Abstract
Z-DNA refers to the left-handed double-helix DNA that has attracted much attention because of its association with some specific biological functions. However, because of its low content and unstable conformation, Z-DNA is normally difficult to observe or identify. Up to now, there has been a lack of unified or standard analytical methods among diverse techniques for probing Z-DNA and its transformation conveniently. In this work, NaCl, MgCl2, and ethanol were utilized to induce d(GC)8 from B-DNA to Z-DNA in vitro, and Fourier transform infrared (FTIR) spectroscopy was employed to monitor the transformation of Z-DNA under different induction conditions. The structural changes during the transformation process were carefully examined, and the DNA chirality alterations were validated by the circular dichroism (CD) measurements. The Z-DNA characteristic signals in the 1450 cm−1–900 cm−1 region of the d(GC)8 infrared (IR) spectrum were observed, which include the peaks at 1320 cm−1, 1125 cm−1 and 925 cm−1, respectively. The intensity ratios of A1320/A970, A1125/A970, and A925/A970 increased with Z-DNA content in the transition process. Furthermore, compared with the CD spectra, the IR spectra showed higher sensitivity to Z-DNA, providing more information about the molecular structure change of DNA. Therefore, this study has established a more reliable FTIR analytical approach to assess BZ DNA conformational changes in solutions, which may help the understanding of the Z-DNA transition mechanism and promote the study of Z-DNA functions in biological systems.
1. Introduction
Deoxyribonucleic acid (DNA), as the hereditary material in biological systems, plays an important role in life. Structural changes of DNA molecules affect the expression of genes inside the nuclei of cells. Unlike Watson–Crick’s B-DNA, Z-DNA is a left-handed double-stranded helix with a zigzag shape that can be transformed from B-DNA [1]. It was found that GC bases are more easily induced to Z-DNA [2,3,4,5], and some base modifications such as methylation of cytosine and guanine and deamination of adenine may increase the stability of Z-DNA [6]. At present, a variety of biological functions of Z-DNA have been revealed. In general, it is involved in gene regulation [7,8]. For example, it may induce genetic instability and cause a variety of diseases [2,9,10,11]. Evidence has shown that Z-DNA appears in the hippocampus of patients with Alzheimer’s disease, and it is speculated that Z-DNA may be related to neurodegenerative diseases [12]. However, currently, there are very few reports on Z-DNA research, not only because of the lack of effective tools for Z-DNA detection but also due to its low content existing in nature.
In order to study the properties of Z-DNA, researchers have induced the formation of Z-DNA in vitro under special conditions such as high ionic strength, chemical modification, and the use of binding proteins and negative superhelices [13,14,15,16,17,18,19,20,21,22,23]. For the detection and analysis of Z-DNA, spectroscopic methods have been proposed and employed, such as X-ray diffraction, circular dichroism (CD), and nuclear magnetic resonance (NMR) spectroscopy [1,13,21,24,25,26,27,28,29]. However, these methods either require expensive instruments and complicated operation or cannot assess the B-Z DNA transition instantly and quantitatively.
Fourier transform infrared (FTIR) spectroscopy can probe the biochemical and structural information of analyte samples. When the composition and/or conformation of DNA change, the peak frequency and intensity of the infrared spectroscopy will change correspondingly. Currently, FTIR spectroscopy has already been applied in DNA structure analysis [30,31]. The application of FTIR spectroscopy in the study of DNA conformation has also gained encouraging attention [8,32,33,34,35,36]. However, previous research on Z-DNA through FTIR had some limitations, such as the requirement of low hydration to achieve the purpose of Z-DNA detection [30,37,38]. This hydration method may lead to changes in the liquid environment and so bring unpredictable effects on the formation of Z-DNA. On the other hand, it is still difficult to conduct a quantitative analysis of Z-DNA because of insufficient identification of the Z-DNA characteristic IR bands [39,40].
In order to establish a more reliable approach for Z-DNA assessment, we tried to induce the transformation of B-DNA into Z-DNA in solutions by a variety of standard inducers (NaCl, MgCl2, and ethanol) without reduction of relative humidity, and then applied FTIR spectroscopy to detect and analyze the gradual change process of the B-Z DNA transition. It is known that NaCl, MgCl2, and ethanol are Z-DNA inducers with good repeatability [20,21,41,42,43]. Especially DNA with multiple GC has a strong tendency to form Z-DNA under these conditions [2,3,4]. Therefore, in the experiment, different concentrations of NaCl, MgCl2, and ethanol solutions were used to induce the dsDNA of d(GC)8 from B-DNA to Z-DNA. The Z-DNA transformation under different conditions was thus observed and evaluated; in particular, the IR band at 970 cm−1 was used as the internal standard for the potential quantitative analysis, and based on the evaluation of the intensity ratios (A1320/A970, A1125/A970, and A925/A970), the B-Z DNA transition process under some specific conditions was inspected and assessed.
2. Materials and Methods
2.1. Materials
The d(GC)8 sequence used in the experiment was purchased from General Biology (Hefei, China). The concentration was 5 OD/tube (the OD value was measured at 260 nm in the ultraviolet absorption spectrum) and the MW was 4885.16 g/mole, equivalent to 181.6 μg/tube. Tm (the temperature at which 50% nucleic acids become denatured [44,45]) was 63.9 °C. The solution was dissolved in buffer (50 mM NaCl, 5 mM Tris-HCl, pH = 8.0), and the A260/A280 = 1.8–2.2 was determined by UV spectrophotometer, confirming that the DNA had high purity. Ultra-pure Milli-Q water was used in all the experiments. The NaCl (analytically pure) used in the experiment was purchased from Tianjin Hengxing Chemical Reagent Manufacturing Co., Ltd. (Tianjin, China), and the MgCl2 (analytically pure) was purchased from Tianjin Zhiyuan Chemical Reagent Co., Ltd. (Tianjin, China). Absolute ethanol was purchased from Beijing Huateng Chemical Co., Ltd. (Beijing, China), and the Tris (analytically pure) was from Solarbio (Beijing, China).
2.2. Induction of Z-DNA
Firstly, the purchased ssDNA was combined into dsDNA using the base complementary pairing principle. The dry nucleic acid powder was first dissolved with buffer solution (50 mM NaCl, 5 mM Tris-HCl, pH = 8.0) and then incubated at 37 °C for 30 min to obtain stable dsDNA [41], used for CD and UV detection. NaCl, MgCl2, and ethanol were prepared in buffer solutions (50 mM NaCl, 5 mM Tris-HCl, pH = 8.0). The prepared inducer solution was added to the dsDNA solution. The final concentrations of NaCl were 1 M, 2 M, 3 M, 4 M, 4.5 M, and 5 M, respectively; the final concentrations of MgCl2 were 0.5 M, 1 M, 1.5 M, 2 M, 2.5 M, and 3 M, respectively; and the volume ratios of ethanol were 0.4, 0.5, 0.55, 0.6, 0.65, and 0.7, respectively. After adding the Z-DNA inducer solution, the DNA samples were incubated at 37 °C for more than 4 h to stabilize the structure of the dsDNA.
2.3. FTIR Spectral Analysis
Before the FTIR spectral measurements, the dry nucleic acid powder was dissolved with ultra-pure Milli-Q water and incubated at 37 °C for 30 min to obtain stable dsDNA. Then the dsDNA was added to a CaF2 window piece drop by drop and put into a drying oven keeping the temperature at 37 °C for about 20 min until the water of the DNA solution evaporated on the window piece. Then, 5 μL of different inducing solutions were added on the film formed by the dsDNA, the film was quickly covered with another clean CaF2 window piece, and the connection of the window piece was sealed with sealing film to reduce the evaporation of water. The final concentrations of NaCl were 50 mM, 1 M, 2 M, 3 M, 4 M, 4.5 M, and 5 M, respectively; the final concentrations of MgCl2 were 0 M, 0.5 M, 1 M, 1.5 M, 2 M, 2.5 M, and 3 M, respectively; and the volume ratios of ethanol were 0, 0.4, 0.5, 0.55, 0.6, 0.65, and 0.7, respectively. The concentration of the DNA solution was 36.32 μg/μL. The structural changes of the dsDNA were detected by a Thermo Fisher Scientific Nicolet iS10 infrared spectrometer.
The parameters of the infrared spectrometer were set as follows: the spectral range was from 4000 cm−1 to 900 cm−1, the spectral resolution was 4 cm−1, and the number of scans was 30. The software Origin 2021 was used for the spectral analysis. At least three replicates were set for each measurement.
2.4. Circular Dichroism Analysis
Circular dichroism was conducted using a ChirascanTM Circular dichroism spectrometer. The spectral scanning range was 200 nm to 350 nm, with the scanning time being 0.25 s per point. The solution sample was put into a quartz colorimetric tube with a diameter of 1 mm. Each CD spectrum was scanned three times on average. Ultra-pure water was used as a blank control.
3. Results
Figure 1 shows the FTIR spectra of d(GC)8 dsDNA, with the spectral assignments given in Table 1. In this work, we mainly focused on the 1450 cm−1−900 cm−1 region of DNA infrared spectra. In addition, the peak at 970 cm−1 is attributed to the deoxyribose C-C stretching mode, and it is often used as an internal reference for normalization to analyze the structural changes of dsDNA [34,46].
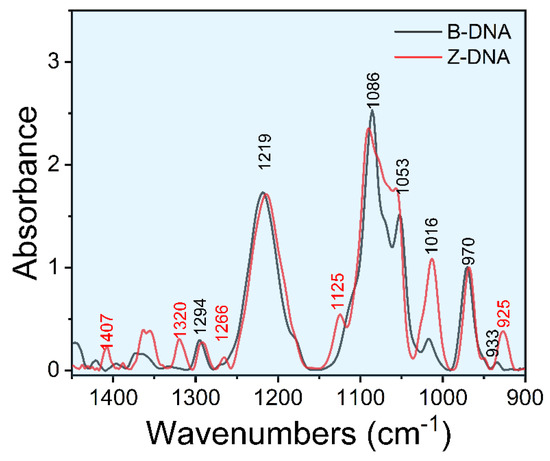
Figure 1.
Fourier transform infrared spectrum of d(GC)8 dsDNA.

Table 1.
Assignment of the relevant vibrational bands used in the second derivative.
3.1. NaCl-Induced Z-DNA
Firstly, the structural changes of dsDNA induced by different concentrations of NaCl were analyzed (Figure 2). The characteristic peaks at ~1125 cm−1 and ~925 cm−1 changed obviously with the increase of NaCl concentration, indicating the transition from B-DNA to Z-DNA. It was found that when the concentration of NaCl reached 3 M, the original 1068 cm−1 of FTIR spectrum disappeared, accompanied by the appearance of 1077 cm−1 and 1066 cm−1 (Figure 2a,b), and the CD spectrum at this concentration showed a trend of Z-DNA transition (Figure 2e). These results indicated that when NaCl was over 3 M, the transformation from B-DNA to Z-DNA occurred.
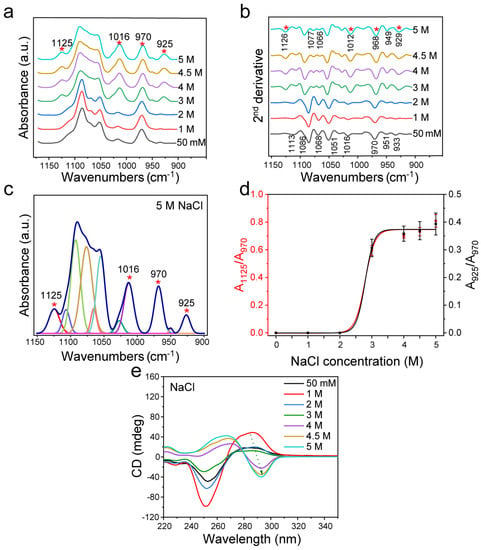
Figure 2.
FTIR spectroscopy analysis of d(GC)8 treated with different concentrations of NaCl. (a) d(GC)8 infrared spectra treated with different concentrations of NaCl. (b) Second derivative spectra of d(GC)8. (c) The curve fitting result under 5 M NaCl (1150 cm−1−900 cm−1). (d) The ratios of the area of Z-DNA characteristic peaks 925 cm−1 and 1125 cm−1 to the area of internal reference 970 cm−1 peak (the value is from the fitting result of the characteristic peak, see the Supplementary Materials). (e) CD was used to detect d(GC)8 chirality after treatment with different concentrations of NaCl.
According to the peak positions indicated by the second derivative spectrum as shown in Figure 2b, the curve fitting of the infrared spectrum from 1150 cm−1 to 900 cm−1 was achieved (Figure 2c and Figure S1 and Table S1). Then, the intensity ratios of the Z-DNA characteristic peak area to the internal reference area (A925/A970 and A1125/A970) were used to evaluate the variation trend of Z-DNA content. The results showed that the relative Z-DNA content increased with the increase of NaCl concentration (Figure 2d), and the changes evaluated based on either A925/A970 or A1125/A970 were the same. It was found that substantial structural transformation of DNA occurred when the NaCl concentration was larger than 3 M. When the concentration reached 4 M, the ratios of A925/A970 and A1125/A970 reached a plateau, indicating that most B-DNA had been transformed into Z-DNA. In addition, I925/I970 and I1125/I970 changed with the same trend as A925/A970 and A1125/A970, both of which changed at 3 M significantly, and reached a plateau above 4 M (Figure S2).
The CD spectrum in the range of 220–350 nm reflects the interaction of base pairs in the helical structure [57]. In fact, Z-DNA has an obvious structural difference from B-DNA as shown by the CD measurement (Figure S3). According to the experimental results of d(GC)8, the CD of Z-DNA showed a positive value near 270 nm and a negative value near 295 nm, while the CD of B-DNA showed a negative value near 255 nm and a positive value near 285 nm [58]. BZ-DNA is the result of the coexistence of Z-DNA and B-DNA, which showed a negative band at 295 nm for Z-DNA and a negative value at 255 nm for B-DNA, and a positive peak between B-DNA and Z-DNA [59].
As shown by the CD spectra in Figure 2e, NaCl could induce the dsDNA transformation, which is consistent with previous studies [21,24,41,42,60]. B-DNA was obviously transformed to Z-DNA after treatment with NaCl with a concentration greater than 4 M, and its structural change was consistent with the concentration change trend of NaCl. Although the structure of dsDNA did not change significantly when the NaCl concentration was below 3 M, the peak intensity of the CD spectrum changed, indicating that the dsDNA conformation was varied.
In addition, as Pohl and Jovin found that the conversion of poly(dG-dC)·poly(dG-dC) from one conformation to another was associated with an increase in the A295/A260 ratio of the UV absorption spectrum, other research groups also applied this method to evaluate conversion [21,61,62,63]. Therefore, we used this for a double check. As shown in Figure S4, when NaCl reached 3 M, the UV absorption ratio A295/A260 gradually increased with the increase of NaCl concentration, further confirming that the Z-DNA structure was gradually transformed.
3.2. MgCl2-Induced Z-DNA
Furthermore, we applied the cation Mg2+ to treat d(GC)8 and analyzed the transformation law of Z-DNA. It was found that the effect of MgCl2 on dsDNA was similar to that caused by NaCl.
The analysis in Figure 3a,b shows that in the transformation from B-DNA to Z-DNA, the characteristic peaks at ~1125 cm−1 and ~925 cm−1 appeared, which are attributed to Z-DNA. When the concentration of MgCl2 reached 1.5 M, the peak at 1068 cm−1 disappeared, and the peaks at 1077 cm−1 and 1066 cm−1 appeared (Figure 3b), similar to the spectrum when the NaCl concentration reached 3 M. When MgCl2 increased to 2 M, the induction effect on dsDNA was further enhanced, and the FTIR spectrum showed new changes. Through curve fitting of the infrared spectrum from 1150 cm−1 to 900 cm−1 (Figure 3c and Figure S5 with the fitting parameters given in Table S2), we found that the ratio of the characteristic peak area to the internal reference area was closely related to the concentration of MgCl2 (Figure 3d), indicating the structural transformation of dsDNA. Combined with the CD spectrum and FTIR second derivative analysis, it was found that when the concentration of MgCl2 exceeded 3 M, the conformation of DNA gradually shifted into a new conformation. In addition to the area ratio of the characteristic peaks, we also analyzed the intensity ratios of the characteristic peaks (Figure S6), and found that the variation trend was the same as that of the area ratio.
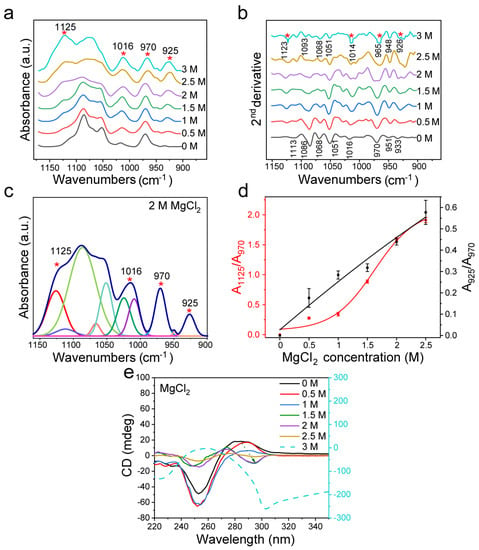
Figure 3.
FTIR detection of d(GC)8 after treatment with different concentrations of MgCl2. (a) Infrared spectra of d(GC)8 treated with different concentrations of MgCl2. (b) Second derivative of the infrared spectrum of d(GC)8 treated with different concentrations of MgCl2. (c) The curve fitting result under 2 M MgCl2 treatment (1150 cm−1−900 cm−1). (d) The ratios of the area of the Z-DNA characteristic peaks 925 cm−1 and 1125 cm−1 to the area of the internal reference 970 cm−1 peak (the value is derived from the fitting results of characteristic peaks, see Supplementary Materials). (e) CD was used to confirm the transformation effect of d(GC)8 treated with different concentrations of MgCl2.
As shown by the CD spectra in Figure 3e, MgCl2 could induce dsDNA transformation, which is consistent with previous studies [20,41,60]. Furthermore, the CD spectral analysis confirmed that the dsDNA structure was gradually transformed into Z-DNA when the concentration of MgCl2 reached more than 1.5 M (Figure 3e). Compared with NaCl, MgCl2 has more electric charge and has a stronger effect on Z-DNA. When the concentration of MgCl2 increased to 3 M, the CD spectrum of dsDNA was quite different from that of Z-DNA, indicating that the structure of DNA had changed again, which was consistent with previous reports [20,64]. Figure S7 shows the UV absorption spectra and A295/A260 ratio after d(GC)8 induction. It shows that A295/A260 increased with the increase of MgCl2 concentration, confirming that an increase of MgCl2 would affect the structure of dsDNA.
3.3. Ethanol-Induced Z-DNA
In addition to the cationic inducers mentioned above, we also investigated Z-DNA induced by ethanol. DNA is insoluble in ethanol and soluble in an aqueous solution. It has been reported that the addition of ethanol could induce the formation of Z-DNA with a low hydration degree by reducing the hydration degree of nucleic acid [65]. In this case, we observed that the induction effect of ethanol on dsDNA conformation was different from that of NaCl and MgCl2. As shown in Figure 4a,b, the peaks at 1125 cm−1 and 925 cm−1 appeared when B-DNA changed to Z-DNA, and then the peaks for dsDNA furanose and phosphate groups at 1100 cm−1−1000 cm−1 changed. This is consistent with the change in MgCl2 treatment. However, when the volume ratio of ethanol reached 0.6, the peak at 1068 cm−1 disappeared in the second derivative spectrum, and the peaks at 1077 cm−1 and 1066 cm−1 appeared. When the volume ratio of ethanol ranged from 0.65 to 0.7, the FTIR spectrum of dsDNA changed obviously, and the peak at 1049 cm−1 disappeared, while the peaks at 1042 cm−1 and 1054 cm−1 appeared (Figure 4b). This change is different from the structural change in the case of MgCl2 treatment. To be noted, the structural changes of dsDNA induced by excessive ethanol were weaker than those induced by excessive MgCl2.
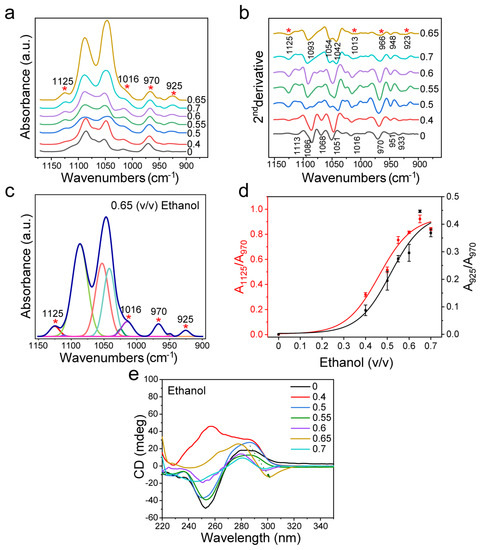
Figure 4.
FTIR spectroscopy analysis of d(GC)8 treated with different volume ratios of ethanol solutions. (a) d(GC)8 infrared spectrum after ethanol treatment at different volume ratios. (b) The second derivative of IR spectrum of d(GC)8 treated with different volume ratios of ethanol. (c) The curve fitting result at ethanol volume ratio 0.65 (1150 cm−1−900 cm−1). (d) The ratio of the area of Z-DNA characteristic peaks 925 cm−1 and 1125 cm−1 to the area of internal reference 970 cm−1 peak (the value is derived from the fitting results of characteristic peaks, see Supplementary Materials). (e) The detection result of d(GC)8 CD after ethanol treatment at different volume ratios.
The relative change of Z-DNA was calculated by curve fitting in the range of 1150 cm−1–900 cm−1 of the infrared spectrum (Figure 4c and Figure S8 with the fitting parameters given in Table S3). It can be seen that the Z-DNA level is positively correlated with ethanol content (Figure 4d). When the volume ratio of ethanol reached 0.6, the ratio of Z-DNA increased. This implies that most B-DNA was converted to Z-DNA at the ethanol volume ratio of 0.6. In addition to the area ratio analysis, we also analyzed the intensity ratio between the characteristic peak and internal reference (Figure S9), and found that its variation trend was the same as that of the area ratio.
As shown by the CD spectra in Figure 4e, ethanol could induce dsDNA transformation, which is consistent with previous studies [42,43]. Furthermore, CD measurements confirmed that the dsDNA structure did change with the increase of ethanol content, and this change was correlated with the ethanol volume ratio (Figure 4e). For the unusual result with 0.4 proportion of ethanol, it was found that the characteristic peaks of B-DNA (i.e., 285 nm peaks) still remained, while the peak intensity at 255 nm was different from that of conventional B-DNA. Based on the literature [66], the special curve for the 0.4 proportion of ethanol might be due to the synergistic effect of ethanol and NaCl in the buffer solution. The ethanol promoted an interaction between NaCl and the anion in nucleic acid, resulting in the CD spectra of Z-DNA at 255 nm different from that of B-DNA. Johan H. van de Sande et al. reported the cationic promotion effect of ethanol and proved that, with an increase of ethanol volume and/or decrease of NaCl concentration, the induction effect of ethanol would be dominant [20]. It can also be seen from the CD spectrum that ethanol showed the strongest effect on dsDNA when the volume ratio of ethanol was 0.65, which is consistent with the changing trend based on the FTIR spectral analysis. Figure S10 shows the changing trend of A295/A260 according to UV absorption statistics. The result shows that when the volume ratio of ethanol reaches 0.6, A295/A260 increases significantly, which further indicates that the ethanol at this content can cause structural changes in dsDNA.
3.4. Sugar Pucker Changes after Treatment with Different Inducers
In addition to the sugar–phosphate backbone structure, the sugar loop folding form is also one of the major differences between B-DNA and Z-DNA. For d(GC)8 in Z-DNA, the G base is in cis conformation with C3′-endo, while the C base is in inverse conformation with C2′-endo [1,67]. Among them, C3′-endo is the main structural difference between Z-DNA and B-DNA. The characteristic signals of C3′-endo in the FTIR spectrum include the peaks at 1411 cm−1, 1355 cm−1, and 1320 cm−1, which are mainly concentrated in 1450 cm−1−1150 cm−1 (Figure 5a−c).
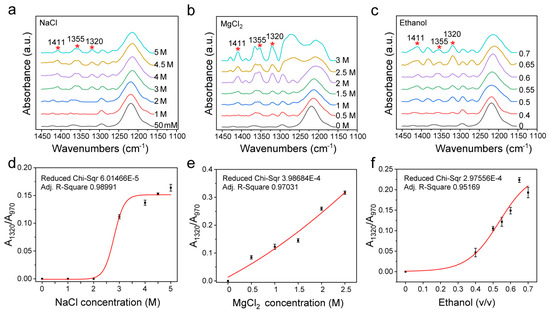
Figure 5.
Changes of infrared spectra of 1450 cm−1−1150 cm−1 and area ratio of C3′-endo to internal reference under different inducers. (a) The infrared spectrum under different concentrations of NaCl. (b) The infrared spectrum under different concentrations of MgCl2. (c) The infrared spectrum of d(GC)8 treated with different volume ratios of ethanol solution. (d) Changes of A1320/A970 ratio under different concentrations of NaCl. (e) Changes of A1320/A970 ratio under different concentrations of MgCl2. (f) Changes of A1320/A970 ratio under different volume ratios of ethanol.
Figure 5b–d show that C3′-endo is closely related to Z-DNA as its content increased with the increase of the content of the Z-DNA inducer, and its change trend is the same as that of A1125/A970 and A925/A970, indicating that Z-DNA and B-DNA can also be distinguished by analyzing the infrared spectral features that are related to sugar loop folding.
3.5. Z-DNA Transition in GC-Rich DNA
B-Z transition may occur in pure GC-DNA and GC-rich DNA. In reality, GC-rich DNA is more prevalent than pure GC-DNA in a genome, so we also further examined GC-rich DNA with the involvement of other bases in the DNA sequence [5]. For this purpose, we also designed d(ATATGCGCGCGCGCGCGCGCATAT), where the actual Z-DNA proportion is less than 66.7% because of the presence of a BZ junction [67,68]. As expected, FTIR spectroscopy could sensitively detect the Z-DNA transformation, and the results of this experiment just verified the B-Z transformation trend. It can be seen in Figure 6 that the characteristic spectral feature of Z-DNA remained, which essentially resembles that of the pure GC-DNA, having the Z-DNA bands such as 925 cm−1, 1016 cm−1, 1125 cm−1, 1320 cm−1, etc. It can be found that the GC proportion is directly associated with the intensities of the characteristic bands of Z-DNA. In addition, we also inspected the IR spectrum of the negative control d(AT)8 sequence (Figure S13 in Supplementary Materials), which showed no obvious characteristic peaks of Z-DNA in all the three induction solutions, confirming the usefulness of FTIR spectroscopy in the identification of Z-DNA conformation and the analysis of B-Z DNA transitions in solutions.
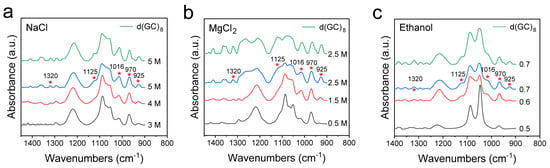
Figure 6.
Comparison of Z-DNA infrared spectra between pure GC-DNA d(GC)8 and GC-rich DNA induced by different conditions. (a) FTIR spectra of d(ATATGCGCGCGCGCGCGCGCATAT) dsDNA treated with different concentrations of NaCl, and FTIR spectra of d(GC)8 treated with 5 M NaCl; (b) FTIR spectra of d(ATATGCGCGCGCGCGCGCGCATAT) dsDNA treated with different concentrations of MgCl2 and FTIR spectra of d(GC)8 treated with 2.5 M MgCl2; (c) FTIR spectra of d(ATATGCGCGCGCGCGCGCGCATAT) dsDNA treated with different volume ratios of ethanol, and FTIR spectra of d(GC)8 treated with 0.7 volume ratio of ethanol.
4. Discussion
First of all, to establish a reliable quantitative analytical method via FTIR spectroscopy, sufficiently stable Z-DNA should be available. In the literature, for the induction of stable Z-DNA, the following interactions or mechanisms may be involved: (1) electrostatic interactions between cations and DNA phosphate residues, such as NaCl, MgCl2, spermine-functionalized C-dots, etc. [21,41,69,70]; (2) reducing the degree of hydration which may promote the formation of Z-DNA conformations with a low degree of hydration, such as non-oriented gel, ethanol, etc. [37,65]; (3) protein Zα domain binding to DNA, such as ADAR1, E3L, etc. [13,71,72]; and (4) construction of negative superhelix structures, such as circular Z-DNA [23,73]. Accordingly, in this work, we applied NaCl, MgCl2, and ethanol to induce Z-DNA, which actually are the typical Z-DNA induction methods in the literature [20,41,42,43].
Our results showed that, compared with CD methods, FTIR spectroscopy could not only detect the structural changes of dsDNA but also provides more precise information on the DNA structure, because it can sensitively reflect the change process of B-Z transition, or even transition to other forms of DNA. For example, in Figure 2, multiple characteristic peaks of Z-DNA were clearly detected by FTIR spectroscopy in the 3 M NaCl solution. However, Z-DNA could not readily be identified from the CD spectrum when the NaCl concentration was below 3 M. In addition, Figure 3 shows that the characteristic peak of Z-DNA could be detected with the 0.5 M MgCl2 treatment, while the CD spectrum at the same concentration could only simply show some difference at the peak value of 285 nm, which was not enough to distinguish the Z-DNA content. This means that FTIR is more sensitive than CD in the identification of Z-DNA. The same is true for the analysis of ethanol, a noncationic inducer. As shown in Figure 4, the characteristic peak of Z-DNA appeared when the ethanol volume ratio was 0.4, while the CD spectrum showed the structural features of Z-DNA only when the ethanol volume ratio was 0.6. When the volume ratio of ethanol increased to 0.65 and 0.7, the FTIR spectra of dsDNA showed many new changes, while the CD spectra showed fewer changes.
By analyzing the second derivative FTIR spectra of B-DNA, it can be recognized clearly that dsDNA of d(GC)8 sequence has a characteristic peak of 1068 cm−1 (Figure 2b). Existing studies refer to a DNA structure as B*-DNA, which is characteristic of GC sequence nucleic acids [30,31,37]. When the concentration of NaCl reached 3 M, the concentration of MgCl2 reached 1.5 M, and the volume ratio of ethanol reached 0.6, this 1068 cm−1 peak disappeared. With the disappearance of the 1068 cm−1 peak, two new peaks of 1077 cm−1 and 1066 cm−1 appeared (Figure 2b, Figure 3b and Figure 4b). This result is consistent with the study of Z-DNA with the infrared spectrum by Christine Rauch et al. [38].
In addition to the characteristic peaks of Z-DNA, such as 1320 cm−1, 1125 cm−1, and 925 cm−1, the intensity of the 1016 cm−1 furanose peak also increased with the increase of inducer content. Different from Z-DNA characteristic peaks, furanose also exists in B-DNA, but is significantly enhanced in Z-DNA [47,48,49]. In this study, the area ratio (Figure S11) and intensity ratio (Figure S12) of the 1016 cm−1 peak to the 970 cm−1 peak in the process of Z-DNA transition showed a consistent variation trend with that of Z-DNA content, suggesting that the 1016 cm−1 peak could also be used as a marker for Z-DNA transition.
Both NaCl and MgCl2 may induce dsDNA to transform into Z-DNA through electrostatic interaction. Due to the large dielectric constant of water, the combination of cation and phosphate residues will be hindered [66]. Therefore, a high salt concentration is often required for the induction of Z-DNA. Na+ and Mg2+ carry different amounts of charge, resulting in different ion concentrations. In addition, when the concentration of MgCl2 is too high, the electrostatic interaction between MgCl2 and nucleic acid exceeds the requirement of Z-DNA structure transformation, which would then cause the DNA structure to change further.
Z-DNA is a conformation with a low degree of hydration. Ethanol solution can increase the degree of environmental hydration to induce the formation of Z-DNA. When the proportion of ethanol in the solution is too high, it will also go beyond the hydration range of Z-DNA. Compared with other inducers, ethanol induced Z-DNA showed significant difference in the FTIR range of 1100 cm−1−1000 cm−1, indicating that the induction mechanism of ethanol is different from that of the cation. In addition, ethanol as a polar organic solvent has a much lower permittivity than water, thus allowing the neutralization of phosphate residues by cations in solution [74]. Johan H. van de Sande et al. also demonstrated the induction of Z-DNA using ethanol with a volume ratio of 0.2 and 1 mM MgCl2 [20].
No matter what kinds of inducers were used in this study, all the above results showed that the use of FTIR spectroscopy could identify Z-DNA readily, monitor the B-Z DNA transition quickly, and evaluate the Z-DNA content quantitatively. Based on the results of this work, we will further continue to study the DNA B-Z transition under more complicated cases such as under the physiological conditions where Z-DNA may be induced with proteins such as ADAR1.
5. Conclusions
This work has established a convenient analytical assessment method based on FTIR spectroscopy for the evaluation of the B-Z transformation of DNA under different induction circumstances. In the experiment, dsDNA (d(GC)8) was treated with different concentrations of NaCl, MgCl2, and ethanol, with different volume ratios, and the Z-DNA transition processes under different conditions were observed and compared using FTIR spectroscopy. It was found that with the increase of NaCl, MgCl2, or ethanol concentration in the induction solution, the characteristic Z-DNA peaks of 1320 cm−1, 1125 cm−1, and 925 cm−1 in the infrared spectra changed during the B-Z DNA transition, and the FTIR intensity ratios of A1320/A970, A1125/A970, and A925/A970 could be used to assess the transition quantitatively. Due to the comparative analysis, we found that MgCl2 had the strongest effect, while NaCl had the weakest effect on the B-Z DNA transition. Compared with other detection approaches such as CD spectroscopy, FTIR spectroscopy showed the advantages of providing not only more information on multiple structural changes of DNA, but also a more sensitive detection of Z-DNA variation. As such, this study has systematically explored the changes of FTIR spectra in the process of Z-DNA transformation, which may provide a more reliable reference for the study of Z-DNA in biological systems in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biom13060964/s1, Figure S1. Fitting results of representative infrared spectra of dsDNA induced by different concentrations of NaCl in the range of 1150 cm−1−900 cm−1; Figure S2. Changes of the ratio of Z-DNA characteristic peaks (925 cm−1 and 1125 cm−1) intensity to 970 cm−1 peak intensity with NaCl treatment; Figure S3. CD spectra for identification of d(GC)8 chiral structures; Figure S4. UV analysis of d(GC)8 dsDNA treated with different concentrations of NaCl; Figure S5. Fitting results of representative infrared spectra of dsDNA induced by different concentrations of MgCl2 in the range of 1150 cm−1−900 cm−1; Figure S6. Changes of the ratio of the Z-DNA characteristic peaks (925 cm−1 and 1125 cm−1) intensity to 970 cm−1 peak intensity after MgCl2 treatment; Figure S7. UV analysis of d(GC)8 dsDNA treated with different concentrations of MgCl2; Figure S8. Fitting results of representative infrared spectra of dsDNA induced by ethanol of different volume ratios in the range of 1150 cm−1−900 cm−1; Figure S9. Changes of the ratio of the Z-DNA characteristic peaks (925 cm−1 and 1125 cm−1) intensity to 970 cm−1 peak intensity after ethanol treatment; Figure S10. UV analysis of d(GC)8 dsDNA after ethanol treatment at different volume ratios; Figure S11. The ratio changes of 1016 cm−1 peak area to 970 cm−1 peak area with the concentration of inducers (data were supplied in the spectral fitting); Figure S12. The ratio changes of 1016 cm−1 peak intensity to 970 cm−1 peak intensity with the concentration of inducers (data were supplied in the spectral fitting); Figure S13. d(AT)8 infrared spectra under different induction conditions; Table S1. Fitting parameters of infrared spectra of dsDNA treated by NaCl; Table S2. Fitting parameters of infrared spectra of dsDNA treated by MgCl2; Table S3. Fitting parameters of infrared spectra of dsDNA treated by ethanol.
Author Contributions
Q.H. and F.Z. conceived the research and designed the experiments. M.D. performed the experiments and collected the data, partly assisted by Y.L. All the authors participated in the research. Q.H., F.Z. and M.D. discussed the results and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported partly by the National Nature Science Foundation of China (Grant No. 11635013), the College Key Research Project in Henan province (22A180030), and the Research Integration Program of Hefei Institutes of Physical Science, Chinese Academy of Sciences.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in this article and its Supplementary Materials.
Acknowledgments
The authors thank Guangchao Zheng of Zhengzhou University for the support in CD analysis, and thank Henan Key Laboratory of Diamond Optoelectronic Materials and Devices, Key Laboratory of Material Physics, Ministry of Education for the support in FTIR analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, A.H.; Quigley, G.J.; Kolpak, F.J.; Crawford, J.L.; van Boom, J.H.; van der Marel, G.; Rich, A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 1979, 282, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, S.; Subramani, V.K.; Kim, K.K. Z-DNA in the genome: From structure to disease. Biophys. Rev. 2019, 11, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Bothe, J.R.; Lowenhaupt, K.; Al-Hashimi, H.M. Incorporation of CC steps into Z-DNA: Interplay between B-Z junction and Z-DNA helical formation. Biochemistry 2012, 51, 6871–6879. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Hur, J.; Han, J.H.; Ha, S.C.; Shin, D.; Lee, S.; Park, S.; Sugiyama, H.; Kim, K.K. Sequence preference and structural heterogeneity of BZ junctions. Nucleic Acids Res. 2018, 46, 10504–10513. [Google Scholar] [CrossRef]
- Doluca, O.; Withers, J.M.; Filichev, V.V. Molecular Engineering of Guanine-Rich Sequences: Z-DNA, DNA Triplexes, and G-Quadruplexes. Chem. Rev. 2013, 113, 3044–3083. [Google Scholar] [CrossRef]
- Wang, A.H.; Hakoshima, T.; van der Marel, G.; van Boom, J.H.; Rich, A. AT base pairs are less stable than GC base pairs in Z-DNA: The crystal structure of d(m5CGTAm5CG). Cell 1984, 37, 321–331. [Google Scholar] [CrossRef]
- Ditlevson, J.V.; Tornaletti, S.; Belotserkovskii, B.P.; Teijeiro, V.; Wang, G.; Vasquez, K.M.; Hanawalt, P.C. Inhibitory effect of a short Z-DNA forming sequence on transcription elongation by T7 RNA polymerase. Nucleic Acids Res. 2008, 36, 3163–3170. [Google Scholar] [CrossRef]
- Li, Y.; Huang, Q.; Yao, G.; Wang, X.; Zhang, F.; Wang, T.; Shao, C.; Zheng, X.; Jing, X.; Zhou, H. Remodeling Chromatin Induces Z-DNA Conformation Detected through Fourier Transform Infrared Spectroscopy. Anal. Chem. 2020, 92, 14452–14458. [Google Scholar] [CrossRef]
- Geng, J.; Zhao, C.; Ren, J.; Qu, X. Alzheimer’s disease amyloid beta converting left-handed Z-DNA back to right-handed B-form. Chem. Commun. 2010, 46, 7187–7189. [Google Scholar] [CrossRef]
- Vasudevaraju, P.; Guerrero, E.; Hegde, M.L.; Collen, T.B.; Britton, G.B.; Rao, K.S. New evidence on alpha-synuclein and Tau binding to conformation and sequence specific GC* rich DNA: Relevance to neurological disorders. J. Pharm. Bioallied Sci. 2012, 4, 112–117. [Google Scholar] [CrossRef]
- Ng, S.K.; Weissbach, R.; Ronson, G.E.; Scadden, A.D. Proteins that contain a functional Z-DNA-binding domain localize to cytoplasmic stress granules. Nucleic Acids Res. 2013, 41, 9786–9799. [Google Scholar] [CrossRef]
- Vasudevaraju, P.; Bharathi; Garruto, R.M.B.; Sambamurti, K.; Rao, K.S.J. Role of DNA dynamics in Alzheimer’s disease. Brain Res. Rev. 2008, 58, 136–148. [Google Scholar] [CrossRef]
- Vongsutilers, V.; Sawaspaiboontawee, K.; Tuesuwan, B.; Shinohara, Y.; Kawai, G. 5-Methylcytosine containing CG decamer as Z-DNA embedded sequence for a potential Z-DNA binding protein probe. Nucleosides Nucleotides Nucleic Acids 2018, 37, 485–497. [Google Scholar] [CrossRef]
- Ferreira, J.M.; Sheardy, R.D. Enthalpy of the B-to-Z conformational transition of a DNA oligonucleotide determined by isothermal titration calorimetry. Biophys. J. 2006, 91, 3383–3389. [Google Scholar] [CrossRef]
- Edgington, S.M.; Stollar, B.D. Immunogenicity of Z-DNA depends on the size of polynucleotide presented in complexes with methylated BSA. Mol. Immunol. 1992, 29, 609–617. [Google Scholar] [CrossRef]
- Chen, H.H.; Charney, E.; Rau, D.C. Length changes in solution accompanying the B-Z transition of poly (dG-m5dC) induced by Co(NH3)63+. Nucleic Acids Res. 1982, 10, 3561–3571. [Google Scholar] [CrossRef]
- Walker, G.T.; Stone, M.P.; Krugh, T.R. Ethidium binding to left-handed (Z) DNAs results in regions of right-handed DNA at the intercalation site. Biochemistry 1985, 24, 7462–7471. [Google Scholar] [CrossRef]
- Thomas, T.J.; Thomas, T. Direct evidence for the presence of left-handed conformation in a supramolecular assembly of polynucleotides. Nucleic Acids Res. 1989, 17, 3795–3810. [Google Scholar] [CrossRef]
- Chaires, J.B.; Norcum, M.T. Structure and stability of Z* DNA. J. Biomol. Struct. Dyn. 1988, 5, 1187–1207. [Google Scholar] [CrossRef]
- van de Sande, J.H.; Jovin, T.M. Z* DNA, the left-handed helical form of poly in MgCl2-ethanol, is biologically active. EMBO J. 1982, 1, 115–120. [Google Scholar] [CrossRef]
- Pohl, F.M.; Jovin, T.M. Salt-induced co-operative conformational change of a synthetic DNA: Equilibrium and kinetic studies with poly (dG-dC). J. Mol. Biol. 1972, 67, 375–396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cui, Y.; An, R.; Liang, X.; Li, Q.; Wang, H.; Wang, H.; Fan, Y.; Dong, P.; Li, J.; et al. Topologically constrained formation of stable Z-DNA from normal sequence under physiological conditions. J. Am. Chem. Soc. 2019, 141, 7758–7764. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, Y.; Ma, W.; Chen, H.; Liu, M.; An, R.; Cheng, B.; Liang, X. Nonalternating purine pyrimidine sequences can form stable left-handed DNA duplex by strong topological constraint. Nucleic Acids Res. 2022, 50, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Lafer, E.M.; Moller, A.; Nordheim, A.; Stollar, B.D.; Rich, A. Antibodies specific for left-handed Z-DNA. Proc. Natl. Acad. Sci. USA 1981, 78, 3546–3550. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yin, C.; Fedorov, A.; Qiao, L.; Bao, H.; Beknazarov, N.; Wang, S.; Gautam, A.; Williams, R.M.; Crawford, J.C.; et al. ADAR1 masks the cancer immunotherapeutic promise of ZBP1-driven necroptosis. Nature 2022, 606, 594–602. [Google Scholar] [CrossRef] [PubMed]
- D’Urso, A.; Choi, J.K.; Shabbir-Hussain, M.; Ngwa, F.N.; Lambousis, M.I.; Purrello, R.; Balaz, M. Recognition of left-handed Z-DNA of short unmodified oligonucleotides under physiological ionic strength conditions. Biochem. Biophys. Res. Commun. 2010, 397, 329–332. [Google Scholar] [CrossRef]
- Feigon, J.; Wang, A.H.; van der Marel, G.A.; Van Boom, J.H.; Rich, A. A one- and two-dimensional NMR study of the B to Z transition of (m5dC-dG)3 in methanolic solution. Nucleic Acids Res. 1984, 12, 1243–1263. [Google Scholar] [CrossRef]
- Patel, D.J.; Kozlowski, S.A.; Nordheim, A.; Rich, A. Right-handed and left-handed DNA: Studies of B- and Z-DNA by using proton nuclear overhauser effect and P NMR. Proc. Natl. Acad. Sci. USA 1982, 79, 1413–1417. [Google Scholar] [CrossRef]
- Jin, H.S.; Kim, N.H.; Choi, S.R.; Oh, K.I.; Lee, J.H. Protein-induced B-Z transition of DNA duplex containing a 2′-OMe guanosine. Biochem. Biophys. Res. Commun. 2020, 533, 417–423. [Google Scholar] [CrossRef]
- Loprete, D.M.; Hartman, K.A. Structures of poly(dG-dC) and poly(dA-dT) stabilized by anions. J. Biomol. Struct. Dyn. 1995, 13, 57–67. [Google Scholar] [CrossRef]
- Loprete, D.M.; Hartman, K.A. The existence of unique B structures in polynucleotides with alternating purine-pyrimidine sequences. Biochem. Biophys. Res. Commun. 1991, 174, 1313–1317. [Google Scholar] [CrossRef]
- Babic, S.D.; Serec, K. Sodium and manganese salt DNA thin films: An infrared spectroscopy study. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 241, 118646. [Google Scholar] [CrossRef]
- Serec, K.; Babic, S.D.; Podgornik, R.; Tomic, S. Effect of magnesium ions on the structure of DNA thin films: An infrared spectroscopy study. Nucleic Acids Res. 2016, 44, 8456–8464. [Google Scholar] [CrossRef]
- Serec, K.; Babic, S.D.; Tomic, S. Magnesium ions reversibly bind to DNA double stranded helix in thin films. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 268, 120663. [Google Scholar] [CrossRef]
- BR, W. The importance of hydration and DNA conformation in interpreting infrared spectra of cells and tissues. Chem. Soc. Rev. 2016, 45, 1980–1998. [Google Scholar] [CrossRef]
- Zhang, F.; Huang, Q.; Yan, J.; Chen, Z. Histone Acetylation Induced Transformation of B-DNA to Z-DNA in Cells Probed through FT-IR Spectroscopy. Anal. Chem. 2016, 88, 4179–4182. [Google Scholar] [CrossRef]
- Loprete, D.M.; Hartman, K.A. Conditions for the stability of the B, C, and Z structural forms of poly(dG-dC) in the presence of lithium, potassium, magnesium, calcium, and zinc cations. Biochemistry 1993, 32, 4077–4082. [Google Scholar] [CrossRef]
- Rauch, C.; Pichler, A.; Trieb, M.; Wellenzohn, B.; Liedl, K.R.; Mayer, E. Z-DNA’s conformer substates revealed by FT-IR difference spectroscopy of nonoriented left-handed double helical poly(dG-dC). J. Biomol. Struct. Dyn. 2005, 22, 595–614. [Google Scholar] [CrossRef]
- Taboury, J.A.; Taillandier, E. Polymorphism of the left-handed helix (Z form) of poly(dG-dC).poly(dG-dC) induced by MgCl2, studied by infrared spectroscopy. Magnesium 1984, 3, 152–158. [Google Scholar]
- Keller, P.B.; Hartman, K.A. Structural forms, stabilities and transitions in double-helical poly(dG-dC) as a function of hydration and NaCl content. An infrared spectroscopic study. Nucleic Acids Res. 1986, 14, 8167–8182. [Google Scholar] [CrossRef]
- Behe, M.; Felsenfeld, G. Effects of methylation on a synthetic polynucleotide: The B-Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc. Natl. Acad. Sci. USA 1981, 78, 1619–1623. [Google Scholar] [CrossRef] [PubMed]
- Revet, B.; Delain, E.; Dante, R.; Niveleau, A. Three dimensional association of double-stranded helices are produced in conditions for Z-DNA formation. J. Biomol. Struct. Dyn. 1983, 1, 857–871. [Google Scholar] [CrossRef] [PubMed]
- Pohl, F.M. Polymorphism of a synthetic DNA in solution. Nature 1976, 260, 365–366. [Google Scholar] [CrossRef] [PubMed]
- Marmur, J.; Doty, P. Heterogeneity in deoxyribonucleic acids. I. Dependence on composition of the configurational stability of deoxyribonucleic acids. Nature 1959, 183, 1427–1429. [Google Scholar] [CrossRef] [PubMed]
- Marmur, J.; Doty, P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J. Mol. Biol. 1962, 5, 109–118. [Google Scholar] [CrossRef]
- Neault, J.F.; Tajmir-Riahi, H.A. Structural analysis of DNA-chlorophyll complexes by Fourier transform infrared difference spectroscopy. Biophys. J. 1999, 76, 2177–2182. [Google Scholar] [CrossRef]
- Taillandier, E.; Liquier, J. Infrared spectroscopy of DNA. Methods Enzymol. 1992, 211, 307–335. [Google Scholar] [CrossRef]
- Taillandier, E.; Peticolas, W.L.; Adam, S.; Huynhdinh, T.; Igolen, J. Polymorphism of the d(CCCGCGGG)2 double helix studied by Ft-I.R. spectroscopy. Spectrochim. Acta Part A-Mol. Biomol. Spectrosc. 1990, 46A, 107–112. [Google Scholar] [CrossRef]
- Banyay, M.; Sarkar, M.; Graslund, A. A library of IR bands of nucleic acids in solution. Biophys. Chem. 2003, 104, 477–488. [Google Scholar] [CrossRef]
- Taboury, J.A.; Liquier, J.; Taillandier, E. Characterization of DNA structures by infrared-spectroscopy-double helical forms of poly(dG-dC)·poly(dG-dC), poly(dD8G-dC)·poly(dD8G-dC), and poly(dG-dm5C)·poly(dG-dm5C). Can. J. Chem.-Rev. Can. Chim. 1985, 63, 1904–1909. [Google Scholar] [CrossRef]
- Taillandier, E.; Liquier, J.; Ghomi, M. Conformational transitions of nucleic-acids studied by Ir and Raman spectroscopies. J. Mol. Struct. 1989, 214, 185–211. [Google Scholar] [CrossRef]
- Notarstefano, V.; Belloni, A.; Sabbatini, S.; Pro, C.; Orilisi, G.; Monterubbianesi, R.; Tosco, V.; Byrne, H.J.; Vaccari, L.; Giorgini, E. Cytotoxic Effects of 5-Azacytidine on Primary Tumour Cells and Cancer Stem Cells from Oral Squamous Cell Carcinoma: An In Vitro FTIRM Analysis. Cells 2021, 10, 2127. [Google Scholar] [CrossRef]
- Gault, N.; Lefaix, J.L. Infrared microspectroscopic characteristics of radiation-induced apoptosis in human lymphocytes. Radiat. Res. 2003, 160, 238–250. [Google Scholar] [CrossRef]
- Notarstefano, V.; Sabbatini, S.; Pro, C.; Belloni, A.; Orilisi, G.; Rubini, C.; Byrne, H.J.; Vaccari, L.; Giorgini, E. Exploiting fourier transform infrared and Raman microspectroscopies on cancer stem cells from oral squamous cells carcinoma: New evidence of acquired cisplatin chemoresistance. Analyst 2021, 145, 8038–8049. [Google Scholar] [CrossRef]
- Tsuboi, M. Application of Infrared Spectroscopy to Structure Studies of Nucleic Acids. Appl. Spectrosc. Rev. 1970, 3, 45–90. [Google Scholar] [CrossRef]
- Batista de Carvalho, A.L.; Pilling, M.; Gardner, P.; Doherty, J.; Cinque, G.; Wehbe, K.; Kelley, C.; Batista de Carvalho, L.A.; Marques, M.P. Chemotherapeutic response to cisplatin-like drugs in human breast cancer cells probed by vibrational microspectroscopy. Faraday Discuss. 2016, 187, 273–298. [Google Scholar] [CrossRef]
- Kypr, J.; Kejnovska, I.; Renciuk, D.; Vorlickova, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef]
- Mazumdar, D.; Nagraj, N.; Kim, H.K.; Meng, X.; Brown, A.K.; Sun, Q.; Li, W.; Lu, Y. Activity, folding and Z-DNA formation of the 8-17 DNAzyme in the presence of monovalent ions. J. Am. Chem. Soc. 2009, 131, 5506–5515. [Google Scholar] [CrossRef]
- Pietropaolo, A.; D’Urso, A.; Purrello, R.; Berova, N. Effect of different Z-inducers on the stabilization of Z portion in BZ-DNA sequence: Correlation between experimental and simulation data. Chirality 2015, 27, 773–778. [Google Scholar] [CrossRef]
- O’Connor, T.; Kilpatrick, M.W.; Klysik, J.; Larson, J.E.; Martin, J.C.; Singleton, C.K.; Stirdivant, S.M.; Zacharias, W.; Wells, R.D. Left-handed Z-DNA helices in polymers, restriction fragments, and recombinant plasmids. J. Biomol. Struct. Dyn. 1983, 1, 999–1009. [Google Scholar] [CrossRef]
- Moller, A.; Nordheim, A.; Nichols, S.R.; Rich, A. 7-Methylguanine in poly(dG-dC)·poly(dG-dC) facilitates Z-DNA formation. Proc. Natl. Acad. Sci. USA 1981, 78, 4777–4781. [Google Scholar] [CrossRef] [PubMed]
- Buzzo, J.R.; Devaraj, A.; Gloag, E.S.; Jurcisek, J.A.; Robledo-Avila, F.; Kesler, T.; Wilbanks, K.; Mashburn-Warren, L.; Balu, S.; Wickham, J.; et al. Z-form extracellular DNA is a structural component of the bacterial biofilm matrix. Cell 2021, 184, 5740–5758. [Google Scholar] [CrossRef] [PubMed]
- Miyahara, T.; Nakatsuji, H.; Sugiyama, H. Similarities and differences between RNA and DNA double-helical structures in circular dichroism spectroscopy: A SAC-CI study. J. Phys. Chem. A 2016, 120, 9008–9018. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Li, Z.; Van DeSande, J.H. Studies on left-handed DNA:B to Z conformation transition of double strand oligo-d(G-C)6. Acta Laser Biol. Sin. 2005, 14, 359–367. [Google Scholar]
- McDonnell, N.B.; Preisler, R.S. Hydrophobic moieties in cations, anions, and alcohols promote the B-to-Z transition in poly[d(G-C)] and poly[d(G-m5C)]. Biochem. Biophys. Res. Commun. 1989, 164, 426–433. [Google Scholar] [CrossRef]
- Arscott, P.G.; Ma, C.; Wenner, J.R.; Bloomfield, V.A. DNA condensation by cobalt hexaammine (III) in alcohol-water mixtures: Dielectric constant and other solvent effects. Biopolymers 1995, 36, 345–364. [Google Scholar] [CrossRef]
- Roy, R.; Chakraborty, P.; Chatterjee, A.; Sarkar, J. Comparative review on left-handed Z-DNA. Front. Biosci. 2021, 26, 29–35. [Google Scholar] [CrossRef]
- Ha, S.C.; Lowenhaupt, K.; Rich, A.; Kim, Y.G.; Kim, K.K. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature 2005, 437, 1183–1186. [Google Scholar] [CrossRef]
- Sun, H.; Ren, J.; Qu, X. Carbon Nanomaterials and DNA: From Molecular Recognition to Applications. Acc. Chem. Res. 2016, 49, 461–470. [Google Scholar] [CrossRef]
- Feng, L.; Zhao, A.; Ren, J.; Qu, X. Lighting up left-handed Z-DNA: Photoluminescent carbon dots induce DNA B to Z transition and perform DNA logic operations. Nucleic Acids Res. 2013, 41, 7987–7996. [Google Scholar] [CrossRef]
- Bae, S.; Kim, D.; Kim, K.K.; Kim, Y.G.; Hohng, S. Intrinsic Z-DNA is stabilized by the conformational selection mechanism of Z-DNA-binding proteins. J. Am. Chem. Soc. 2011, 133, 668–671. [Google Scholar] [CrossRef]
- Wang, G.; Vasquez, K.M. Z-DNA, an active element in the genome. Front. Biosci. 2007, 12, 4424–4438. [Google Scholar] [CrossRef]
- Yi, J.; Yeou, S.; Lee, N.K. DNA Bending Force Facilitates Z-DNA Formation under Physiological Salt Conditions. J. Am. Chem. Soc. 2022, 144, 13137–13145. [Google Scholar] [CrossRef]
- Hoopes, B.C.; McClure, W.R. Studies on the selectivity of DNA precipitation by spermine. Nucleic Acids Res. 1981, 9, 5493–5504. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).