, J.M.; Kim, B.; Suh, E.; Papatriantafyllou, J.D.; Changolkar, L.; Nguyen, A.T.; O’Rourke, C.M.; He, Z.; Porta, S.; et al. Autosomal Dominant VCP Hypomorph Mutation Impairs Disaggregation of PHF-Tau. Science 2020, 370, eaay8826. [Google Scholar] [CrossRef]
- Wong, T.H.; Pottier, C.; Hondius, D.; Meeter, L.H.H.; van Rooij, J.G.J.; Melhem, S.; Bank, T.N.B.; van Minkelen, R.; van Duijn, C.M.; Rozemuller, A.J.M.; et al. Three VCP Mutations in Patients with Frontotemporal Dementia. J. Alzheimer’s Dis. 2018, 65, 1139–1146. [Google Scholar] [CrossRef]
- Al-Saif, A.; Al-Mohanna, F.; Bohlega, S. A Mutation in Sigma-1 Receptor Causes Juvenile Amyotrophic Lateral Sclerosis. Ann. Neurol. 2011, 70, 913–919. [Google Scholar] [CrossRef]
- Skibinski, G.; Parkinson, N.J.; Brown, J.M.; Chakrabarti, L.; Lloyd, S.L.; Hummerich, H.; Nielsen, J.E.; Hodges, J.R.; Spillantini, M.G.; Thusgaard, T.; et al. Mutations in the Endosomal ESCRTIII-Complex Subunit CHMP2B in Frontotemporal Dementia. Nat. Genet. 2005, 37, 806–808. [Google Scholar] [CrossRef]
- Parkinson, N.; Ince, P.G.; Smith, M.O.; Highley, R.; Skibinski, G.; Andersen, P.M.; Morrison, K.E.; Pall, H.S.; Hardiman, O.; Collinge, J.; et al. ALS Phenotypes with Mutations in CHMP2B (Charged Multivesicular Body Protein 2B). Neurology 2006, 67, 1074–1077. [Google Scholar] [CrossRef]
- van der Zee, J.; Urwin, H.; Engelborghs, S.; Bruyland, M.; Vandenberghe, R.; Dermaut, B.; Pooter, T.D.; Peeters, K.; Santens, P.; Deyn, P.P.D.; et al. CHMP2B C-Truncating Mutations in Frontotemporal Lobar Degeneration Are Associated with an Aberrant Endosomal Phenotype in Vitro. Hum. Mol. Genet. 2007, 17, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, N.C.; Wang, Y.-D.; Scarborough, E.A.; Moore, J.; Diaz, Z.; MacLea, K.S.; Freibaum, B.; Li, S.; Molliex, A.; et al. Mutations in Prion-like Domains in HnRNPA2B1 and HnRNPA1 Cause Multisystem Proteinopathy and ALS. Nature 2013, 495, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.N.; Ticozzi, N.; Fallini, C.; Gkazi, A.S.; Topp, S.; Kenna, K.P.; Scotter, E.L.; Kost, J.; Keagle, P.; Miller, J.W.; et al. Exome-Wide Rare Variant Analysis Identifies TUBA4A Mutations Associated with Familial ALS. Neuron 2014, 84, 324–331. [Google Scholar] [CrossRef]
- Strassel, C.; Magiera, M.M.; Dupuis, A.; Batzenschlager, M.; Hovasse, A.; Pleines, I.; Guéguen, P.; Eckly, A.; Moog, S.; Mallo, L.; et al. An Essential Role for A4A-Tubulin in Platelet Biogenesis. Life Sci. Alliance 2019, 2, e201900309. [Google Scholar] [CrossRef]
- Reid, E.; Kloos, M.; Ashley-Koch, A.; Hughes, L.; Bevan, S.; Svenson, I.K.; Graham, F.L.; Gaskell, P.C.; Dearlove, A.; Pericak-Vance, M.A.; et al. A Kinesin Heavy Chain (KIF5A) Mutation in Hereditary Spastic Paraplegia (SPG10). Am. J. Hum. Genet. 2002, 71, 1189–1194. [Google Scholar] [CrossRef]
- Fichera, M.; Giudice, M.L.; Falco, M.; Sturnio, M.; Amata, S.; Calabrese, O.; Bigoni, S.; Calzolari, E.; Neri, M. Evidence of Kinesin Heavy Chain (KIF5A) Involvement in Pure Hereditary Spastic Paraplegia. Neurology 2004, 63, 1108–1110. [Google Scholar] [CrossRef] [PubMed]
- Goizet, C.; Boukhris, A.; Mundwiller, E.; Tallaksen, C.; Forlani, S.; Toutain, A.; Carriere, N.; Paquis, V.; Depienne, C.; Durr, A.; et al. Complicated Forms of Autosomal Dominant Hereditary Spastic Paraplegia Are Frequent in SPG10. Hum. Mutat. 2009, 30, E376–E385. [Google Scholar] [CrossRef] [PubMed]
- Crimella, C.; Baschirotto, C.; Arnoldi, A.; Tonelli, A.; Tenderini, E.; Airoldi, G.; Martinuzzi, A.; Trabacca, A.; Losito, L.; Scarlato, M.; et al. Mutations in the Motor and Stalk Domains of KIF5A in Spastic Paraplegia Type 10 and in Axonal Charcot–Marie–Tooth Type 2. Clin. Genet. 2012, 82, 157–164. [Google Scholar] [CrossRef]
- Liu, Y.-T.; Laurá, M.; Hersheson, J.; Horga, A.; Jaunmuktane, Z.; Brandner, S.; Pittman, A.; Hughes, D.; Polke, J.M.; Sweeney, M.G.; et al. Extended Phenotypic Spectrum of KIF5A Mutations. Neurology 2014, 83, 612–619. [Google Scholar] [CrossRef]
- Dawkins, J.L.; Hulme, D.J.; Brahmbhatt, S.B.; Auer-Grumbach, M.; Nicholson, G.A. Mutations in SPTLC1, Encoding Serine Palmitoyltransferase, Long Chain Base Subunit-1, Cause Hereditary Sensory Neuropathy Type I. Nat. Genet. 2001, 27, 309–312. [Google Scholar] [CrossRef]
- Bejaoui, K.; Wu, C.; Scheffler, M.D.; Haan, G.; Ashby, P.; Wu, L.; de Jong, P.; Brown, R.H. SPTLC1 is Mutated in Hereditary Sensory Neuropathy, Type 1. Nat. Genet. 2001, 27, 261–262. [Google Scholar] [CrossRef] [PubMed]
- Rotthier, A.; Baets, J.; Vriendt, E.D.; Jacobs, A.; Auer-Grumbach, M.; Lévy, N.; Bonello-Palot, N.; Kilic, S.S.; Weis, J.; Nascimento, A.; et al. Genes for Hereditary Sensory and Autonomic Neuropathies: A Genotype–Phenotype Correlation. Brain 2009, 132, 2699–2711. [Google Scholar] [CrossRef] [PubMed]
- Auer-Grumbach, M.; Bode, H.; Pieber, T.R.; Schabhüttl, M.; Fischer, D.; Seidl, R.; Graf, E.; Wieland, T.; Schuh, R.; Vacariu, G.; et al. Mutations at Ser331 in the HSN Type I Gene SPTLC1 are Associated with a Distinct Syndromic Phenotype. Eur. J. Med. Genet. 2013, 56, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Mohassel, P.; Donkervoort, S.; Lone, M.A.; Nalls, M.; Gable, K.; Gupta, S.D.; Foley, A.R.; Hu, Y.; Saute, J.A.M.; Moreira, A.L.; et al. Childhood Amyotrophic Lateral Sclerosis Caused by Excess Sphingolipid Synthesis. Nat. Med. 2021, 27, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Bannwarth, S.; Ait-El-Mkadem, S.; Chaussenot, A.; Genin, E.C.; Lacas-Gervais, S.; Fragaki, K.; Berg-Alonso, L.; Kageyama, Y.; Serre, V.; Moore, D.G.; et al. A Mitochondrial Origin for Frontotemporal Dementia and Amyotrophic Lateral Sclerosis through CHCHD10 Involvement. Brain 2014, 137, 2329–2345. [Google Scholar] [CrossRef] [PubMed]
- Penttilä, S.; Jokela, M.; Bouquin, H.; Saukkonen, A.M.; Toivanen, J.; Udd, B. Late Onset Spinal Motor Neuronopathy is Caused by Mutation in CHCHD10. Ann. Neurol. 2015, 77, 163–172. [Google Scholar] [CrossRef]
- Ajroud-Driss, S.; Fecto, F.; Ajroud, K.; Lalani, I.; Calvo, S.E.; Mootha, V.K.; Deng, H.-X.; Siddique, N.; Tahmoush, A.J.; Heiman-Patterson, T.D.; et al. Mutation in the Novel Nuclear-Encoded Mitochondrial Protein CHCHD10 in a Family with Autosomal Dominant Mitochondrial Myopathy. Neurogenetics 2015, 16, 1–9. [Google Scholar] [CrossRef]
- Williams, K.L.; Topp, S.; Yang, S.; Smith, B.; Fifita, J.A.; Warraich, S.T.; Zhang, K.Y.; Farrawell, N.; Vance, C.; Hu, X.; et al. CCNF Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Nat. Commun. 2016, 7, 11253. [Google Scholar] [CrossRef]
- Lord, S.J.; Velle, K.B.; Mullins, R.D.; Fritz-Laylin, L.K. SuperPlots: Communicating Reproducibility and Variability in Cell Biology. J. Cell Biol. 2020, 219, e202001064. [Google Scholar] [CrossRef]
- Gkogkas, C.; Wardrope, C.; Hannah, M.; Skehel, P. The ALS8-Associated Mutant VAPBP56S Is Resistant to Proteolysis in Neurons. J. Neurochem. 2011, 117, 286–294. [Google Scholar] [CrossRef]
- Barnes, G.; Hansen, W.J.; Holcomb, C.L.; Rine, J. Asparagine-Linked Glycosylation in Saccharomyces cerevisiae: Genetic Analysis of an Early Step. Mol. Cell Biol. 1984, 4, 2381–2388. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.; Baryshnikova, A.; Brown, G.W. Unification of Protein Abundance Datasets Yields a Quantitative Saccharomyces cerevisiae Proteome. Cell Syst. 2018, 6, 192–205.e3. [Google Scholar] [CrossRef] [PubMed]
- Ghaemmaghami, S.; Huh, W.-K.; Bower, K.; Howson, R.W.; Belle, A.; Dephoure, N.; O’Shea, E.K.; Weissman, J.S. Global Analysis of Protein Expression in Yeast. Nature 2003, 425, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, Encapsulated Proteomic-Sample Processing Applied to Copy-Number Estimation in Eukaryotic Cells. Nat. Meth. 2014, 11, 319–324. [Google Scholar] [CrossRef]
- Nagaraj, N.; Kulak, N.A.; Cox, J.; Neuhauser, N.; Mayr, K.; Hoerning, O.; Vorm, O.; Mann, M. System-Wide Perturbation Analysis with Nearly Complete Coverage of the Yeast Proteome by Single-Shot Ultra HPLC Runs on a Bench Top Orbitrap. Mol. Cell. Proteom. 2012, 11, M111.013722. [Google Scholar] [CrossRef]
- Peng, M.; Taouatas, N.; Cappadona, S.; van Breukelen, B.; Mohammed, S.; Scholten, A.; Heck, A.J.R. Protease Bias in Absolute Protein Quantitation. Nat. Meth. 2012, 9, 524–525. [Google Scholar] [CrossRef]
- Webb, K.J.; Xu, T.; Park, S.K.; Yates, J.R. Modified MuDPIT Separation Identified 4488 Proteins in a System-Wide Analysis of Quiescence in Yeast. J. Proteome Res. 2013, 12, 2177–2184. [Google Scholar] [CrossRef]
- Yofe, I.; Weill, U.; Meurer, M.; Chuartzman, S.; Zalckvar, E.; Goldman, O.; Ben-Dor, S.; Schütze, C.; Wiedemann, N.; Knop, M.; et al. One Library to Make Them All: Streamlining the Creation of Yeast Libraries via a SWAp-Tag Strategy. Nat. Meth. 2016, 13, 371–378. [Google Scholar] [CrossRef]
- Treusch, S.; Hamamichi, S.; Goodman, J.L.; Matlack, K.E.S.; Chung, C.Y.; Baru, V.; Shulman, J.M.; Parrado, A.; Bevis, B.J.; Valastyan, J.S.; et al. Functional Links between Aβ Toxicity, Endocytic Trafficking, and Alzheimer’s Disease Risk Factors in Yeast. Science 2011, 334, 1241–1245. [Google Scholar] [CrossRef]
- Cooper, A.A.; Gitler, A.D.; Cashikar, A.; Haynes, C.M.; Hill, K.J.; Bhullar, B.; Liu, K.; Xu, K.; Strathearn, K.E.; Liu, F.; et al. Alpha-Synuclein Blocks ER-Golgi Traffic and Rab1 Rescues Neuron Loss in Parkinson’s Models. Science 2006, 313, 324–328. [Google Scholar] [CrossRef]
- Bevis, B.J.; Hammond, A.T.; Reinke, C.A.; Glick, B.S. De Novo Formation of Transitional ER Sites and Golgi Structures in Pichia Pastoris. Nat. Cell Biol. 2002, 4, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Rosen, D.R.; Siddique, T.; Patterson, D.; Figlewicz, D.A.; Sapp, P.; Hentati, A.; Donaldson, D.; Goto, J.; O’Regan, J.P.; Deng, H.-X.; et al. Mutations in Cu/Zn Superoxide Dismutase Gene Are Associated with Familial Amyotrophic Lateral Sclerosis. Nature 1993, 362, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Nikawa, J.; Murakami, A.; Esumi, E.; Hosaka, K. Cloning and Sequence of the SCS2 Gene, Which Can Suppress the Defect of INO1 Expression in an Inositol Auxotrophic Mutant of Saccharomyces cerevisiae. J. Biochem. 1995, 118, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.H.; Laurent, J.M.; Yellman, C.M.; Meyer, A.G.; Wilke, C.O.; Marcotte, E.M. Systematic Humanization of Yeast Genes Reveals Conserved Functions and Genetic Modularity. Science 2015, 348, 921–925. [Google Scholar] [CrossRef] [PubMed]
- Tudor, E.L.; Galtrey, C.M.; Perkinton, M.S.; Lau, K.-F.; Vos, K.J.D.; Mitchell, J.C.; Ackerley, S.; Hortobágyi, T.; Vámos, E.; Leigh, P.N.; et al. Amyotrophic Lateral Sclerosis Mutant Vesicle-Associated Membrane Protein-Associated Protein-B Transgenic Mice Develop TAR-DNA-Binding Protein-43 Pathology. Neuroscience 2010, 167, 774–785. [Google Scholar] [CrossRef]
- Kabashi, E.; Oussini, H.E.; Bercier, V.; Gros-Louis, F.; Valdmanis, P.N.; McDearmid, J.; Mejier, I.A.; Dion, P.A.; Dupre, N.; Hollinger, D.; et al. Investigating the Contribution of VAPB/ALS8 Loss of Function in Amyotrophic Lateral Sclerosis. Hum. Mol. Genet. 2013, 22, 2350–2360. [Google Scholar] [CrossRef]
- Zhang, W.; Colavita, A.; Ngsee, J.K. Mitigating Motor Neuronal Loss in C. Elegans Model of ALS8. Sci. Rep. 2017, 7, 11582. [Google Scholar] [CrossRef]
- Chai, A.; Withers, J.; Koh, Y.H.; Parry, K.; Bao, H.; Zhang, B.; Budnik, V.; Pennetta, G. HVAPB, the Causative Gene of a Heterogeneous Group of Motor Neuron Diseases in Humans, is Functionally Interchangeable with Its Drosophila Homologue DVAP-33A at the Neuromuscular Junction. Hum. Mol. Genet. 2008, 17, 266–280. [Google Scholar] [CrossRef]

| Strain | Genotype | Source |
|---|---|---|
| W303 1 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 his3-11,15 | Laboratory strain |
| DPY2271 | MATa leu2-3,112::LEU2 | This study |
| DPY2275 | MATa leu2-3,112::LEU2 Sec61-GFP::HIS3 | This study |
| DPY2287 | MATa scs2::KANMX6 leu2-3,112::LEU2 Sec61-GFP::HIS3 | This study |
| DPY2290 | MATa scs22::HPHMX4 leu2-3,112::LEU2 | This study |
| DPY2295 | MATa scs22::HPHMX4 leu2-3,112::LEU2 Sec61-GFP::HIS3 | This study |
| DPY2298 | MATa scs2::KANMX6 scs22::HPHMX4 leu2-3,112::LEU2 | This study |
| DPY2301 | MATa scs2::KANMX6 scs22::HPHMX4 leu2-3,112::LEU2 Sec61-GFP::HIS3 | This study |
| DPY2309 | MATa scs2::KANMX6 scs22::HPHMX4 leu2:SCS2::LEU2 | This study |
| DPY2311 | MATa scs2::KANMX6 scs22::HPHMX4 leu2:SCS2::LEU2 Sec61-GFP::HIS3 | This study |
| DPY2317 | MATa scs2::KANMX6 scs22::HPHMX4 leu2:scs2P51S, P58S::LEU2 | This study |
| DPY2319 | MATa scs2::KANMX6 scs22::HPHMX4 leu2:scs2P51S, P58S::LEU2 Sec61-GFP::HIS3 | This study |
| DPY2339 | MATa scs2::KANMX6 leu2-3,112::LEU2 | This study |
| DPY2822 | MATa scs2::KANMX6 scs22::HPHMX4 leu2:hsVAPB::LEU2 Sec61-GFP::HIS3 | This study |
| DPY2823 | MATa scs2::KANMX6 scs22::HPHMX4 leu2:hsVAPBP56S::LEU2 Sec61-GFP::HIS3 | This study |
| DPY2854 | MATa scs2::KANMX6 scs22::HPHMX4 leu2:hsVAPB::LEU2 | This study |
| DPY2855 | MATa scs2::KANMX6 scs22::HPHMX4 leu2:hsVAPBP56S::LEU2 | This study |
| DPY2952 | MATa leu2-3,112::LEU2 Sec61-GFP::HIS3 Rtn1-mScarlet1::NATMX4 | This study |
| DPY2953 | MATa scs2::KANMX6 leu2-3,112::LEU2 Sec61-GFP::HIS3 Rtn1-mScarlet1::NATMX4 | This study |
| DPY2954 | MATa scs22::HPHMX4 leu2-3,112::LEU2 Sec61-GFP::HIS3 Rtn1-mScarlet1::NATMX4 | This study |
| DPY2955 | MATa scs2::KANMX6 scs22::HPHMX4 leu2-3,112::LEU2 Sec61-GFP::HIS3 Rtn1-mScarlet1::NATMX4 | This study |
| DPY2956 | MATa scs2::KANMX6 scs22::HPHMX4 leu2:SCS2::LEU2 Sec61-GFP::HIS3 Rtn1-mScarlet1::NATMX4 | This study |
| DPY2957 | MATa scs2::KANMX6 scs22::HPHMX4 leu2:scs2P51S, P58S::LEU2 Sec61-GFP::HIS3 Rtn1-mScarlet1::NATMX4 | This study |
| DPY2958 | MATa scs2::KANMX6 scs22::HPHMX4 leu2:hsVAPB::LEU2 Sec61-GFP::HIS3 Rtn1-mScarlet1::NATMX4 | This study |
| DPY2959 | MATa scs2::KANMX6 scs22::HPHMX4 leu2:hsVAPBP56S::LEU2 Sec61-GFP::HIS3 Rtn1-mScarlet1::NATMX4 | This study |
| Plasmid | Description | Source |
|---|---|---|
| pRS405 | LEU2 integrating plasmid | [42] |
| pRS413 | CEN HIS3 empty vector | [42] |
| pRS416 | CEN URA3 empty vector | [42] |
| pDP0558 | SCS2.413 [CEN HIS3] | This study |
| pDP0559 | scs2P51S, P58S.413 [CEN HIS3] | This study |
| pDP0562 | SCS2.405 [LEU2] | This study |
| pDP0563 | scs2P51S, P58S.405 [LEU2] | This study |
| pDP0900 | PSCS2-hsVAPBWT-TSCS2.416 [CEN URA3] | This study |
| pDP0901 | PSCS2-hsVAPBP56S-TSCS2.416 [CEN URA3] | This study |
| pDP0911 | PSCS2-hsVAPBWT-TSCS2.405 [LEU2] | This study |
| pDP0912 | PSCS2-hsVAPBP56SS-TSCS2.405 [LEU2] | This study |
| pHDEL | pRS416.DsRed-HDEL [CEN URA3] | [43] |
| ALS Type | Human Gene (Accession) | Yeast Homolog | Identity (%) a | Human Mutation (Yeast Analog) a | Suppl. File | Refs. |
|---|---|---|---|---|---|---|
| ALS1 | SOD1 (NP_000445.1) | SOD1 | 54.9 | A4T/V(A4); C6F(A6); G12R(A12); G16S(G16); E21K(E21); G37R(G37); G41D/S(A42); H43R(R44); F45C(F46); H46R(H47); G72S(G73); H80R(H81); L84V(M85); G85R(G86); D90A(D91); G93A/C/R(G94); D95N(K97); E100G(K101); I104F(I105); L106V(L107); I113T(V114); V119∆ or V120∆(V118 or V119) b; L126T(L127); S134N(S135); A145T(A146); I151T(L152) | 2-1 | [45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66] |
| ALS4 | SETX (AAI37351.1) | SEN1 | 23.1 | No conserved ALS-linked mutations SCA-linked c: M271I(I137); E343*(K230) d; Q653K(Q404); Q868*(N587); R1363*(N895); L1976R(I1370); L1977F(I1371); P2213L(P1622) | 2-2 | [67,68,69,70,71,72] |
| ALS8 | VAPB (NP_004729.1) | SCS2 SCS22 | 26.34 28.57 | T46I(T41); P56S(P51) T46I(T38); P56S(P48) | 2-3 | [28,73] |
| ALS11 | FIG4 (NP055660.1) | FIG4 | 38.03 | D53Y(E72); R183*(K195) BTOP-linked e: D783V(H776) CMT4J-linked f: L17P(L35); I41T(I59); F98fsTer102(F109); G253fsTer261(G244); E302K(E293) YVS-linked g: L175P(L187); K278WfsTer6(H269); T422NfsTer6(S422) | 2-4 | [74,75,76,77,78,79] |
| ALS13 | ATXN2 (AAB19200.1) | PBP1 | 24.06 | No conserved ALS-linked mutations; (CAG)n repeat expansion (n = 27–33 in ALS13) | 2-5 | [80] |
| ALS14/ FTD-ALS6 | VCP (AAI10914.1) | CDC48 | 69.48 | R155H/C(R165); R159G/S(R169); D395G(D405); D592N(D602) IBMPFD1-linked h: N91Y(N101); R95G(R105); R155P(R165); R159H(R169); R191Q(R201); A232E(A242) CMT2Y-linked i: G97E(G107); E185K(E195) | 2-6 | [81,82,83,84,85,86,87,88] |
| ALS15 | UBQLN2 (NP_038472.2) | DSK2 | 30.31 | No conserved ALS-linked mutations | 2-7 | |
| ALS16 | SIGMAR1 (NP_005857.1) | ERG2 | 31.6 | E102Q(E104) | 2-8 | [89] |
| ALS17/ FTD-ALS7 | CHMP2B (NP_054762.2) | DID4 | 28.17 | D148Y(N152); Q165*(K170); Q206H(R224) | 2-9 | [90,91,92] |
| ALS18 | PFN1 (AAA36486.1) | PFY1 | 29.03 | No conserved ALS-linked mutations | 2-10 | |
| ALS20 | HNRNPA1 (NP_112420.1) | HRP1 | 33.33 | D314N(D490); N319S(N497) IBMPFD3-linked j: D314V(D490) | 2-11 | [93] |
| ALS22 | TUBA4A (AAH09238.1) | TUB1 TUB3 | 74.44 72.3 | T145P(T146); R215C(R216); R320C/H(R321); A383T(S384); W407*(W408) Macrothrombocytopenia: V181M(V182), E183Q(E184) T145P(T146); R215C(R216); R320C/H(R321); A383T(S384); W407*(W408) Macrothrombocytopenia: V181M(V182), E183Q(E184) | 2-12 | [94,95] |
| ALS25 | KIF5A (AAA20231.1) | SMY1 | 26.49 | No conserved ALS-linked mutations Spastic paraplegia 10: R204Q(R234); D232N(D262); G235E(G265); N256S(N286); R280C/H(R319) | 2-13 | [96,97,98,99,100] |
| ALS26 | TIA1 (NP_071505.2) | PUB1 | 34.82 | No conserved ALS-linked mutations | 2-14 | |
| ALS27 | SPTLC1 (NP_006406.1) | LCB1 | 35.32 | Y23F(Y55); F40∆, S41∆ (L72, S73); S331Y(G378) HSAN1A k: C133Y/W(C180); V144D(V191); S331F(G379); A352V(V399) | 2-15 | [101,102,103,104,105] |
| FTD-ALS2 | CHCHD10 (AAH65232.1) | MIX17 | 35.25 | S59L(S70) SMAJ-linked l: G66V(G77) IMMD-linked m: G58R(G69) | 2-16 | [106,107,108] |
| FTD-ALS5 | CCNF (AAH12349.1) | CLN1 CLN2 CLN3 | 18.14 19.66 22.75 | R392T(Q337) R392T(H136) S195R(A2); S621G(S423) | 2-17 | [108] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stump, A.L.; Rioux, D.J.; Albright, R.; Melki, G.L.; Prosser, D.C. Yeast Models of Amyotrophic Lateral Sclerosis Type 8 Mimic Phenotypes Seen in Mammalian Cells Expressing Mutant VAPBP56S. Biomolecules 2023, 13, 1147. https://doi.org/10.3390/biom13071147
Stump AL, Rioux DJ, Albright R, Melki GL, Prosser DC. Yeast Models of Amyotrophic Lateral Sclerosis Type 8 Mimic Phenotypes Seen in Mammalian Cells Expressing Mutant VAPBP56S. Biomolecules. 2023; 13(7):1147. https://doi.org/10.3390/biom13071147
Chicago/Turabian StyleStump, AnnaMari L., Daniel J. Rioux, Richard Albright, Guiliano L. Melki, and Derek C. Prosser. 2023. "Yeast Models of Amyotrophic Lateral Sclerosis Type 8 Mimic Phenotypes Seen in Mammalian Cells Expressing Mutant VAPBP56S" Biomolecules 13, no. 7: 1147. https://doi.org/10.3390/biom13071147
APA StyleStump, A. L., Rioux, D. J., Albright, R., Melki, G. L., & Prosser, D. C. (2023). Yeast Models of Amyotrophic Lateral Sclerosis Type 8 Mimic Phenotypes Seen in Mammalian Cells Expressing Mutant VAPBP56S. Biomolecules, 13(7), 1147. https://doi.org/10.3390/biom13071147






