Effects of the Combination of the C1473G Mutation in the Tph2 Gene and Lethal Yellow Mutations in the Raly-Agouti Locus on Behavior, Brain 5-HT and Melanocortin Systems in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Open Field Test
2.3. Elevated Plus-Maze Test
2.4. Forced Swim Test
2.5. Tail Suspension Test
2.6. Genotyping for C1473G Polymorphism
2.7. Tissue Preparation
2.8. Assay of 5-HT and 5-HIAA Levels
2.9. Assay of TPH Activity
2.10. mRNA Level Assay by qPCR
2.11. Statistics
3. Results
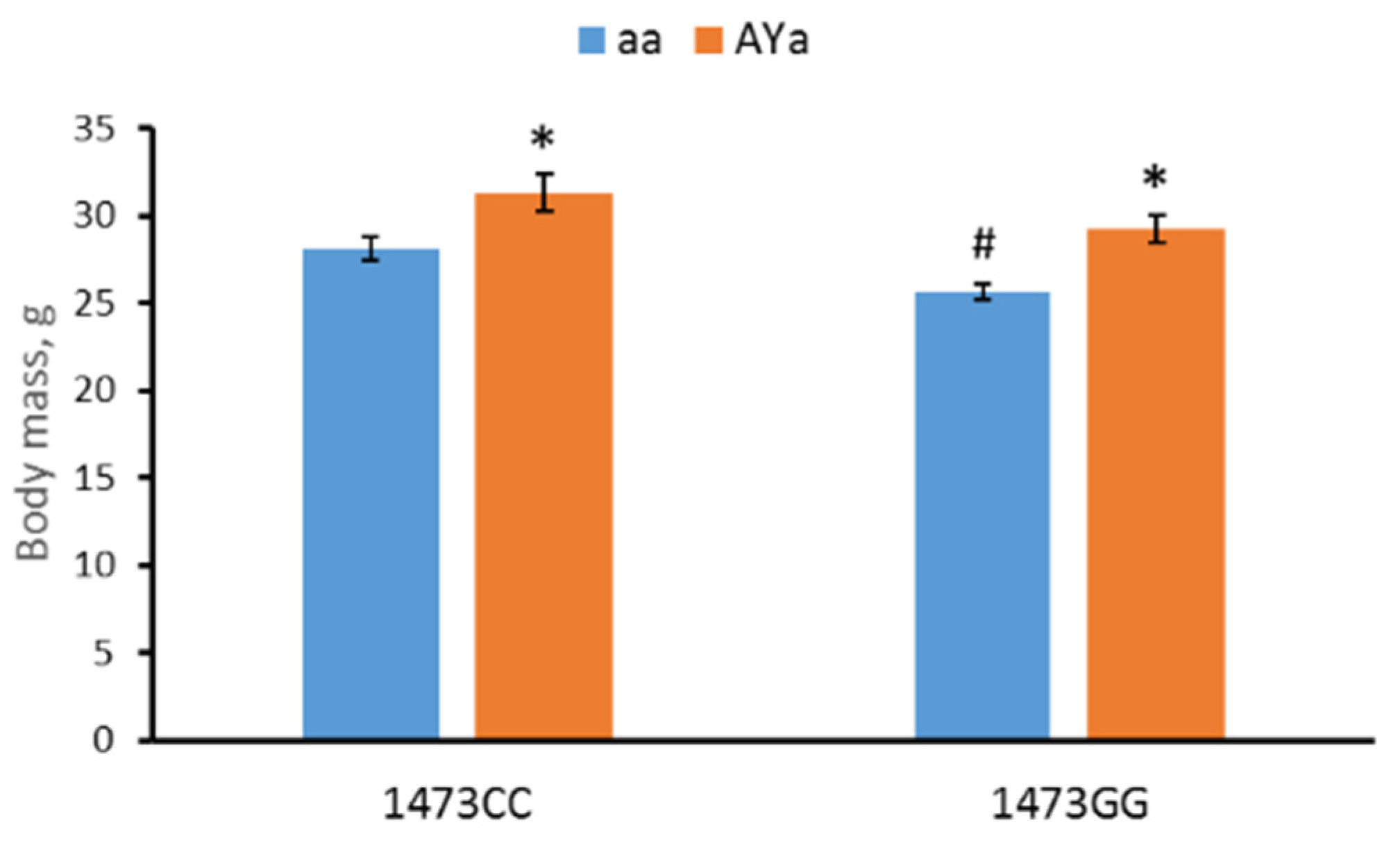
3.1. Effects of C1473G and AY Mutations on Body Mass
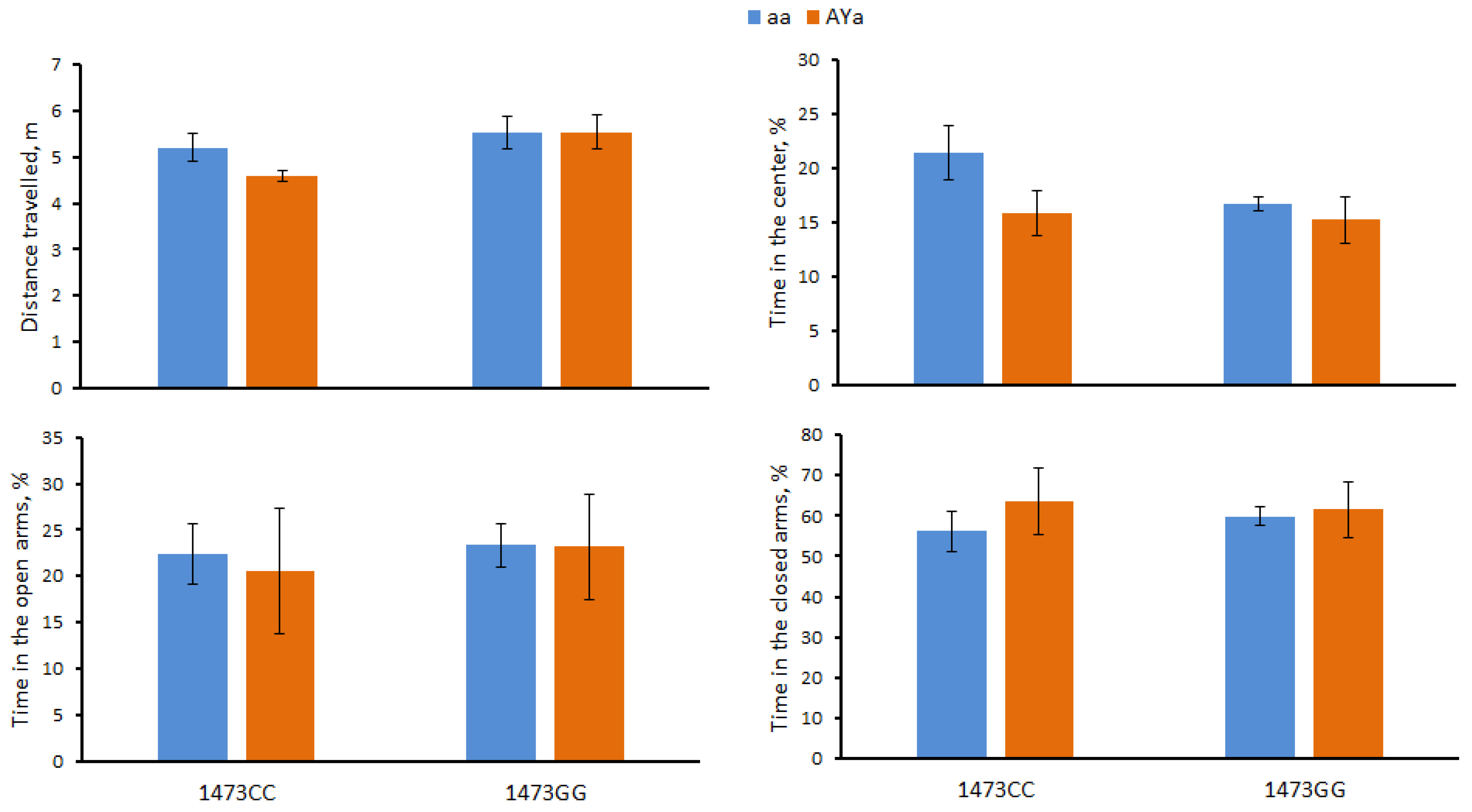
3.2. Effects of C1473G and AY Mutations on Mouse Behavior in the Open Field Test
3.3. Effects of C1473G and AY Mutations on Mouse Behavior in the Elevated Plus-Maze Test
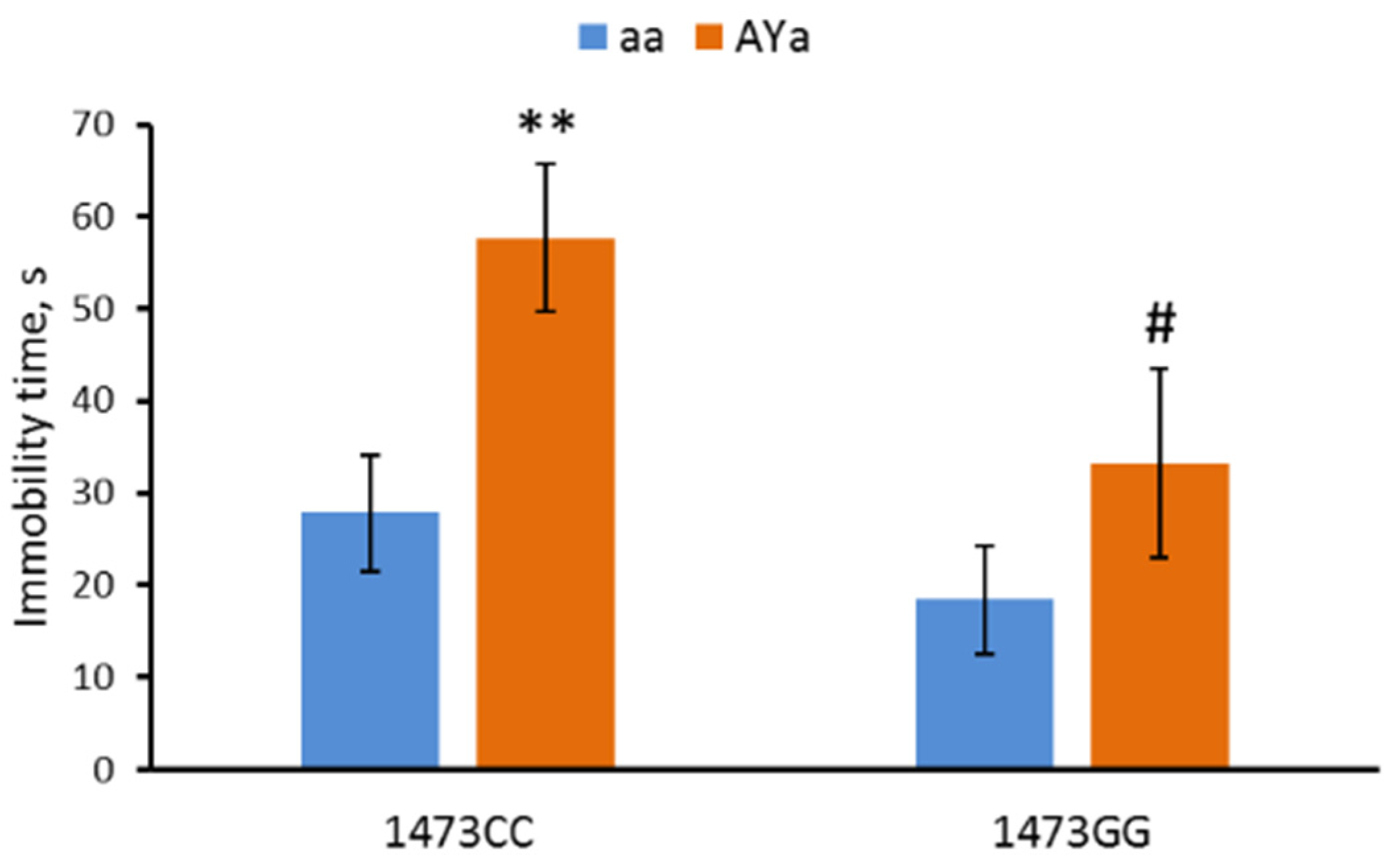
3.4. Effects of C1473G and AY Mutations on Mouse Behavior in the Forced Swim Test
3.5. Effects of C1473G and AY Mutations on the Hind Limb Clasping in the Tail Suspension Test
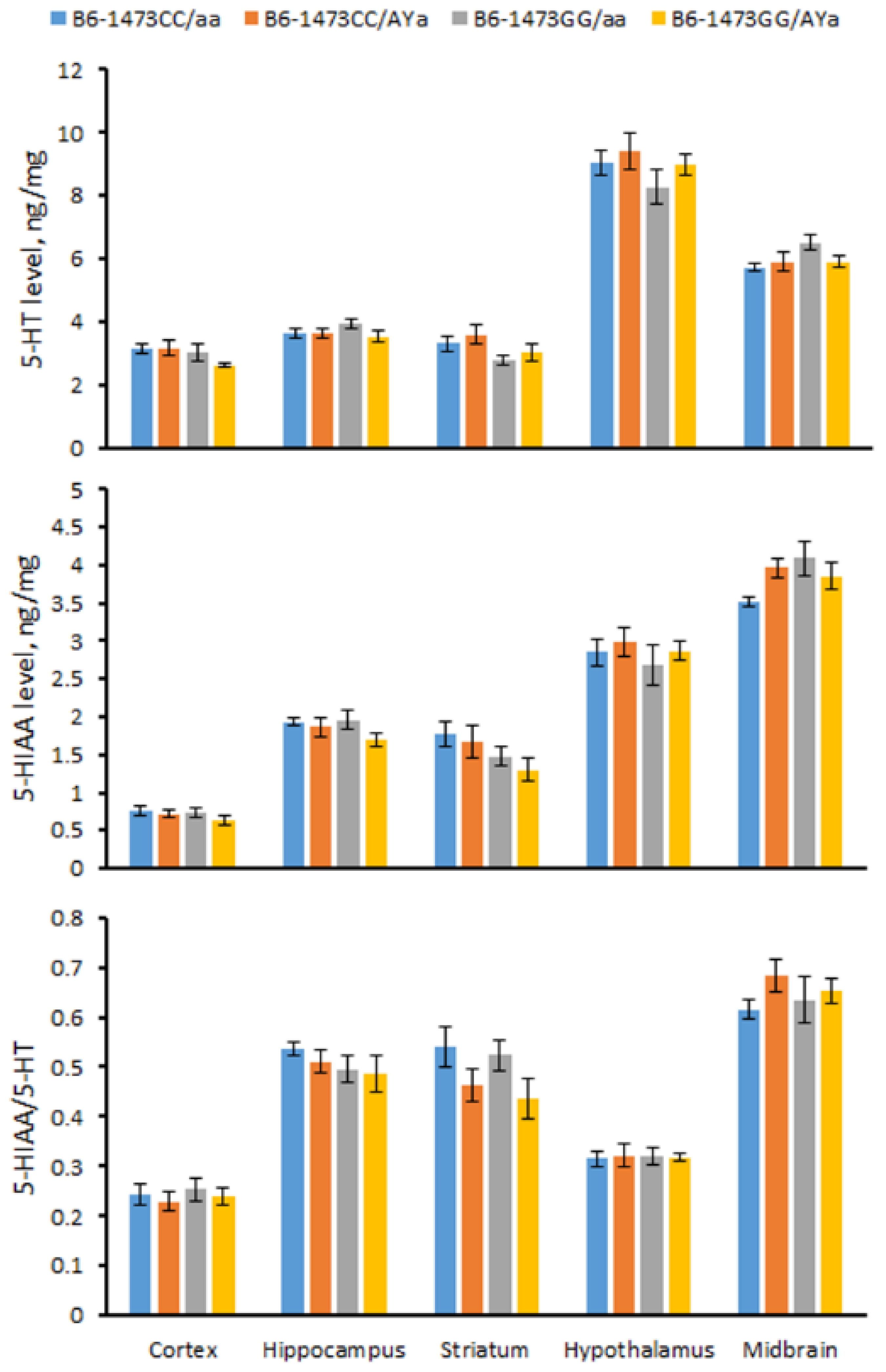
3.6. Effects of C1473G and AY Mutations on 5-HT, 5-HIAA Levels and the 5-HIAA/5-HT Turnover Rate in the Brain
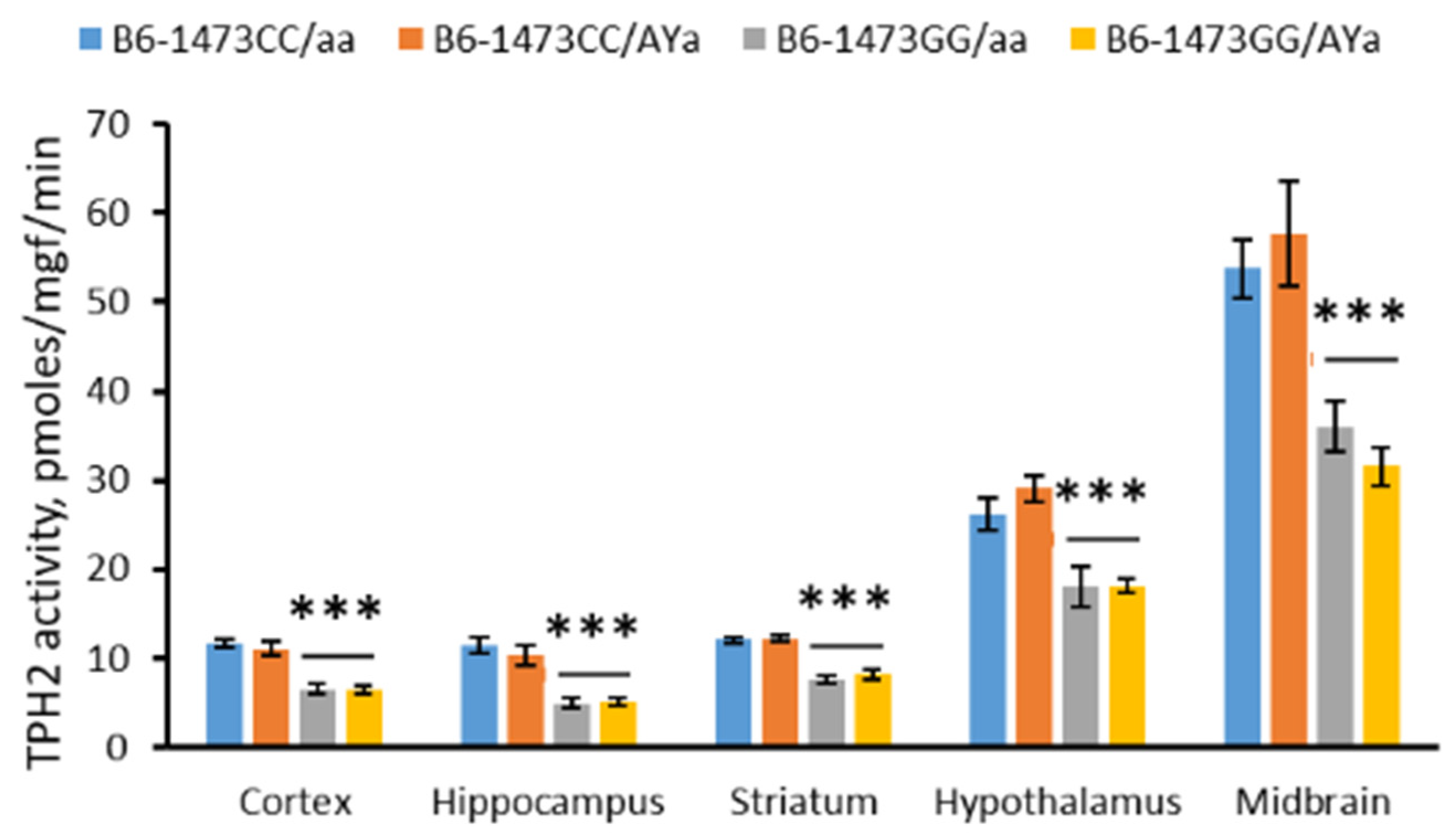
3.7. Effects of C1473G and AY Mutations on the TPH2 Activity in the Brain
3.8. Effects of C1473G and AY Mutations on mRNA Levels of Tph2, Maoa, Slc6a4, Htr1a, Htr2a, Agouti, Mc3r and Mc4r Genes in the Brain
4. Discussion
5. Conclusions
- As expected, the AY allele increased body mass and depressive-like immobility in the forced swim test.
- The 1473G allele decreased body mass and depressive-like immobility and increased locomotor activity in the open field test.
- No effect of the AY allele on the 5-HT system and no effect of the 1473G allele on the melanocortin system in the brain were observed.
- A marked effect of the combination of the AY and 1473G alleles on hind limb dystonia was revealed. Although these alleles do not cause hind limb dystonia separately, their combination results in a high level of expression of this trait in adult B6-1473GG/AYa mice.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Walther, D.J.; Peter, J.U.; Bashammakh, S.; Hörtnagl, H.; Voits, M.; Fink, H.; Bader, M. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science 2003, 299, 76. [Google Scholar] [CrossRef] [PubMed]
- Walther, D.J.; Bader, M. A unique central tryptophan hydroxylase isoform. Biochem. Pharmacol. 2003, 66, 1673–1680. [Google Scholar] [CrossRef] [PubMed]
- Gutknecht, L.; Waider, J.; Kraft., S.; Kriegebaum, C.; Holtmann, B.; Reif, A.; Schmitt, A.; Lesch, K.P. Deficiency of brain 5-HT synthesis but serotonergic neuron formation in Tph2 knockout mice. J. Neural. Transm. 2008, 115, 1127–1132. [Google Scholar] [CrossRef]
- Savelieva, K.V.; Zhao, S.; Pogorelov, V.M.; Rajan, I.; Yang, Q.; Cullinan, E.; Lanthorn, T.H. Genetic disruption of both tryptophan hydroxylase genes dramatically reduces serotonin and affects behavior in models sensitive to antidepressants. PLoS ONE 2008, 3, e3301. [Google Scholar] [CrossRef] [PubMed]
- Alenina, N.; Kikic, D.; Todiras, M.; Mosienko, V.; Qadri, F.; Plehm, R.; Boyé, P.; Vilianovitch, L.; Sohr, R.; Tenner, K.; et al. Growth retardation and altered autonomic control in mice lacking brain serotonin. Proc. Natl. Acad. Sci. USA 2009, 106, 10332–10337. [Google Scholar] [CrossRef]
- Kulikov, A.V.; Osipova, D.V.; Naumenko, V.S.; Terenina, E.; Mormède, P.; Popova, N.K. A pharmacological evidence of positive association between mouse intermale aggression and brain serotonin metabolism. Behav. Brain Res. 2012, 233, 113–119. [Google Scholar] [CrossRef]
- Zhang, X.; Beaulieu, J.M.; Sotnikova, T.D.; Gainetdinov, R.R.; Caron, M.G. Tryptophan hydroxylase-2 controls brain serotonin synthesis. Science 2004, 305, 217. [Google Scholar] [CrossRef] [PubMed]
- Kulikov, A.V.; Osipova, D.V.; Naumenko, V.S.; Popova, N.K. Association between Tph2 gene polymorphism, brain tryptophan hydroxylase activity and aggressiveness in mouse strains. Genes Brain Behav. 2005, 4, 482–485. [Google Scholar] [CrossRef]
- Osipova, D.V.; Kulikov, A.V.; Popova, N.K. C1473G polymorphism in mouse tph2 gene is linked to tryptophan hydroxylase-2 activity in the brain, intermale aggression, and depressive-like behavior in the forced swim test. J. Neurosci. Res. 2009, 87, 1168–1174. [Google Scholar] [CrossRef]
- Popova, N.K.; Kulikov, A.V. Targeting tryptophan hydroxylase 2 in affective disorder. Expert Opin. Ther. Targets 2010, 14, 1259–1271. [Google Scholar] [CrossRef]
- Ottenhof, K.W.; Sild, M.; Lévesque, M.L.; Ruhé, H.G.; Booij, L. TPH2 polymorphisms across the spectrum of psychiatric morbidity: A systematic review and meta-analysis. Neurosci. Biobehav. Rev. 2018, 92, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Kulikova, E.A.; Kulikov, A.V. Tryptophan hydroxylase 2 as a therapeutic target for psychiatric disorders: Focus on animal models. Expert Opin. Ther. Targets 2019, 23, 655–667. [Google Scholar] [CrossRef]
- Koshimizu, H.; Hirata, N.; Takao, K.; Toyama, K.; Ichinose, T.; Furuya, S.; Miyakawa, T. Comprehensive behavioral analysis and quantification of brain free amino acids of C57BL/6J congenic mice carrying the 1473G allele in tryptophan hydroxylase-2. Neuropsychopharmacol. Rep. 2019, 39, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Osipova, D.V.; Kulikov, A.V.; Mekada, K.; Yoshiki, A.; Moshkin, M.P.; Kotenkova, E.V.; Popova, N.K. Distribution of the C1473G polymorphism in tryptophan hydroxylase 2 gene in laboratory and wild mice. Genes Brain Behav. 2010, 9, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.C.; Scherer, T.; Pey, A.L.; Ying, M.; Winge, I.; McKinney, J.; Haavik, J.; Thöny, B.; Martinez, A. Effect of pharmacological chaperones on brain tyrosine hydroxylase and tryptophan hydroxylase 2. J. Neurochem. 2010, 114, 853–863. [Google Scholar] [CrossRef]
- Perry, W.L.; Copeland, N.G.; Jenkins, N.A. The molecular basis for dominant yellow agouti coat color mutations. BioEssays 1994, 16, 705–707. [Google Scholar] [CrossRef]
- Boston, B.A.; Blaydon, K.M.; Varnerin, J.; Cone, R.D. Independent and additive effects of central POMC and leptin pathways on murine obesity. Science 1997, 278, 1641–1644. [Google Scholar] [CrossRef]
- Miller, M.W.; Duhl, D.M.; Vrieling, H.; Cordes, S.P.; Ollmann, M.M.; Winkes, B.M.; Barsh, G.S. Cloning of the mouse agouti gene predicts a secreted protein ubiquitously expressed in mice carrying the lethal yellow mutation. Genes Dev. 1993, 7, 454–467. [Google Scholar] [CrossRef]
- Klebig, M.L.; Wilkinson, J.E.; Geisler, J.G.; Woychik, R.P. Ectopic expression of the agouti gene in transgenic mice causes obesity, features of type II diabetes, and yellow fur. Proc. Natl. Acad. Sci. USA 1995, 92, 4728–4732. [Google Scholar] [CrossRef]
- Izyurov, A.E.; Plyusnina, A.V.; Kulikova, E.A.; Kulikov, A.V.; Khotskin, N.V. Lethal yellow mutation causes anxiety, obsessive-compulsive behavior and affects the brain melanocortin system in males and females of mice. Curr. Protein Pept. Sci. 2023, 24, 329–338. [Google Scholar] [CrossRef]
- Lu, D.; Willard, D.; Patel, I.R.; Kadwell, S.; Overton, L.; Kost, T.; Luther, M.; Chen, W.; Woychik, R.P.; Wilkison, W.O.; et al. Agouti protein is an antagonist of the melanocyte-stimulating-hormone receptor. Nature 1994, 371, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Sohn, J.W. Network of hypothalamic neurons that control appetite. BMB Rep. 2015, 48, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Bazhan, N.M.; Yakovleva, T.V.; Kazantseva, A.Y.; Makarova, E.N. Exaggerated anorexigenic response to restraint stress in A(y) mice is associated with elevated CRFR2 mRNA expression in the hypothalamus. Physiol. Behav. 2013, 120, 19–25. [Google Scholar] [CrossRef]
- Khotskin, N.V.; Plyusnina, A.V.; Kulikova, E.A.; Bazhenova, E.Y.; Fursenko, D.V.; Sorokin, I.E.; Kolotygin, I.; Mormede, P.; Terenina, E.E.; Shevelev, O.B.; et al. On association of the lethal yellow (AY) mutation in the agouti gene with the alterations in mouse brain and behavior. Behav. Brain Res. 2019, 359, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Paylor, R.; Spencer, C.M.; Yuva-Paylor, L.A.; Pieke-Dahl, S. The use of behavioral test batteries, II: Effect of test interval. Physiol. Behav. 2006, 87, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Võikar, V.; Vasar, E.; Rauvala, H. Behavioral alterations induced by repeated testing in C57BL/6J and 129S2/Sv mice: Implications for phenotyping screens. Genes Brain Behav. 2004, 3, 27–38. [Google Scholar] [CrossRef]
- Kulikov, A.V.; Tikhonova, M.A.; Kulikov, V.A. Automated measurement of spatial preference in the open field test with transmitted lighting. J. Neurosci. Methods 2008, 170, 345–351. [Google Scholar] [CrossRef]
- Kulikov, V.A.; Khotskin, N.V.; Nikitin, S.V.; Lankin, V.S.; Kulikov, A.V.; Trapezov, O.V. Application of 3-D imaging sensor for tracking minipigs in the open field test. J. Neurosci. Methods 2014, 235, 219–225. [Google Scholar] [CrossRef]
- Kulikov, A.V.; Morozova, M.V.; Kulikov, V.A.; Kirichuk, V.S.; Popova, N.K. Automated analysis of antidepressants’ effect in the forced swim test. J. Neurosci. Methods 2010, 191, 26–31. [Google Scholar] [CrossRef]
- Sato, K.; Sumi-Ichinose, C.; Kaji, R.; Ikemoto, K.; Nomura, T.; Nagatsu, I.; Ichinose, H.; Ito, M.; Sako, W.; Nagahiro, S.; et al. Differential involvement of striosome and matrix dopamine systems in a transgenic model of dopa-responsive dystonia. Proc. Natl. Acad. Sci. USA 2008, 105, 12551–12556. [Google Scholar] [CrossRef]
- Kulikov, A.V.; Naumenko, V.S.; Voronova, I.P.; Tikhonova, M.A.; Popova, N.K. Quantitative RT-PCR assay of 5-HT1A and 5-HT2A serotonin receptor mRNAs using genomic DNA as an external standard. J. Neurosci. Methods 2005, 141, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Naumenko, V.S.; Osipova, D.V.; Kostina, E.V.; Kulikov, A.V. Utilization of a two- standard system in real-time PCR for quantification of gene expression in the brain. J. Neurosci. Methods 2008, 170, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Fursenko, D.V.; Bazhenova, E.Y.; Khotskin, N.V.; Sorokin, I.E.; Kulikova, E.A.; Kulikov, A.V. Effect of Photoperiod and Lethal Yellow Mutation on Depression-Like Behavior and Expression of Proinflammatory Cytokines in the Hypothalamus in Mice. Bull. Exp. Biol. Med. 2019, 167, 100–103. [Google Scholar] [CrossRef]
- Bazhenova, E.Y.; Fursenko, D.V.; Khotskin, N.V.; Sorokin, I.E.; Kulikov, A.V. Effect of lethal yellow (AY) mutation and photoperiod alterations on mouse behavior. Vavilovskii Zhurnal Genet. I Sel. Vavilov J. Genet. Breed. 2019, 23, 55–61. [Google Scholar] [CrossRef]
- Siesser, W.B.; Zhang, X.; Jacobsen, J.P.; Sotnikova, T.D.; Gainetdinov, R.R.; Caron, M.G. Tryptophan hydroxylase 2 genotype determines brain serotonin synthesis but not tissue content in C57Bl/6J and BALB/cJ congenic mice. Neurosci. Lett. 2010, 481, 6–11. [Google Scholar] [CrossRef]
- Anderson, E.J.; Çakir, I.; Carrington, S.J.; Cone, R.D.; Ghamari-Langroudi, M.; Gillyard, T.; Gimenez, L.E.; Litt, M.J. 60 YEARS OF POMC: Regulation of feeding and energy homeostasis by α-MSH. J. Mol. Endocrinol. 2016, 56, T157–T174. [Google Scholar] [CrossRef]
- Yang, Y. Structure, function and regulation of the melanocortin receptors. Eur. J. Pharmacol. 2011, 660, 125–130. [Google Scholar] [CrossRef]
- Begriche, K.; Sutton, G.M.; Butler, A.A. Homeostastic and non-homeostatic functions of melanocortin-3 receptors in the control of energy balance and metabolism. Physiol. Behav. 2011, 104, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Begriche, K.; Girardet, C.; McDonald, P.; Butler, A.A. Melanocortin-3 receptors and metabolic homeostasis. Prog. Mol. Biol. Transl. Sci. 2013, 114, 109–146. [Google Scholar] [CrossRef]
- Girardet, C.; Butler, A.A. Neural melanocortin receptors in obesity and related metabolic disorders. Biochim. Biophys. Acta 2014, 1842, 482–494. [Google Scholar] [CrossRef]
- Yadav, V.K.; Oury, F.; Suda, N.; Liu, Z.W.; Gao, X.B.; Confavreux, C.; Klemenhagen, K.C.; Tanaka, K.F.; Gingrich, J.A.; Guo, X.E.; et al. A serotonin-dependent mechanism explains the leptin regulation of bone mass, appetite, and energy expenditure. Cell 2009, 138, 976–989. [Google Scholar] [CrossRef] [PubMed]
- Gutknecht, L.; Araragi, N.; Merker, S.; Waider, J.; Sommerlandt, F.M.; Mlinar, B.; Baccini, G.; Mayer, U.; Proft, F.; Hamon, M.; et al. Impacts of brain serotonin deficiency following Tph2 inactivation on development and raphe neuron serotonergic specification. PLoS ONE 2012, 7, e43157. [Google Scholar] [CrossRef] [PubMed]
- Mosienko, V.; Beis, D.; Pasqualetti, M.; Waider, J.; Matthes, S.; Qadri, F.; Bader, M.; Alenina, N. Life without brain serotonin: Reevaluation of serotonin function with mice deficient in brain serotonin synthesis. Behav. Brain Res. 2015, 277, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Stunkard, A.J.; Faith, M.S.; Allison, K.C. Depression and obesity. Biol. Psychiatry 2003, 54, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Simon, G.E.; Von Korff, M.; Saunders, K.; Miglioretti, D.L.; Crane, P.K.; van Belle, G.; Kessler, R.C. Association between obesity and psychiatric disorders in the US adult population. Arch. Gen. Psychiatry 2006, 63, 824–830. [Google Scholar] [CrossRef]
- Luppino, F.S.; de Wit, L.M.; Bouvy, P.F.; Stijnen, T.; Cuijpers, P.; Penninx, B.W.; Zitman, F.G. Overweight, obesity, and depression: A systematic review and metaanalysis of longitudinal studies. Arch. Gen. Psychiatry 2010, 67, 220–229. [Google Scholar] [CrossRef]
- Łojko, D.; Buzuk, G.; Owecki, M.; Ruchała, M.; Rybakowski, J.K. Atypical features indepression: Association with obesity and bipolar disorder. J. Affect. Disord. 2015, 185, 76–80. [Google Scholar] [CrossRef]
- Sharma, A.N.; Elased, K.M.; Garrett, T.L.; Lucot, J.B. Neurobehavioral deficits in db/db diabetic mice. Physiol. Behav. 2010, 101, 381–388. [Google Scholar] [CrossRef]
- Guo, M.; Lu, X.Y. Leptin receptor deficiency confers resistance to behavioral effects of fluoxetine and desipramine via separable substrates. Transl. Psychiatry 2014, 4, e486. [Google Scholar] [CrossRef]
- Dinel, A.L.; André, C.; Aubert, A.; Ferreira, G.; Layé, S.; Castanon, N. Cognitive and emotional alterations are related to hippocampal inflammation in a mouse model of metabolic syndrome. PLoS ONE 2011, 6, e24325. [Google Scholar] [CrossRef]
- Cryan, J.F.; Mombereau, C. In search of a depressed mouse: Utility of models for studying depression-related behavior in genetically modified mice. Mol. Psychiatry 2004, 9, 326–357. [Google Scholar] [CrossRef] [PubMed]
- Mosienko, V.; Bert, B.; Beis, D.; Matthes, S.; Fink, H.; Bader, M.; Alenina, N. Exaggerated aggression and decreased anxiety in mice deficient in brain serotonin. Transl. Psychiatry 2012, 2, e122. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, J.M.; Zhang, X.; Rodriguiz, R.M.; Sotnikova, T.D.; Cools, M.J.; Wetsel, W.C.; Gainetdinov, R.R.; Caron, M.G. Role of GSK3 beta in behavioral abnormalities induced by serotonin deficiency. Proc. Natl. Acad. Sci. USA 2008, 105, 1333–1338. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, J.P.; Medvedev, I.O.; Caron, M.G. The 5-HT deficiency theory of depression: Perspectives from a naturalistic 5-HT deficiency model, the tryptophan hydroxylase 2 Arg439His knockin mouse. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 2444–2459. [Google Scholar] [CrossRef]
- Jacobsen, J.P.; Siesser, W.B.; Sachs, B.D.; Peterson, S.; Cools, M.J.; Setola, V.; Folgering, J.H.; Flik, G.; Caron, M.G. Deficient serotonin neurotransmission and depression-like serotonin biomarker alterations in tryptophan hydroxylase 2 (Tph2) loss-of-function mice. Mol. Psychiatry 2012, 17, 694–704. [Google Scholar] [CrossRef]


| Gene | Primer Sequences | Annealing Temperature, °C | Amplicon Size, bp |
|---|---|---|---|
| Polr2a | 5′-TGTGACAACTCCATACAATGC 5′-CTCTCTTACTGAATTTGCGTACT | 60 | 194 |
| Tph2 | 5′-CATTCCTCGCACAATTCCAGTCG′ 5′-AGTCTACATCCATCCCAACTGCTG | 61 | 239 |
| Maoa | 5′AATGAGGATGTTAAATGGGTAGATGTTGGT 5′-CTTGACATATTCAACTAGACGCTC | 62 | 138 |
| Slc6a4 | 5′-AAGCCCCACCTTGACTCCTCC 5′-CTCCTTCCTCTCCTCACATATCC | 57 | 198 |
| Htr1a | 5′-GACTGCCACCCTCTGCCCTATATC 5′-TCAGCAAGGCAAACAATTCCAG | 62 | 200 |
| Htr2a | 5′-AGAAGCCACCTTGTGTGTGA 5′-TTGCTCATTGCTGATGGACT | 61 | 169 |
| Agouti | 5’-GGATGTCACCCGCCTACT 5’-GTTACTCCGCAGACTCCT | 62 | 150 |
| Mc3r | 5′-TCCGATGCTGCCTAACCT 5′-TGCAGGTTGCCATTCCT | 62 | 130 |
| Mc4r | 5′-GTCGGAAACCATCGTCATTACC 5′-GCAAATGGATGCGAGCAAG | 62 | 150 |
| Gene | B6-1473CC/aa | B6-1473CC/AYa | B6-1473CC/aa | B6-1473CC/AYa | F(3,28) |
|---|---|---|---|---|---|
| Cortex | |||||
| Maoa | 650.3 ± 128.3 | 556.2 ± 50.1 | 632.2 ± 82.8 | 602.0 ± 52.4 | F < 1 |
| Htr1a | 34.0 ± 6.6 | 33.5 ± 4.6 | 37.0 ± 3.7 | 57.0 ± 19.2 | F = 1.2, p = 0.3 |
| Htr2a | 126.0 ± 16.1 | 119.6 ± 11.0 | 140.5 ± 23.5 | 138.5 ± 11.4 | F < 1 |
| Agouti | 0.56 ± 0.42 | 36.3 ± 11.5 *** | 0.07 ± 0.03 | 24.1 ± 6.4 ** | F = 7.4, p < 0.001 |
| Mc3r | 0.72 ± 0.32 | 0.46 ± 0.09 | 0.52 ± 0.09 | 1.14 ± 0.47 | F = 1.1, p = 0.4 |
| Mc4r | 12.20 ± 2.57 | 8.81 ± 1.37 | 11.25 ± 2.29 | 9.09 ± 1.54 | F < 1 |
| Hippocampus | |||||
| Maoa | 200.0 ± 20.6 | 185.4 ± 17.0 | 185.6 ± 18.0 | 187.0 ± 23.6 | F < 1 |
| Htr1a | 53.5 ± 7.0 | 45.2 ± 4.6 | 40.1 ± 6.4 | 59.9 ± 7.2 | F < 1 |
| Htr2a | 10.5 ± 0.86 | 10.3 ± 1.30 | 10.3 ± 2.17 | 11.5 ± 1.59 | F < 1 |
| Agouti | 1.25 ± 0.56 | 30.8 ± 3.3 *** | 2.23 ± 1.95 | 31.3 ± 3.7 *** | F = 40.6, p < 0.001 |
| Mc3r | 3.58 ± 1.33 | 1.68 ± 1.09 | 1.10 ± 0.54 | 3.31 ± 1.87 | F < 1 |
| Mc4r | 17.29 ± 2.58 | 14.84 ± 2.27 | 18.83 ± 7.06 | 16.39 ± 2.61 | F < 1 |
| Striatum | |||||
| Maoa | 106.1 ± 9.9 | 114.5 ± 8.8 | 120.4 ± 13.9 | 127.2 ± 13.8 | F < 1 |
| Htr1a | 2.61 ± 0.57 | 3.79 ± 0.53 | 3.82 ± 0.54 | 3.51 ± 0.45 | F = 1.2, p = 0.3 |
| Htr2a | 28.7 ± 2.54 | 30.5 ± 5.62 | 25.2 ± 3.35 | 32.5 ± 3.61 | F < 1 |
| Agouti | 0.14 ± 0.12 | 42.1 ± 7.6 *** | 0.03 ± 0.01 | 61.1 ± 7.5 *** | F = 33.0, p < 0.001 |
| Mc3r | 0.50 ± 0.14 | 1.46 ± 0.49 | 0.66 ± 0.10 | 1.06 ± 0.29 | F = 2.1, p = 0.1 |
| Mc4r | 6.90 ± 1.15 | 6.61 ± 1.07 | 6.91 ± 1.12 | 7.36 ± 1.05 | F < 1 |
| Hypothalamus | |||||
| Maoa | 242.7 ± 21.3 | 228.7 ± 18.9 | 249.9 ± 16.8 | 294.3 ± 46.4 | F < 1 |
| Htr1a | 22.8 ± 1.6 | 20.9 ± 2.2 | 20.4 ± 2.3 | 24.8 ± 4.7 | F < 1 |
| Htr2a | 16.81 ± 1.08 | 18.74 ± 2.43 | 18.94 ± 1.86 | 21.88 ± 4.36 | F < 1 |
| Agouti | 0.21 ± 0.19 | 40.5 ± 4.2 *** | 0.02 ± 0.01 | 45.6 ± 9.2 *** | F = 24.2, p = 0.001 |
| Mc3r | 12.41 ± 1.18 | 17.67 ± 7.48 | 13.54 ± 2.31 | 13.91 ± 3.94 | F < 1 |
| Mc4r | 10.55 ± 0.62 | 9.91 ± 1.72 | 10.29 ± 1.09 | 11.63 ± 2.48 | F < 1 |
| Midbrain | |||||
| Tph2 | 337.2 ± 59.1 | 261.2 ± 51.3 | 375.1 ± 19.2 | 283.6 ± 35.8 | F < 1 |
| Maoa | 811.73.1 | 809.9 ± 70.6 | 847.9 ± 86.0 | 787.8 ± 50.0 | F < 1 |
| Slc6a4 | 78.4 ± 9.9 | 57.8 ± 4.7 | 64.3 ± 6.9 | 81.0 ± 19.2 | F < 1 |
| Htr1a | 40.7 ± 3.6 | 34.2 ± 2.5 | 30.8 ± 2.8 | 34.4 ± 2.7 | F = 1.8, p = 0.2 |
| Htr2a | 36.0 ± 3.84 | 30.8 ± 2.32 | 29.3 ± 2.45 | 40.9 ± 9.73 | F < 1 |
| Agouti | 5.54 ± 1.24 | 46.4 ± 2.4 *** | 3.45 ± 0.83 | 48.2 ± 4.7 *** | F = 81.0, p = 0.001 |
| Mc3r | 10.21 ± 1.61 | 3.93 ± 1.43 | 6.42 ± 1.65 | 6.49 ± 3.06 | F = 2.0, p = 0.1 |
| Mc4r | 20.13 ± 4.97 | 16.38 ± 2.51 | 15.36 ± 2.33 | 15.60 ± 3.19 | F < 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komleva, P.D.; Alhalabi, G.; Izyurov, A.E.; Khotskin, N.V.; Kulikov, A.V. Effects of the Combination of the C1473G Mutation in the Tph2 Gene and Lethal Yellow Mutations in the Raly-Agouti Locus on Behavior, Brain 5-HT and Melanocortin Systems in Mice. Biomolecules 2023, 13, 963. https://doi.org/10.3390/biom13060963
Komleva PD, Alhalabi G, Izyurov AE, Khotskin NV, Kulikov AV. Effects of the Combination of the C1473G Mutation in the Tph2 Gene and Lethal Yellow Mutations in the Raly-Agouti Locus on Behavior, Brain 5-HT and Melanocortin Systems in Mice. Biomolecules. 2023; 13(6):963. https://doi.org/10.3390/biom13060963
Chicago/Turabian StyleKomleva, Polyna D., Ghofran Alhalabi, Arseniy E. Izyurov, Nikita V. Khotskin, and Alexander V. Kulikov. 2023. "Effects of the Combination of the C1473G Mutation in the Tph2 Gene and Lethal Yellow Mutations in the Raly-Agouti Locus on Behavior, Brain 5-HT and Melanocortin Systems in Mice" Biomolecules 13, no. 6: 963. https://doi.org/10.3390/biom13060963
APA StyleKomleva, P. D., Alhalabi, G., Izyurov, A. E., Khotskin, N. V., & Kulikov, A. V. (2023). Effects of the Combination of the C1473G Mutation in the Tph2 Gene and Lethal Yellow Mutations in the Raly-Agouti Locus on Behavior, Brain 5-HT and Melanocortin Systems in Mice. Biomolecules, 13(6), 963. https://doi.org/10.3390/biom13060963