Spatio-Temporal Expression Pattern of CAKUT Candidate Genes DLG1 and KIF12 during Human Kidney Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Procurement and Processing
2.2. Immunohistochemistry with Chromogenic Detection
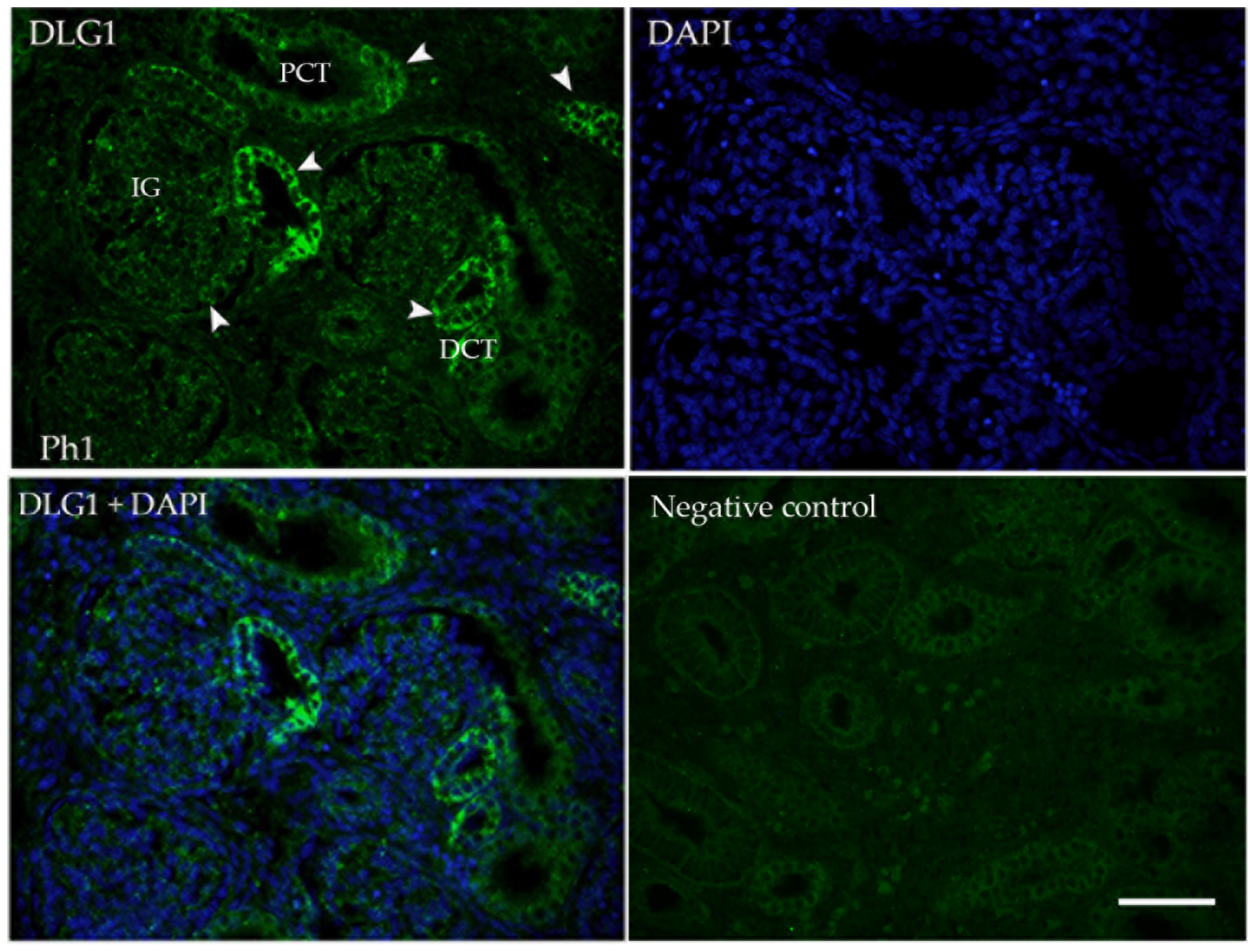
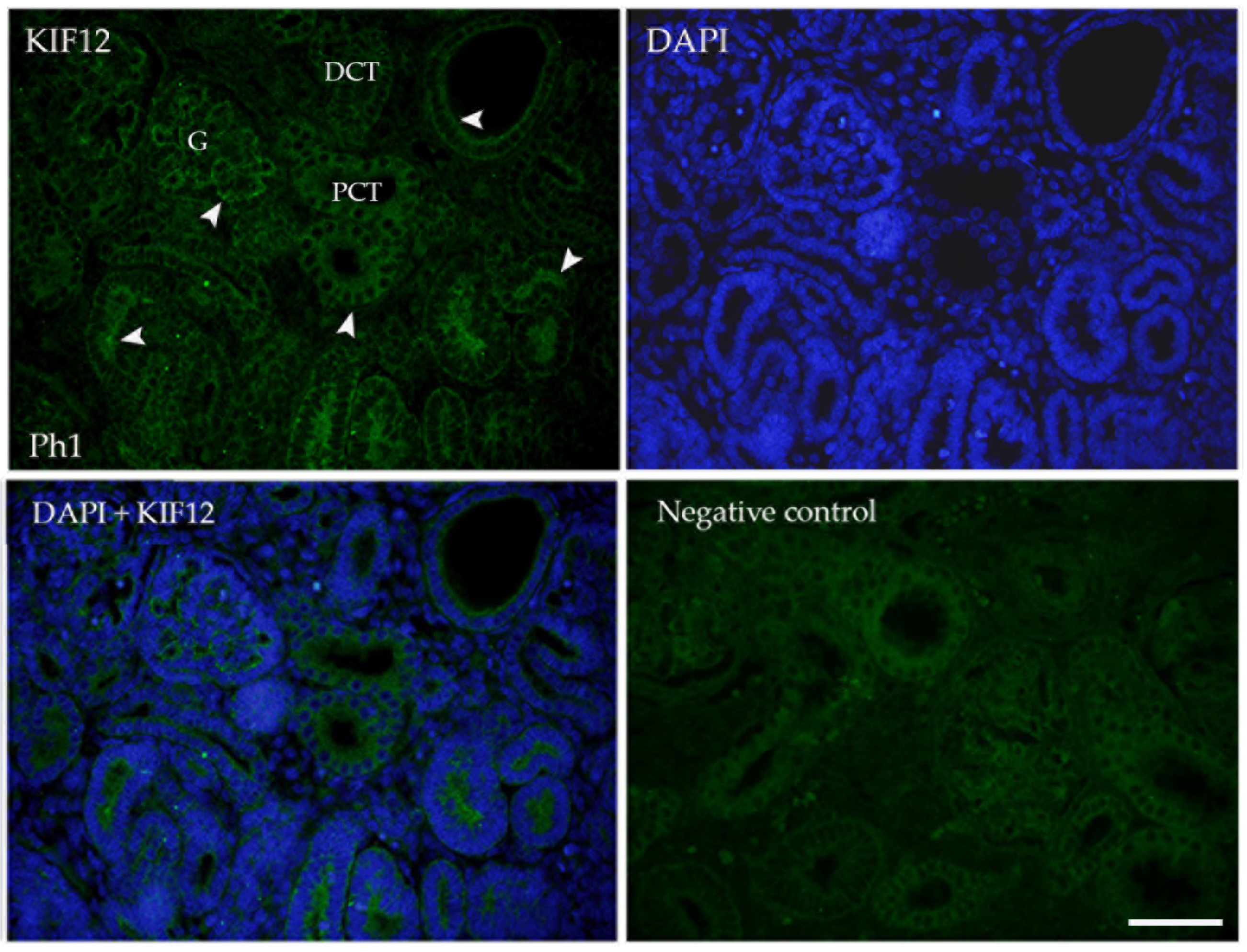
2.3. Immunofluorescence
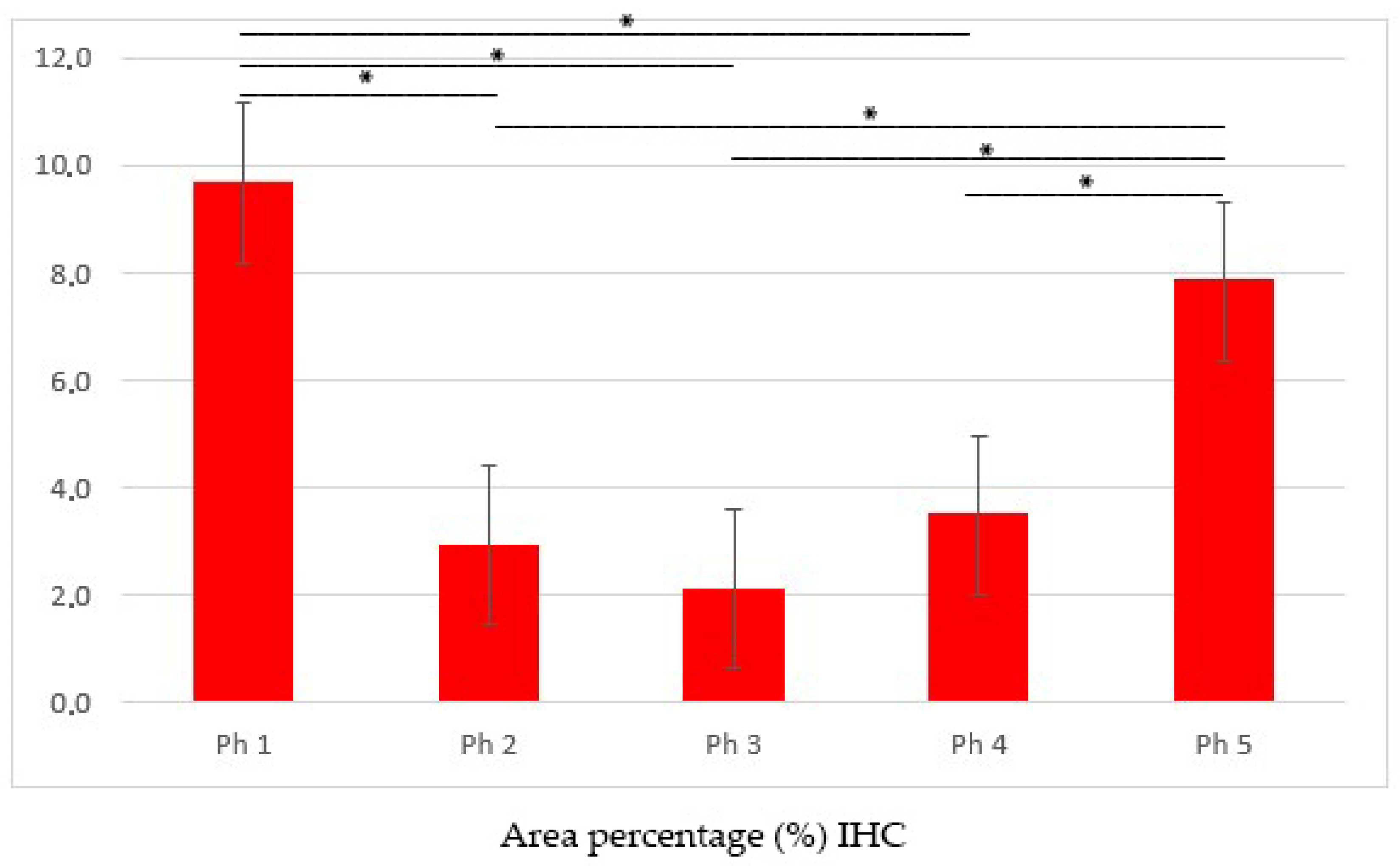
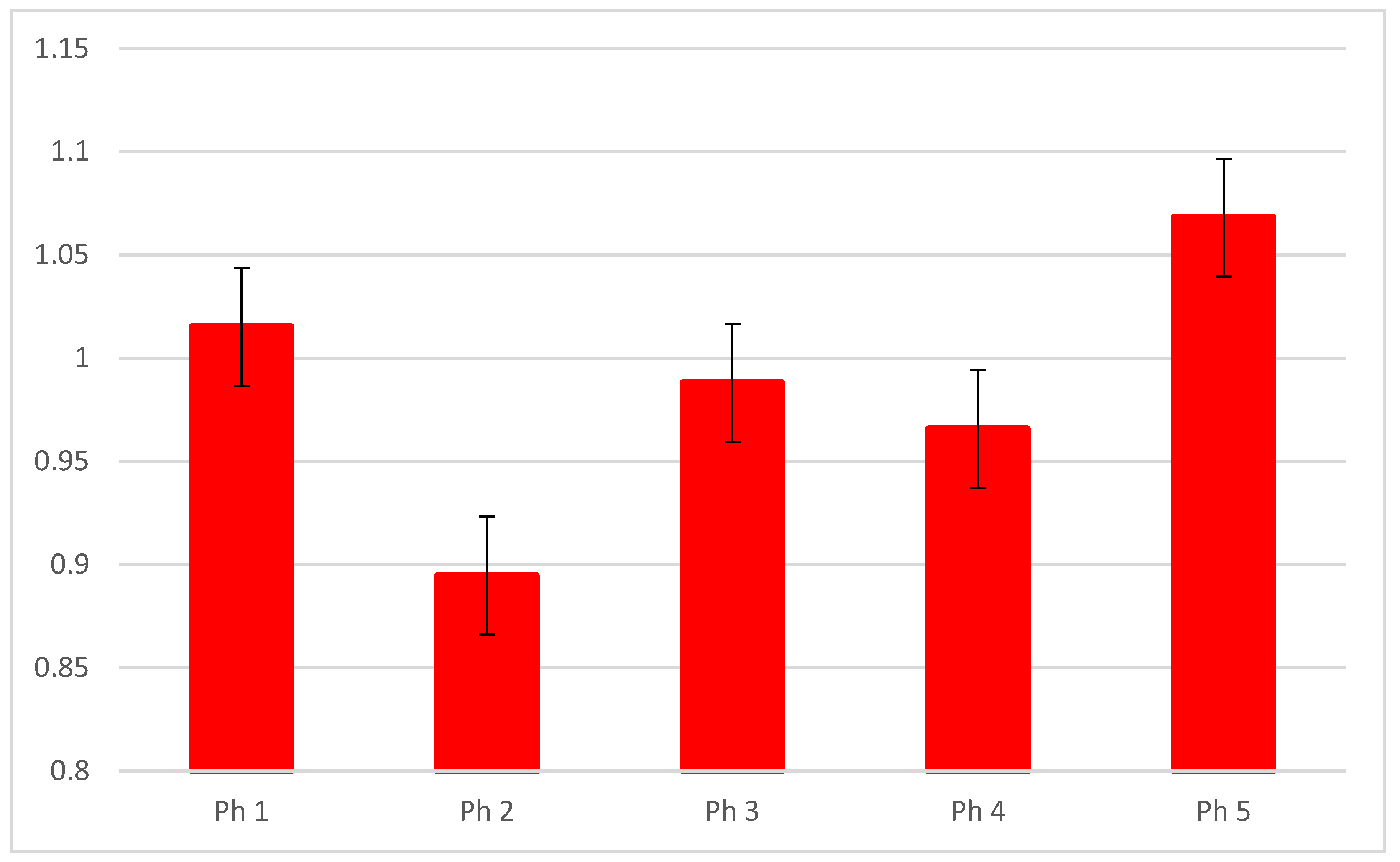
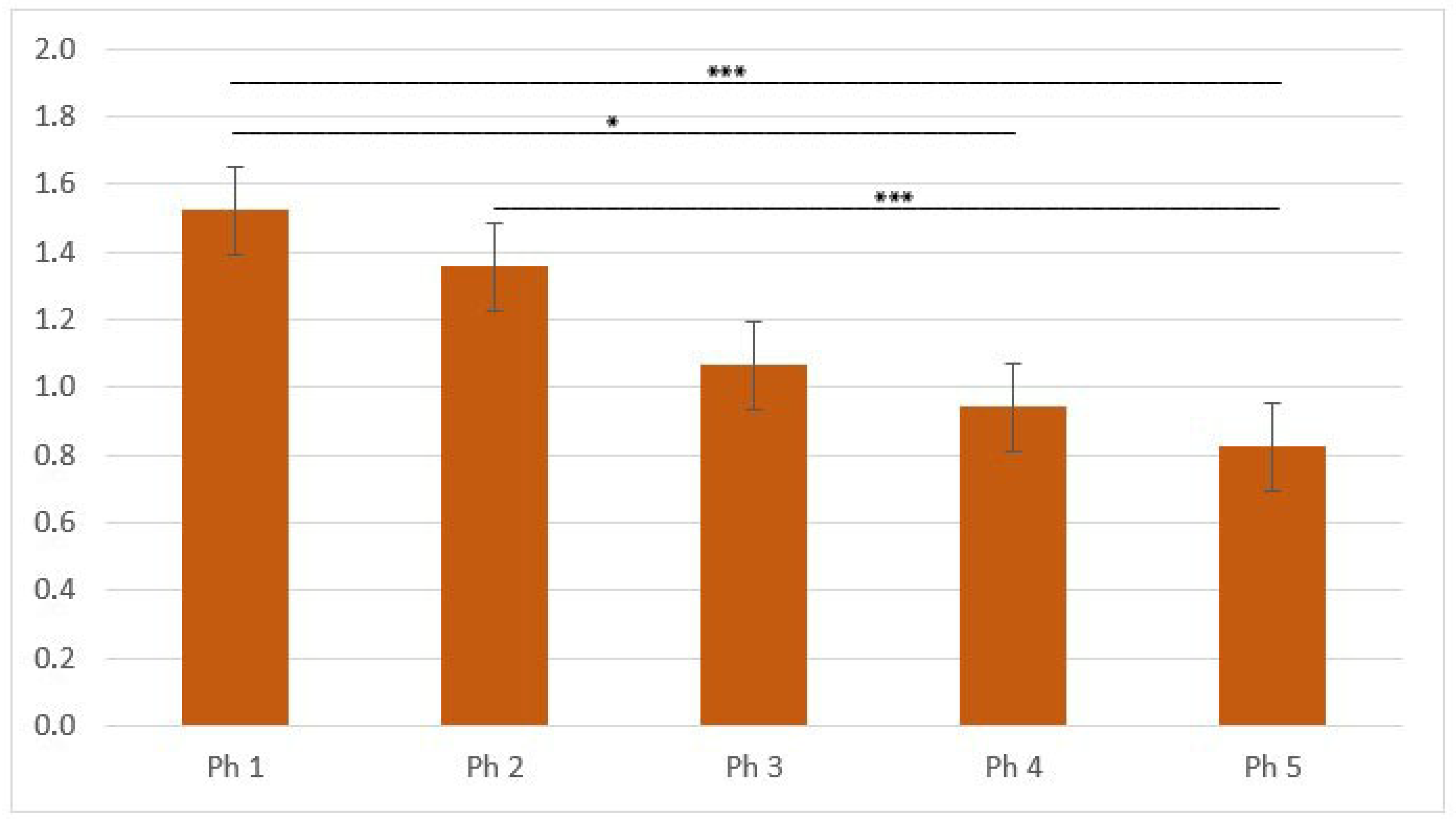
2.4. Data Acquisition, Quantification, and Statistical Analysis of Area Percentages
2.5. RNA Isolation and RT-qPCR
2.6. Gel Electrophoresis
3. Results
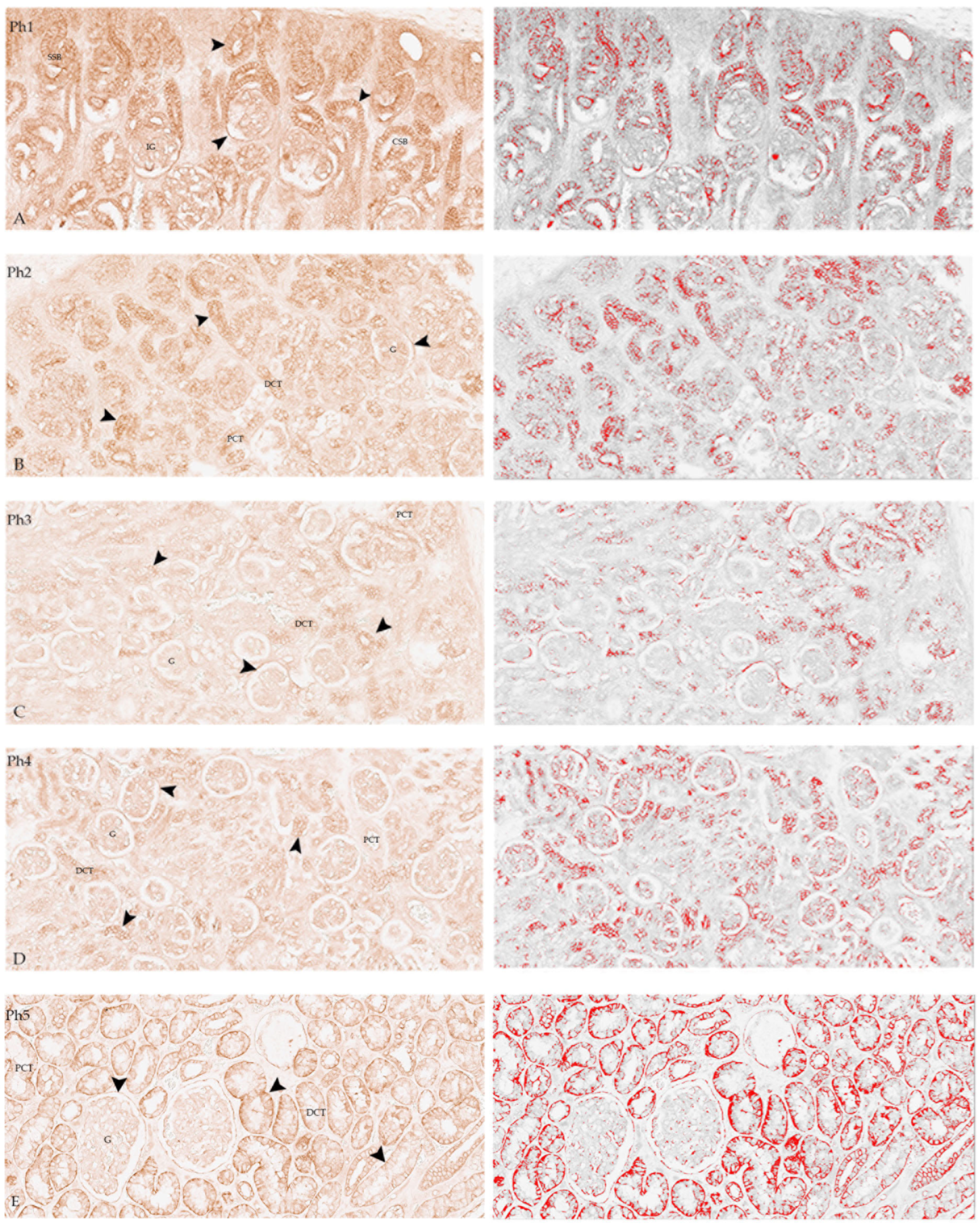
3.1. DLG1
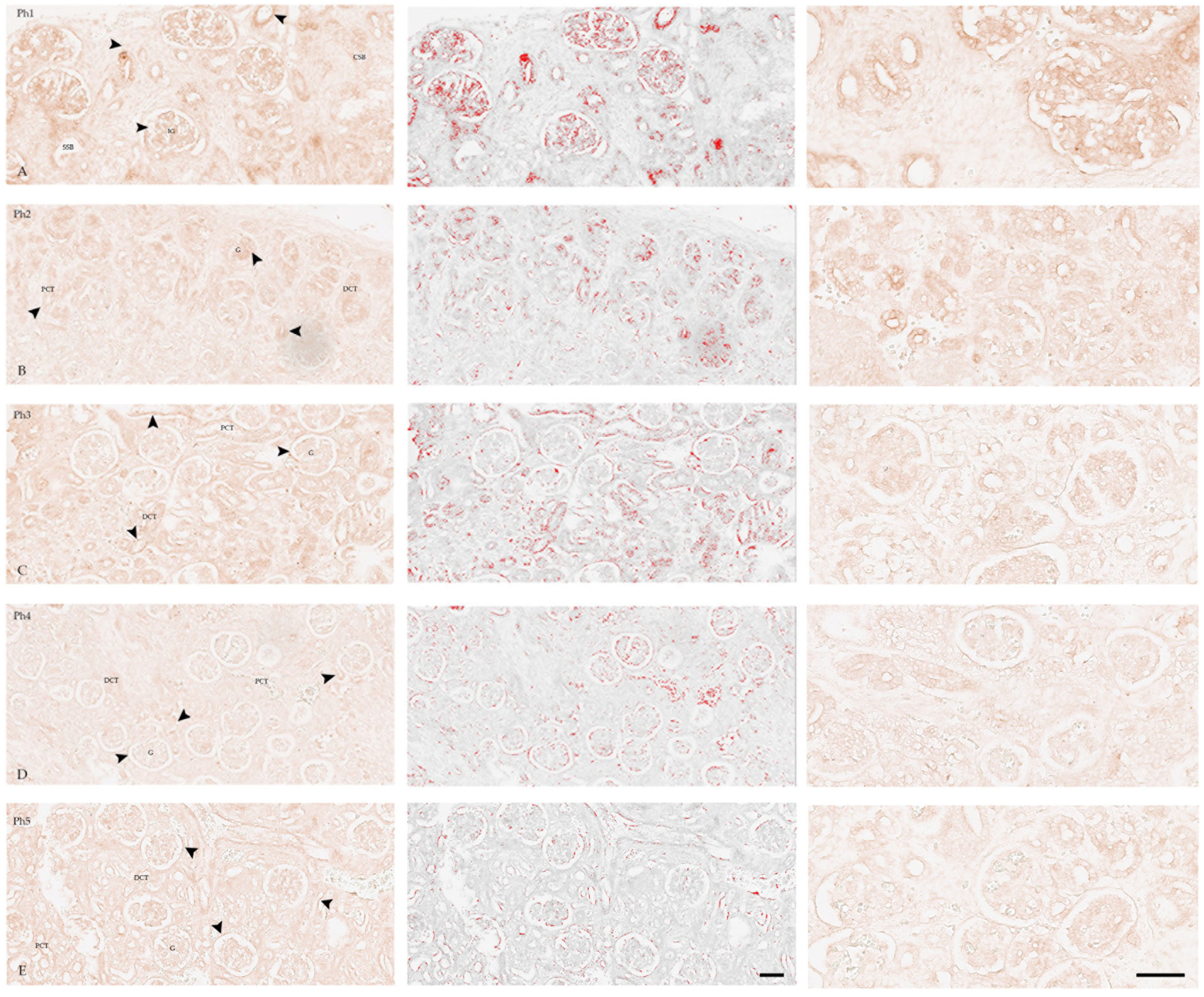
3.2. KIF12
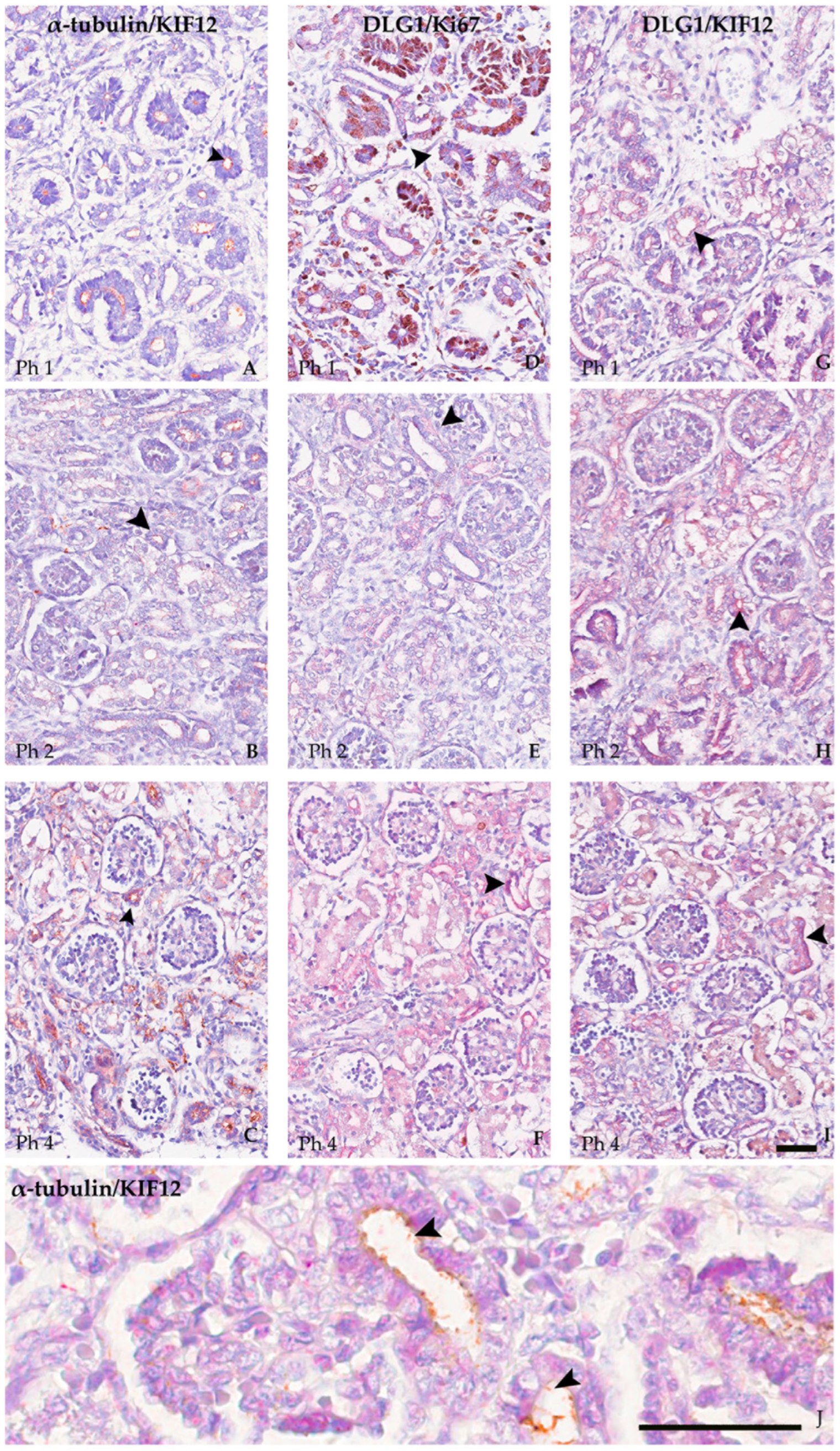
3.3. Co-Localisation in Double IHC
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Racetin, A.; Jurić, M.; Filipović, N.; Šolić, I.; Kosović, I.; Durdov, M.G.; Kunac, N.; Tomaš, S.Z.; Saraga, M.; Šoljić, V.; et al. Expression and Localization of DAB1 and Reelin during Normal Human Kidney Development. Croat. Med. J. 2019, 60, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Jeličić, I.; Vukojević, K.; Racetin, A.; Čarić, D.; Durdov, M.G.; Saraga-Babić, M.; Filipović, N. Expression of Pannexin 1 in the Human Kidney during Embryonal, Early Fetal and Postnatal Development and Its Prognostic Significance in Diabetic Nephropathy. Biomedicines 2022, 10, 944. [Google Scholar] [CrossRef] [PubMed]
- Potter, E.L. Normal and Abnormal Development of the Kidney; Year Book Medical Publishers: Chicago, IL, USA, 1972. [Google Scholar]
- Moore, K.L.; Persaud, T.V.N.; Torchia, M.G. The Developing Human: Clinically Oriented Embryology; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Rehman, S.; Ahmed, D. Embryology, Kidney, Bladder, and Ureter; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Racetin, A.; Raguž, F.; Durdov, M.G.; Kunac, N.; Saraga, M.; Sanna-Cherchi, S.; Šoljić, V.; Martinović, V.; Petričević, J.; Kostić, S.; et al. Immunohistochemical Expression Pattern of RIP5, FGFR1, FGFR2 and HIP2 in the Normal Human Kidney Development. Acta Histochem. 2019, 121, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Quigley, R. Developmental Changes in Renal Function. Curr. Opin. Pediatr. 2012, 24, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Pleniceanu, O.; Harari-Steinberg, O.; Dekel, B. Concise Review: Kidney Stem/Progenitor Cells: Differentiate, Sort out, or Reprogram? Stem Cells 2010, 28, 1649–1660. [Google Scholar] [CrossRef]
- Racetin, A.; Filipović, N.; Lozić, M.; Ogata, M.; Ensor, L.G.; Kelam, N.; Kovačević, P.; Watanabe, K.; Katsuyama, Y.; Saraga-Babić, M.; et al. A Homozygous Dab1−/− Is a Potential Novel Cause of Autosomal Recessive Congenital Anomalies of the Mice Kidney and Urinary Tract. Biomolecules 2021, 11, 609. [Google Scholar] [CrossRef]
- Sanna-Cherchi, S.; Westland, R.; Ghiggeri, G.M.; Gharavi, A.G. Genetic Basis of Human Congenital Anomalies of the Kidney and Urinary Tract. J. Clin. Investig. 2018, 128, 4–15. [Google Scholar] [CrossRef]
- Murugapoopathy, V.; Gupta, I.R. A Primer on Congenital Anomalies of the Kidneys and Urinary Tracts (CAKUT). Clin. J. Am. Soc. Nephrol. 2020, 15, 723–731. [Google Scholar] [CrossRef]
- Capone, V.P.; Morello, W.; Taroni, F.; Montini, G. Genetics of Congenital Anomalies of the Kidney and Urinary Tract: The Current State of Play. Int. J. Mol. Sci. 2017, 18, 796. [Google Scholar] [CrossRef]
- Westland, R.; Verbitsky, M.; Vukojevic, K.; Perry, B.J.; Fasel, D.A.; Zwijnenburg, P.J.G.; Bökenkamp, A.; Gille, J.J.P.; Saraga-Babic, M.; Ghiggeri, G.M.; et al. Copy Number Variation Analysis Identifies Novel CAKUT Candidate Genes in Children with a Solitary Functioning Kidney. Kidney Int. 2015, 88, 1402–1410. [Google Scholar] [CrossRef]
- Azim, A.C.; Knoll, J.H.; Marfatia, S.M.; Peel, D.J.; Bryant, P.J.; Chishti, A.H. DLG1: Chromosome location of the closest human homologue of the Drosophila discs large tumor suppressor gene. Genomics 1995, 30, 613–616. [Google Scholar] [CrossRef]
- Caruana, G.; Bernstein, A. Craniofacial Dysmorphogenesis Including Cleft Palate in Mice with an Insertional Mutation in the Discs Large Gene. Mol. Cell Biol. 2001, 21, 1475–1483. [Google Scholar] [CrossRef]
- Naim, E.; Bernstein, A.; Bertram, J.F.; Caruana, G. Mutagenesis of the Epithelial Polarity Gene, Discs Large 1, Perturbs Nephrogenesis in the Developing Mouse Kidney. Kidney Int. 2005, 68, 955–965. [Google Scholar] [CrossRef]
- Iizuka-Kogo, A.; Ishidao, T.; Akiyama, T.; Senda, T. Abnormal Development of Urogenital Organs in Dlgh1-Deficient Mice. Development 2007, 134, 1799–1807. [Google Scholar] [CrossRef]
- Iizuka-Kogo, A.; Akiyama, T.; Senda, T. Decreased Apoptosis and Persistence of the Common Nephric Duct during the Development of an Aberrant Vesicoureteral Junction in Dlg1 Gene-Targeted Mice. Anat. Rec. 2013, 296, 1936–1942. [Google Scholar] [CrossRef]
- Willatt, L.; Cox, J.; Barber, J.; Dachs Cabanas, E.; Collins, A.; Donnai, D.; Fitzpatrick, D.R.; Maher, E.; Martin, H.; Parnau, J.; et al. 3q29 Microdeletion Syndrome: Clinical and Molecular Characterization of a NewSyndrome. Am. J. Hum. Genet. 2005, 77, 154–160. [Google Scholar] [CrossRef]
- Riva, A.; Gambadauro, A.; Dipasquale, V.; Casto, C.; Ceravolo, M.D.; Accogli, A.; Scala, M.; Ceravolo, G.; Iacomino, M.; Zara, F.; et al. Biallelic Variants in Kif17 Associated with Microphthalmia and Coloboma Spectrum. Int. J. Mol. Sci. 2021, 22, 4471. [Google Scholar] [CrossRef]
- Miki, H.; Setou, M.; Kaneshiro, K.; Hirokawa, N. All Kinesin Superfamily Protein, KIF, Genes in Mouse and Human. Proc. Natl. Acad. Sci. USA 2001, 98, 7004–7011. [Google Scholar] [CrossRef]
- Hirokawa, N.; Noda, Y.; Tanaka, Y.; Niwa, S. Kinesin Superfamily Motor Proteins and Intracellular Transport. Nat. Rev. Mol. Cell Biol. 2009, 10, 682–696. [Google Scholar] [CrossRef]
- Nakagawa, T.; Tanaka, Y.; Matsuoka, E.; Kondo, S.; Okada, Y.; Noda, Y.; Kanai, Y.; Hirokawa, N. Identification and Classification of 16 New Kinesin Superfamily (KIF) Proteins in Mouse Genome. Proc. Natl. Acad. Sci. USA 1997, 94, 9654–9659. [Google Scholar] [CrossRef]
- Katoh, M.K.M. Characterization of KIF12 Gene in Silico. Oncol. Rep. 2005, 13, 367–370. [Google Scholar] [PubMed]
- Mrug, M.; Zhou, J.; Yang, C.; Aronow, B.J.; Cui, X.; Schoeb, T.R.; Siegal, G.P.; Yoder, B.K.; Guay-Woodford, L.M. Genetic and Informatic Analyses Implicate Kif12 as a Candidate Gene within the Mpkd2 Locus That Modulates Renal Cystic Disease Severity in the Cys1cpk Mouse. PLoS ONE 2015, 10, e0135678. [Google Scholar] [CrossRef] [PubMed]
- Lozić, M.; Filipović, N.; Jurić, M.; Kosović, I.; Benzon, B.; Šolić, I.; Kelam, N.; Racetin, A.; Watanabe, K.; Katsuyama, Y.; et al. Alteration of Cx37, Cx40, Cx43, Cx45, PanX1, and Renin Expression Patterns in Postnatal Kidneys of Dab1-/-(Yotari) Mice. Int. J. Mol. Sci. 2021, 22, 1284. [Google Scholar] [CrossRef] [PubMed]
- Lozic, M.; Minarik, L.; Racetin, A.; Filipovic, N.; Babic, M.S.; Vukojevic, K. CRKL, AIFM3, AIF, BCL2 and UBASH3A during Human Kidney Development. Int. J. Mol. Sci. 2021, 22, 9183. [Google Scholar] [CrossRef]
- Solic, I.; Racetin, A.; Filipovic, N.; Mardesic, S.; Bocina, I.; Galesic-Ljubanovic, D.; Durdov, M.G.; Saraga-Babić, M.; Vukojevic, K. Expression Pattern of α-Tubulin, Inversin and Its Target Dishevelled-1 and Morphology of Primary Cilia in Normal Human Kidney Development and Diseases. Int. J. Mol. Sci. 2021, 22, 3500. [Google Scholar] [CrossRef]
- Carev, D.; Krnić, D.; Saraga, M.; Sapunar, D.; Saraga-Babić, M. Role of Mitotic, pro-Apoptotic and Anti-Apoptotic Factors in Human Kidney Development. Pediatr. Nephrol. 2006, 21, 627–636. [Google Scholar] [CrossRef]
- Hinchliffe, S.A.; Sargent, P.H.; Howard, C.V.; Chan, Y.F.; van Velzen, D. Human Intrauterine Renal Growth Expressed in Absolute Number of Glomeruli Assessed by the Disector Method and Cavalieri Principle. Lab. Investig. 1991, 64, 777–784. [Google Scholar]
- Siller, K.H.; Doe, C.Q. Spindle Orientation during Asymmetric Cell Division. Nat. Cell Biol. 2009, 14, 365–374. [Google Scholar] [CrossRef]
- Albertson, R.; Doe, C.Q. Dlg, Scrib and Lgl Regulate Neuroblast Cell Size and Mitotic Spindle Asymmetry. Nat. Cell Biol. 2003, 5, 166–170. [Google Scholar] [CrossRef]
- Gönczy, P. Mechanisms of Asymmetric Cell Division: Flies and Worms Pave the Way. Nat. Rev. Mol. Cell Biol. 2008, 9, 355–366. [Google Scholar] [CrossRef]
- Williams, S.E.; Ratliff, L.A.; Postiglione, M.P.; Knoblich, J.A.; Fuchs, E. Par3-MInsc and Gα I3 Cooperate to Promote Oriented Epidermal Cell Divisions through LGN. Nat. Cell Biol. 2014, 16, 758–769. [Google Scholar] [CrossRef]
- Knoblich, J.A. Asymmetric Cell Division: Recent Developments and Their Implications for Tumour Biology. Nat. Rev. Mol. Cell Biol. 2010, 11, 849–860. [Google Scholar] [CrossRef]
- Williams, S.E.; Fuchs, E. Oriented Divisions, Fate Decisions. Curr. Opin. Cell Biol. 2013, 25, 749–758. [Google Scholar] [CrossRef]
- Stanzick, K.J.; Li, Y.; Schlosser, P.; Gorski, M.; Wuttke, M.; Thomas, L.F.; Rasheed, H.; Rowan, B.X.; Graham, S.E.; Vanderweff, B.R.; et al. Discovery and Prioritization of Variants and Genes for Kidney Function in >1.2 Million Individuals. Nat. Commun. 2021, 12, 4350. [Google Scholar] [CrossRef]
- Fuja, T.J.; Lin, F.; Osann, K.E.; Bryant, P.J. Somatic Mutations and Altered Expression of the Candidate Tumor Suppressors CSNK1, DLG1, and EDD/HHYD in Mammary Ductal Carcinoma. Cancer Res. 2004, 64, 942–951. [Google Scholar] [CrossRef]
- Hou, Y.Y.; Cao, W.W.; Li, L.; Li, S.P.; Liu, T.; Wan, H.Y.; Liu, M.; Li, X.; Tang, H. MicroRNA-519d Targets MKi67 and Suppresses Cell Growth in the Hepatocellular Carcinoma Cell Line QGY-7703. Cancer Lett. 2011, 307, 182–190. [Google Scholar] [CrossRef]
- Combs, H.L.; Shankland, S.J.; Setzer, S.V.; Hudkins, K.L.; Alpers, C.E. Expression of the Cyclin Kinase Inhibitor, P27(Kip1), in Developing and Mature Human Kidney. Kidney Int. 1998, 53, 892–896. [Google Scholar] [CrossRef]
- Aksu, A.Ü.; Das, S.K.; Nelson-Williams, C.; Jain, D.; Hoşnut, F.Ö.; Şahin, G.E.; Lifton, R.P.; Vilarinho, S. Recessive Mutations in KIF12 Cause High Gamma-Glutamyltransferase Cholestasis. Hepatol. CommuniCations 2019, 3, 471–477. [Google Scholar] [CrossRef]
- Stalke, A.; Sgodda, M.; Cantz, T.; Skawran, B.; Lainka, E.; Hartleben, B.; Baumann, U.; Pfister, E.D. KIF12 Variants and Disturbed Hepatocyte Polarity in Children with a Phenotypic Spectrum of Cholestatic Liver Disease. J. Pediatr. 2022, 240, 284–291. [Google Scholar] [CrossRef]
- Mirvis, M.; Stearns, T.; Nelson, W.J. Cilium Structure, Assembly, and Disassembly Regulated by the Cytoskeleton. Biochem. J. 2018, 475, 2329–2353. [Google Scholar] [CrossRef]
- Lienkamp, S.; Ganner, A.; Boehlke, C.; Schmidt, T.; Arnold, S.J.; Schäfer, T.; Romaker, D.; Schuler, J.; Hoff, S.; Powelske, C.; et al. Inversin Relays Frizzled-8 Signals to Promote Proximal Pronephros Development. Proc. Natl. Acad. Sci. USA 2010, 107, 20388–20393. [Google Scholar] [CrossRef] [PubMed]
- Papakrivopoulou, E.; Dean, C.H.; Copp, A.J.; Long, D.A. Planar Cell Polarity and the Kidney. Nephrol. Dial. Transplant. 2014, 29, 1320–1326. [Google Scholar] [CrossRef] [PubMed]
- Saraga, M.; Vukojević, K.; Krželj, V.; Puretić, Z.; Bočina, I.; Durdov, M.G.; Weber, S.; Dworniczak, B.; Ljubanović, D.G.; Saraga-Babić, M. Mechanism of Cystogenesis in Nephrotic Kidneys: A Histopathological Study. BMC Nephrol. 2014, 15, 3. [Google Scholar] [CrossRef] [PubMed]
- Mrug, M.; Li, R.; Cui, X.; Schoeb, T.R.; Churchill, G.A.; Guay-Woodford, L.M. Kinesin Family Member 12 Is a Candidate Polycystic Kidney Disease Modifier in the Cpk Mouse. J. Am. Soc. Nephrol. 2005, 16, 905–916. [Google Scholar] [CrossRef]
- Gong, Y.; Ma, Z.; Patel, V.; Fischer, E.; Hiesberger, T.; Pontoglio, M.; Igarashi, P. HNF-1β Regulates Transcription of the PKD Modifier Gene Kif12. J. Am. Soc. Nephrol. 2009, 20, 41–47. [Google Scholar] [CrossRef]
- Porter, A.P.; White, G.R.M.; Mack, N.A.; Malliri, A. The Interaction between CASK and the Tumour Suppressor Dlg1 Regulates Mitotic Spindle Orientation in Mammalian Epithelia. J. Cell Sci. 2019, 132, jcs230086. [Google Scholar] [CrossRef]
- Hu, M.C.; Mo, R.; Bhella, S.; Wilson, C.W.; Chuang, P.T.; Hui, C.C.; Rosenblum, N.D. GLI3-Dependent Transcriptional Repression of Gli1, Gli2 and Kidney Patterning Genes Disrupts Renal Morphogenesis. Development 2006, 133, 569–578. [Google Scholar] [CrossRef]
- Gill, P.S.; Rosenblum, N.D. Control of Murine Kidney Development by Sonic Hedgehog and Its GLI Effectors. Cell Cycle 2006, 5, 1426–1430. [Google Scholar] [CrossRef]
- Skalická, K.; Hrčková, G.; Vaská, A.; Baranyaiová, A.; Janega, P.; Žilinská, Z.; Daniš, D.; Kovács, L. Pilot Study of the Occurrence of Somatic Mutations in Ciliary Signalling Pathways as a Contribution Factor to Autosomal Dominant Polycystic Kidney Development. Folia Biol. 2017, 63, 174–181. [Google Scholar]
- He, M.; Agbu, S.; Anderson, K.v. Microtubule Motors Drive Hedgehog Signaling in Primary Cilia. Trends Cell Biol. 2017, 27, 110–125. [Google Scholar] [CrossRef]
- Schou, K.B.; Mogensen, J.B.; Morthorst, S.K.; Nielsen, B.S.; Aleliunaite, A.; Serra-Marques, A.; Fürstenberg, N.; Saunier, S.; Bizet, A.A.; Veland, I.R.; et al. KIF13B Establishes a CAV1-Enriched Microdomain at the Ciliary Transition Zone to Promote Sonic Hedgehog Signalling. Nat. Commun. 2017, 8, 14177. [Google Scholar] [CrossRef]
- Asaba, N.; Hanada, T.; Takeuchi, A.; Chishti, A.H. Direct Interaction with a Kinesin-Related Motor Mediates Transport of Mammalian Discs Large Tumor Suppressor Homologue in Epithelial Cells. J. Biol. Chem. 2003, 278, 8395–8400. [Google Scholar] [CrossRef]
- Kanai, Y.; Wang, D.; Hirokawa, N. KIF13B Enhances the Endocytosis of LRP1 by Recruiting LRP1 to Caveolae. J. Cell Biol. 2014, 204, 395–408. [Google Scholar] [CrossRef]
- Hanada, T.; Lin, L.; Tibaldi, E.V.; Reinherz, E.L.; Chishti, A.H. GAKIN, a Novel Kinesin-like Protein Associates with the Human Homologue of the Drosophila Discs Large Tumor Suppressor in T Lymphocytes. J. Biol. Chem. 2000, 275, 28774–28784. [Google Scholar] [CrossRef]
- Bolis, A.; Coviello, S.; Visigalli, I.; Taveggia, C.; Bachi, A.; Chishti, A.H.; Hanada, T.; Quattrini, A.; Previtali, S.C.; Biffi, A.; et al. Dlg1, Sec8, and Mtmr2 Regulate Membrane Homeostasis in Schwann Cell Myelination. J. Neurosci. 2009, 29, 8858–8870. [Google Scholar] [CrossRef]
- Zhu, J.; Shang, Y.; Xia, Y.; Zhang, R.; Zhang, M. An Atypical MAGUK GK Target Recognition Mode Revealed by the Interaction between DLG and KIF13B. Structure 2016, 24, 1876–1885. [Google Scholar] [CrossRef]


| Gene | Primary Antibody |
|---|---|
| DLG1 | Mouse anti-DLG1 sc-9961, 1:50; Santa Cruz, CA, USA |
| KIF12 | Mouse anti-KIF12 sc-376766, 1:50; Santa Cruz, CA, USA |
| α-tubulin | Rabbit anti-α-tubulin ab179484, 1:50, Abcam, Cambridge, UK |
| Ki-67 | Mouse anti-Ki-67 M7240, 1:100, Agilent Technologies, CA, USA |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| DLG1 | 5′-TCC ACC CAG GCA AAT CCT CC-3′ | 5′-GGG TTG TCC GTA CCT CCT GC-3′ |
| KIF12 | 5′-CCG GAC TCT GCA GTT TCT CCT G-3′ | 5′-GGC GAA GGT CCT CTG CAT GAT-3′ |
| RPS9 | 5′-GGA TTT CTT AGA GAG ACG CCT G-3′ | 5′-GGA CAA TGA AGG ACG GGA TG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veljačić Visković, D.; Lozić, M.; Vukoja, M.; Šoljić, V.; Vukojević, K.; Glavina Durdov, M.; Filipović, N.; Lozić, B. Spatio-Temporal Expression Pattern of CAKUT Candidate Genes DLG1 and KIF12 during Human Kidney Development. Biomolecules 2023, 13, 340. https://doi.org/10.3390/biom13020340
Veljačić Visković D, Lozić M, Vukoja M, Šoljić V, Vukojević K, Glavina Durdov M, Filipović N, Lozić B. Spatio-Temporal Expression Pattern of CAKUT Candidate Genes DLG1 and KIF12 during Human Kidney Development. Biomolecules. 2023; 13(2):340. https://doi.org/10.3390/biom13020340
Chicago/Turabian StyleVeljačić Visković, Daniela, Mirela Lozić, Martina Vukoja, Violeta Šoljić, Katarina Vukojević, Merica Glavina Durdov, Natalija Filipović, and Bernarda Lozić. 2023. "Spatio-Temporal Expression Pattern of CAKUT Candidate Genes DLG1 and KIF12 during Human Kidney Development" Biomolecules 13, no. 2: 340. https://doi.org/10.3390/biom13020340
APA StyleVeljačić Visković, D., Lozić, M., Vukoja, M., Šoljić, V., Vukojević, K., Glavina Durdov, M., Filipović, N., & Lozić, B. (2023). Spatio-Temporal Expression Pattern of CAKUT Candidate Genes DLG1 and KIF12 during Human Kidney Development. Biomolecules, 13(2), 340. https://doi.org/10.3390/biom13020340







