Cowpox Viruses: A Zoo Full of Viral Diversity and Lurking Threats
Abstract
1. Introduction
2. Variola Virus, Variolation, Cowpox Virus, and Vaccination
3. Post Jenner: Vaccinia and Cow-Pox Intertwined
4. From Poison to Intercellular Obligate Parasite
5. DNA and Genomics: Cowpox Virus to Cowpox Viruses

6. CPXV Genes and Host Interactions
7. Animal Models of CPXV Infection
8. Pathology of Modern CPXVs
9. Reservoir Hosts and Geographic Range
10. Modern CPXV Hosts and Outbreaks
11. The Future of CPXVs
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schramm, B.; Locker, J.K. Cytoplasmic organization of POXvirus DNA replication. Traffic 2005, 6, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Henderson, D.A. The eradication of smallpox—An overview of the past, present, and future. Vaccine 2011, 29, D7–D9. [Google Scholar] [CrossRef]
- Strassburg, M.A. The global eradication of smallpox. Am. J. Infect. Control 1982, 10, 53–59. [Google Scholar] [CrossRef]
- Hammarlund, E.; Lewis, M.W.; Carter, S.V.; Amanna, I.; Hansen, S.G.; Strelow, L.I.; Wong, S.W.; Yoshihara, P.; Hanifin, J.M.; Slifka, M.K. Multiple diagnostic techniques identify previously vaccinated individuals with protective immunity against monkeypox. Nat. Med. 2005, 11, 1005–1011. [Google Scholar] [CrossRef]
- Gilchuk, I.; Gilchuk, P.; Sapparapu, G.; Lampley, R.; Singh, V.; Kose, N.; Blum, D.L.; Hughes, L.J.; Satheshkumar, P.S.; Townsend, M.B.; et al. Cross-neutralizing and protective human antibody specificities to poxvirus infections. Cell 2016, 167, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Trindade, G.S.; Emerson, G.L.; Carroll, D.S.; Kroon, E.G.; Damon, I.K. Brazilian vaccinia viruses and their origins. Emerg. Infect. Dis. 2007, 13, 965–972. [Google Scholar] [CrossRef] [PubMed]
- Abrahão, J.S.; Campos, R.K.; Trindade, G.e.S.; Guimarães da Fonseca, F.; Ferreira, P.C.; Kroon, E.G. Outbreak of severe zoonotic vaccinia virus infection, Southeastern Brazil. Emerg. Infect. Dis. 2015, 21, 695–698. [Google Scholar] [CrossRef]
- Roy, P.; Chandramohan, A. Buffalopox disease in livestock and milkers, India. Emerg. Infect. Dis. 2021, 27, 1989–1991. [Google Scholar] [CrossRef]
- CDC. 2022 Monkeypox Outbreak Global Map. Available online: https://www.cdc.gov/poxvirus/monkeypox/response/2022/world-map.html (accessed on 6 February 2023).
- Jenner, E. On the origin of the vaccine inoculation. Med. Phys. J. 1801, 5, 505–508. [Google Scholar]
- Jenner, E. An Inquiry into the Causes and Effects of the Variolae Vaccinae, a Disease Discovered in Some of the Western Counties of England, Particularly Gloucestershire and Known by the Name of Cow Pox; Sampson Low: London, UK, 1798. [Google Scholar]
- Bailey, I. Edward Jenner (1749–1823): Naturalist, scientist, country doctor, benefactor to mankind. J. Med. Biogr. 1996, 4, 63–70. [Google Scholar] [CrossRef]
- Baxby, D. Jenner, Edward. 2009. Available online: https://www.oxforddnb.com/display/10.1093/ref:odnb/9780198614128.001.0001/odnb-9780198614128-e-14749 (accessed on 6 February 2023).
- Franke, A.; Pfaff, F.; Jenckel, M.; Hoffmann, B.; Höper, D.; Antwerpen, M.; Meyer, H.; Beer, M.; Hoffmann, D. Classification of cowpox viruses into several distinct clades and identification of a novel lineage. Viruses 2017, 9, 142. [Google Scholar] [CrossRef] [PubMed]
- ICTV. Current ICTV Taxonomy Release. 2022. Available online: https://ictv.global/taxonomy (accessed on 6 February 2023).
- Gubser, C.; Hué, S.; Kellam, P.; Smith, G.L. Poxvirus genomes: A phylogenetic analysis. J. Gen. Virol. 2004, 85, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Downie, A.W. A study of the lesion produced experimentally by cowpox virus. J. Pathol. Bacteriol. 1939, 48, 361–379. [Google Scholar]
- Kurth, A.; Straube, M.; Kuczka, A.; Dunsche, A.J.; Meyer, H.; Nitsche, A. Cowpox virus outbreak in banded mongooses (Mungos mungo) and jaguarundis (Herpailurus yagouaroundi) with a time-delayed infection to humans. PLoS ONE 2009, 4, e6883. [Google Scholar] [CrossRef] [PubMed]
- Baxby, D.; Bennett, M.; Getty, B. Human cowpox 1969–93: A review based on 54 cases. Br. J. Dermatol. 1994, 131, 598–607. [Google Scholar] [CrossRef]
- Campe, H.; Zimmermann, P.; Glos, K.; Bayer, M.; Bergemann, H.; Dreweck, C.; Graf, P.; Weber, B.K.; Meyer, H.; Büttner, M.; et al. Cowpox virus transmission from pet rats to humans, Germany. Emerg. Infect. Dis. 2009, 15, 777–780. [Google Scholar] [CrossRef]
- Antwerpen, M.H.; Georgi, E.; Nikolic, A.; Zoeller, G.; Wohlsein, P.; Baumgärtner, W.; Peyrefitte, C.; Charrel, R.; Meyer, H. Use of next generation sequencing to study two cowpox virus outbreaks. PeerJ 2019, 7, e6561. [Google Scholar] [CrossRef]
- Babkin, I.V.; Babkina, I.N. The origin of the variola virus. Viruses 2015, 7, 1100–1112. [Google Scholar] [CrossRef]
- Littman, R.J.; Littman, M.L. Galen and the Antonine plague. Am. J. Philol. 1973, 94, 243–255. [Google Scholar] [CrossRef]
- Fenner, F. Smallpox and its eradication; World Health Organization: Geneva, Switzerland, 1988; Volume 6, p. 1460. [Google Scholar]
- Berche, P. Life and death of smallpox. Presse Med. 2022, 51, 104117. [Google Scholar] [CrossRef]
- Suzuki, A. Smallpox and the epidemiological heritage of modern Japan: Towards a total history. Med. Hist. 2011, 55, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Patterson, K.B.; Runge, T. Smallpox and the native American. Am. J. Med. Sci 2002, 323, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Slifka, M.K.; Hanifin, J.M. Smallpox: The basics. Dermatol. Clin. 2004, 22, 263–274. [Google Scholar] [CrossRef]
- McFadden, G. Poxvirus tropism. Nat. Rev. Microbiol. 2005, 3, 201–213. [Google Scholar] [CrossRef]
- Gross, C.P.; Sepkowitz, K.A. The myth of the medical breakthrough: Smallpox, vaccination, and Jenner reconsidered. Int. J. Infect. Dis. 1998, 3, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Macgowan, D. Report on the health of Wenchow for the half-year ended 31 March 1884. Shanghai Chin. Imp. Marit. Cust. Med. Rep. 1884, 27, 9–18. [Google Scholar]
- Boylston, A. The origins of inoculation. J. R. Soc. Med. 2012, 105, 309–313. [Google Scholar] [CrossRef]
- Needham, J.; Wang, L.; Bollingen Foundation Collection (Library of Congress). Science and Civilisation in China; University Press: Cambridge, UK, 2000. [Google Scholar]
- Holwell, J.Z.; Becket, T.; De Hondt, P.A. An Account of The Manner Of Inoculating For The Small Pox in The East Indies: With Some Observations on the Practice and Mode of Treating That Disease in those Parts; T. Becket: London, UK, 1767. [Google Scholar]
- Bird, A. James Jurin and the avoidance of bias in collecting and assessing evidence on the effects of variolation. J. R. Soc. Med. 2019, 112, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Mercer, A.A.; Schmidt, A.; Weber, O.F. Poxviruses; Birkhäuser: Basel, Switzerland, 2007. [Google Scholar]
- Toledo-Pereyra, L.H. Zabdiel Boylston. First American surgeon of the English colonies in North America. J. Investig. Surg. 2006, 19, 5–10. [Google Scholar] [CrossRef]
- Riedel, S. Edward Jenner and the history of smallpox and vaccination. Bayl. Univ. Med. Cent. Proc. 2005, 18, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.C. The historic evolution of variolation. BJH Hosp. 1913, 24, 72. [Google Scholar]
- Jenner, E. Letter to Dr Heberden, the life of Edward Jenner, MD, with Illustrations of his doctrines and selections from his correspondence. Baron J. Lond. Henry Colburn 1838, 1, 39–40. [Google Scholar]
- Boylston, A.W. The myth of the milkmaid. N. Engl. J. Med. 2018, 378, 414–415. [Google Scholar] [CrossRef] [PubMed]
- Plett, P.C. Peter Plett and other discoverers of cowpox vaccination before Edward Jenner. Sudhoffs Arch. 2006, 90, 219–232. [Google Scholar] [PubMed]
- Pearson, G. An Inquiry Concerning the History of the Cowpox: Principally with a View to Supersede and Extinguish the Smallpox; J. Johnson: London, UK, 1798; pp. 102–104. [Google Scholar]
- Creighton, C. Jenner and Vaccination: A Strange Chapter of Medical History; Sonnenschein: London, UK, 1889. [Google Scholar]
- Essbauer, S.; Pfeffer, M.; Meyer, H. Zoonotic poxviruses. Vet. Microbiol. 2010, 140, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Franco-Paredes, C.; Lammoglia, L.; Santos-Preciado, J.I. The Spanish royal philanthropic expedition to bring smallpox vaccination to the New World and Asia in the 19th century. Clin. Infect. Dis. 2005, 41, 1285–1289. [Google Scholar] [CrossRef]
- ‘Vaccine’: The Word’s History Ain’t Pretty. Available online: https://www.merriam-webster.com/words-at-play/vaccine-the-words-history-aint-pretty (accessed on 6 February 2023).
- Berche, P. Louis Pasteur, from crystals of life to vaccination. Clin. Microbiol. Infect. 2012, 18, 1–6. [Google Scholar] [CrossRef]
- Duggan, A.T.; Klunk, J.; Porter, A.F.; Dhody, A.N.; Hicks, R.; Smith, G.L.; Humphreys, M.; McCollum, A.M.; Davidson, W.B.; Wilkins, K.; et al. The origins and genomic diversity of american civil war era smallpox vaccine strains. Genome Biol. 2020, 21, 175. [Google Scholar] [CrossRef]
- Brinkmann, A.; Souza, A.R.V.; Esparza, J.; Nitsche, A.; Damaso, C.R. Re-assembly of nineteenth-century smallpox vaccine genomes reveals the contemporaneous use of horsepox and horsepox-related viruses in the USA. Genome Biol. 2020, 21, 286. [Google Scholar] [CrossRef]
- Schrick, L.; Tausch, S.H.; Dabrowski, P.W.; Damaso, C.R.; Esparza, J.; Nitsche, A. An early american smallpox vaccine based on horsepox. N. Engl. J. Med. 2017, 377, 1491–1492. [Google Scholar] [CrossRef]
- Esparza, J.; Nitsche, A.; Damaso, C.R. Beyond the myths: Novel findings for old paradigms in the history of the smallpox vaccine. PLoS Pathog. 2018, 14, e1007082. [Google Scholar] [CrossRef]
- Fields, B.N.; Knipe, D.M.; Howley, P.M. Fields virology, 4th ed.; Fields, B., Knipe, D., Howley, P., Griffin, D., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2002. [Google Scholar]
- Huygelen, C. Jenner’s cowpox vaccine in light of current vaccinology. Verh. K. Acad. Voor Geneeskd. Van Belg. 1996, 58, 479–536. [Google Scholar]
- Sympson, E.M. Notes of a case of accidental cow-pox. Br. Med. J. 1892, 1, 115–116. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.J. An outbreak of cowpox. Am. J. Public Health 1915, 5, 1036–1037. [Google Scholar] [CrossRef]
- Crookshank, E. An Investigation of an outbreak of cow-pox in wiltshire, with a comparative account of some previous outbreaks in England, Germany, and France. Br. Med. J. 1888, 2, 1–5. [Google Scholar]
- Damaso, C.R. Revisiting Jenner’s mysteries, the role of the Beaugency lymph in the evolutionary path of ancient smallpox vaccines. Lancet Infect. Dis. 2018, 18, e55–e63. [Google Scholar] [CrossRef]
- Hime, T.W. Successful transformation of small-pox into cow-pox. Br. Med. J. 1892, 2, 116–120. [Google Scholar] [CrossRef]
- Reece, R.J. An account of the circumstances associated with an outbreak of disease among milch cows, horses, and their attendants, believed to be of the nature of “cow-pox” in the county of somersetshire in the year 1909, and considerations arising thereform. Proc. R. Soc. Med. 1909, 15, 13–16. [Google Scholar]
- Downie, A.W. Jenner’s cowpox inoculation. Br. Med. J. 1951, 2, 251–256. [Google Scholar] [CrossRef]
- Horgan, E.S. The experimental transformation of variola to vaccinia. J. Hyg. 1938, 38, 702–715. [Google Scholar] [CrossRef]
- Ivanowski, D. Concerning the Mosaic Disease of the Tobacco Plant. Am. Phytopathol. Soc. 1892, 7, 24–30. [Google Scholar]
- Beijerinck, M. Concerning a contagium vivum fluidium as a cause of the spot-disease of tobacco leaves. In Phytopathology Classic; APS: St. Paul, MN, USA, 1898; Volume 7, pp. 33–52. [Google Scholar]
- Lecoq, H. Discovery of the first virus, the tobacco mosaic virus: 1892 or 1898? C R Acad. Sci. III 2001, 324, 929–933. [Google Scholar] [CrossRef] [PubMed]
- Buist, J.B. Vaccinia and Variola: A Study of their Life History; J. Churchill: London, UK, 1887. [Google Scholar]
- Smadel, J.E.; Hoagland, C.L. Elementary bodies of vaccinia. Bacteriol. Rev. 1942, 6, 79–110. [Google Scholar] [CrossRef] [PubMed]
- Dawson, I.M.; McFarlane, A.S. Structure of an animal virus. Nature 1948, 161, 464–466. [Google Scholar] [CrossRef]
- Peters, D. Morphology of resting vaccinia virus. Nature 1956, 178, 1453–1455. [Google Scholar] [CrossRef]
- Davies Twiston, J.H.; Janes, L.R.; Downie, A.W. Cowpox infection in farmworkers. Lancet 1938, 232, 1534–1538. [Google Scholar]
- Cheville, N.F. Cytopathology in Viral Diseases; S. Karger: Basel, Switzerland, 1975; p. 4. [Google Scholar]
- Fenner, F. The biological characters of several strains of vaccinia, cowpox and rabbitpox viruses. Virology 1958, 5, 502–529. [Google Scholar] [CrossRef] [PubMed]
- Baxby, D. Variability in the characteristics of pocks produced on the chick chorioallantois by white pock mutants of cowpox and other poxviruses. J. Hyg. 1969, 67, 637–647. [Google Scholar] [CrossRef]
- Pickup, D.J.; Ink, B.S.; Parsons, B.L.; Hu, W.; Joklik, W.K. Spontaneous deletions and duplications of sequences in the genome of cowpox virus. Proc. Natl. Acad. Sci. USA 1984, 81, 6817–6821. [Google Scholar] [CrossRef]
- Howard, A.R.; Moss, B. Formation of orthopoxvirus cytoplasmic A-type inclusion bodies and embedding of virions are dynamic processes requiring microtubules. J. Virol. 2012, 86, 5905–5914. [Google Scholar] [CrossRef]
- Buddingh, G.J. The nomenclature and classification of the pox group of viruses. Ann. N. Y. Acad. Sci. 1953, 56, 561–566. [Google Scholar] [CrossRef]
- Downie, A.W. The immunological relationship of the virus of spontaneous cowpox to vaccinia virus. Br. J. Exp. Pathol. 1939, 20, 158–167. [Google Scholar]
- Matthews, R.E.F. Classification and nomenclature of viruses. Fourth report of the International committee on taxonomy of viruses. Intervirology 1982, 17, 1–199. [Google Scholar] [CrossRef]
- Nakano, J.H. Evaluation of virological laboratory methods for smallpox diagnosis. Bull. World Health Organ. 1973, 48, 529. [Google Scholar] [PubMed]
- Turner, A.; Baxby, D. Structural polypeptides of orthopoxvirus: Their distribution in various members and location within the virion. J. Gen. Virol. 1979, 45, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, J.; Baxby, D.; Bennett, M.; Gaskell, R.M.; Gaskell, C.J. Characterization of orthopoxviruses isolated from feline infections in Britain. Arch. Virol. 1992, 125, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Kurth, A.; Nitsche, A. Fowler’s Zoo and Wild Animal Medicine; Elsevier Health Sciences: London, UK, 2011; pp. 32–37. [Google Scholar]
- Pilaski, J.; Rösen-Wolff, A. Virus Infection in Zoo-Kept Mammals; Springer: Boston, MA, USA, 1988; pp. 83–100. [Google Scholar]
- Zwart, P.; Gispen, R.; Peters, J.C. Cowpox in okapis okapia johnstoni at rotterdam zoo. Br. Vet. J. 1971, 127, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Marennikova, S.S.; Maltseva, N.N.; Korneeva, V.I.; Garanina, N. Outbreak of pox disease among carnivora (felidae) and edentata. J. Infect. Dis. 1977, 135, 358–366. [Google Scholar] [CrossRef]
- Baxby, D.; Ghaboosi, B. Laboratory characteristics of poxviruses isolated from captive elephants in Germany. J. Gen. Virol 1977, 37, 407–414. [Google Scholar] [CrossRef]
- Pilaski, J.; Rösen, A.; Darai, G. Comparative analysis of the genomes of orthopoxviruses isolated from elephant, rhinoceros, and okapi by restriction enzymes. Brief report. Arch. Virol. 1986, 88, 135–142. [Google Scholar] [CrossRef]
- Alcam, A.; Khanna, A.; Paul, N.L.; Smith, G.L. Vaccinia virus strains lister, USSR and evans express soluble and cell-surface tumour necrosis factor receptors. J. Gen. Virol. 1999, 80, 949–959. [Google Scholar] [CrossRef] [PubMed]
- ICTV. Current ICTV Taxonomy Release. 2023. Available online: https://ictv.global/ (accessed on 6 February 2023).
- Motley, R.J.; Holt, P.J. Cowpox presenting with sporotrichoid spread: A case report. Br. J. Dermatol. 1990, 122, 705–708. [Google Scholar] [CrossRef] [PubMed]
- Marennikova, S.S.; Shelukhina, E.M.; Efremova, E.V. New outlook on the biology of cowpox virus. Acta Virol. 1984, 28, 437–444. [Google Scholar] [PubMed]
- Wildy, P. Classification and Nomenclature of Viruses: First Report of the International Committee on Nomenclature of Viruses; Karger Publishers: Basel, Switzerland, 1971. [Google Scholar]
- Fenner, F.; Pereira, H.G.; Porterfield, J.S.; Joklik, W.K.; Downie, A.W. Family and generic names for viruses approved by the international committee on taxonomy of viruses. Intervirology 1974, 3, 193–198. [Google Scholar] [PubMed]
- Fenner, F. The Orthopoxviruses; Academic Press Inc.: San Diego, CA, USA, 1989; pp. 8–10. [Google Scholar]
- Lwoff, A.; Horne, R.; Tournier, P. A system of viruses. Cold Spring Harb. Symp. Quant. Biol. 1962, 27, 51–55. [Google Scholar] [CrossRef]
- Holland, J.J.; De La Torre, J.C.; Steinhauer, D.A. RNA virus populations as quasispecies. Curr. Top. Microbiol. Immunol. 1992, 176, 1–20. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H. Genetical implications of the structure of deoxyribonucleic acid. Nature 1953, 171, 964–967. [Google Scholar] [CrossRef]
- Avery, O.T.; Macleod, C.M.; McCarty, M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 1944, 79, 137–158. [Google Scholar] [CrossRef]
- Judson, H.F. The Eighth Day of Creation: Makers of the Revolution in Biology; CSHL Press: Plainview, NY, USA, 1996. [Google Scholar]
- Hoagland, C.L.; Lavin, G.I.; Smadel, J.E.; Rivers, T.M. Constituents of elementary bodies of vaccinia: II. Properties of nucleic acid obtained from vaccine virus. J. Exp. Med. 1940, 72, 139–147. [Google Scholar] [CrossRef]
- Hyde, J.M.; Gafford, L.G.; Randall, C.C. Molecular weight determination of fowlpox virus DNA by electron microscopy. Virology 1967, 33, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Sarov, I.; Becker, Y. Studies on vaccinia virus DNA. Virology 1967, 33, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Woodson, B. Recent progress in poxvirus research. Bacteriol. Rev. 1968, 32, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Baroudy, B.M.; Venkatesan, S.; Moss, B. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell 1982, 28, 315–324. [Google Scholar] [CrossRef]
- Luria, S.E.; Human, M.L. A nonhereditary, host-induced variation of bacterial viruses. J. Bacteriol. 1952, 64, 557–569. [Google Scholar] [CrossRef]
- Lederberg, S.; Meselson, M. Degradation of non-replicating bacteriophage DNA in non-accepting cells. J. Mol. Biol. 1964, 8, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Meselson, M.; Yuan, R. DNA restriction enzyme from E. coli. Nature 1968, 217, 1110–1114. [Google Scholar] [CrossRef]
- Loenen, W.A.; Dryden, D.T.; Raleigh, E.A.; Wilson, G.G.; Murray, N.E. Highlights of the DNA cutters: A short history of the restriction enzymes. Nucleic Acids Res. 2014, 42, 3–19. [Google Scholar] [CrossRef]
- Helling, R.B.; Goodman, H.M.; Boyer, H.W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J. Virol. 1974, 14, 1235–1244. [Google Scholar] [CrossRef]
- Esposito, J.J.; Obijeski, J.F.; Nakano, J.H. Orthopoxvirus DNA: Strain differentiation by electrophoresis of restriction endonuclease fragmented virion DNA. Virology 1978, 89, 53–66. [Google Scholar] [CrossRef]
- Archard, L.C.; Mackett, M. Restriction endonuclease analysis of red cowpox virus and its white pock variant. J. Gen. Virol. 1979, 45, 51–63. [Google Scholar] [CrossRef]
- Esposito, J.J.; Knight, J.C. Orthopoxvirus DNA: A comparison of restriction profiles and maps. Virology 1985, 143, 230–251. [Google Scholar] [CrossRef]
- Rösen, A.; Pilaski, J.; Darai, G. Genomic characterization of a poxvirus isolated from a child. Med. Microbiol. Immunol. 1987, 176, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, A.; Fenner, F. Methods for comparing sequence data such as restriction endonuclease maps or nucleotide sequences of viral nucleic acid molecules. J. Virol. Methods 1984, 9, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Saiki, R.K.; Gelfand, D.H.; Stoffel, S.; Scharf, S.J.; Higuchi, R.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 1988, 239, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.; Schay, C.; Mahnel, H.; Pfeffer, M. Characterization of orthopoxviruses isolated from man and animals in Germany. Arch. Virol. 1999, 144, 491–501. [Google Scholar] [CrossRef]
- Tryland, M.; Sandvik, T.; Hansen, H.; Haukenes, G.; Holtet, L.; Bennett, M.; Mehl, R.; Moens, U.; Olsvik, O.; Traavik, T. Characteristics of four cowpox virus isolates from Norway and Sweden. APMIS 1998, 106, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Coulson, A.R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 1975, 94, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Pickup, D.J.; Bastia, D.; Stone, H.O.; Joklik, W.K. Sequence of terminal regions of cowpox virus DNA: Arrangement of repeated and unique sequence elements. Proc. Natl. Acad. Sci. USA 1982, 79, 7112–7116. [Google Scholar] [CrossRef]
- Pickup, D.J.; Ink, B.S.; Hu, W.; Ray, C.A.; Joklik, W.K. Hemorrhage in lesions caused by cowpox virus is induced by a viral protein that is related to plasma protein inhibitors of serine proteases. Proc. Natl. Acad. Sci. USA 1986, 83, 7698–7702. [Google Scholar] [CrossRef]
- Kaysser, P.; von Bomhard, W.; Dobrzykowski, L.; Meyer, H. Genetic diversity of feline cowpox virus, Germany 2000–2008. Vet. Microbiol. 2010, 141, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Ninove, L.; Domart, Y.; Vervel, C.; Voinot, C.; Salez, N.; Raoult, D.; Meyer, H.; Capek, I.; Zandotti, C.; Charrel, R.N. Cowpox virus transmission from pet rats to humans, France. Emerg. Infect. Dis. 2009, 15, 781–784. [Google Scholar] [CrossRef]
- Shchelkunov, S.N.; Safronov, P.F.; Totmenin, A.V.; Petrov, N.A.; Ryazankina, O.I.; Gutorov, V.V.; Kotwal, G.J. The genomic sequence analysis of the left and right species-specific terminal region of a cowpox virus strain reveals unique sequences and a cluster of intact ORFs for immunomodulatory and host range proteins. Virology 1998, 243, 432–460. [Google Scholar] [CrossRef]
- Mauldin, M.R.; Antwerpen, M.; Emerson, G.L.; Li, Y.; Zoeller, G.; Carroll, D.S.; Meyer, H. Cowpox virus: What’s in a Name? Viruses 2017, 9, 101. [Google Scholar] [CrossRef] [PubMed]
- Emerson, G.L.; Li, Y.; Frace, M.A.; Olsen-Rasmussen, M.A.; Khristova, M.L.; Govil, D.; Sammons, S.A.; Regnery, R.L.; Karem, K.L.; Damon, I.K.; et al. The phylogenetics and ecology of the orthopoxviruses endemic to North America. PLoS ONE 2009, 4, e7666. [Google Scholar] [CrossRef]
- Hendrickson, R.C.; Wang, C.; Hatcher, E.L.; Lefkowitz, E.J. Orthopoxvirus genome evolution: The role of gene loss. Viruses 2010, 2, 1933–1967. [Google Scholar] [CrossRef]
- Qin, L.; Favis, N.; Famulski, J.; Evans, D.H. Evolution of and evolutionary relationships between extant vaccinia virus strains. J. Virol. 2015, 89, 1809–1824. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.G.; Guagliardo, S.A.J.; Nakazawa, Y.J.; Doty, J.B.; Mauldin, M.R. Understanding orthopoxvirus host range and evolution: From the enigmatic to the usual suspects. Curr. Opin. Virol. 2018, 28, 108–115. [Google Scholar] [CrossRef]
- Goebel, S.J.; Johnson, G.P.; Perkus, M.E.; Davis, S.W.; Winslow, J.P.; Paoletti, E. The complete DNA sequence of vaccinia virus. Virology 1990, 179, 247–266. [Google Scholar] [CrossRef]
- Upton, C.; Slack, S.; Hunter, A.L.; Ehlers, A.; Roper, R.L. Poxvirus orthologous clusters: Toward defining the minimum essential poxvirus genome. J. Virol. 2003, 77, 7590–7600. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, A.; Osborne, J.; Slack, S.; Roper, R.L.; Upton, C. Poxvirus orthologous clusters (POCs). Bioinformatics 2002, 18, 1544–1545. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D.S.; Emerson, G.L.; Li, Y.; Sammons, S.; Olson, V.; Frace, M.; Nakazawa, Y.; Czerny, C.P.; Tryland, M.; Kolodziejek, J.; et al. Chasing Jenner’s vaccine: Revisiting cowpox virus classification. PLoS ONE 2011, 6, e23086. [Google Scholar] [CrossRef]
- Dabrowski, P.W.; Radonić, A.; Kurth, A.; Nitsche, A. Genome-wide comparison of cowpox viruses reveals a new clade related to Variola virus. PLoS ONE 2013, 8, e79953. [Google Scholar] [CrossRef]
- Hoffmann, D.; Franke, A.; Jenckel, M.; Tamošiūnaitė, A.; Schluckebier, J.; Granzow, H.; Hoffmann, B.; Fischer, S.; Ulrich, R.G.; Höper, D.; et al. Out of the reservoir: Phenotypic and genotypic characterization of a novel cowpox virus isolated from a common vole. J. Virol. 2015, 89, 10959–10969. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.; Carstens, E.B. Recently agreed changes to the international code of virus classification and nomenclature. Arch. Virol. 2013, 158, 2633–2639. [Google Scholar] [CrossRef]
- Diaz-Cánova, D.; Moens, U.L.; Brinkmann, A.; Nitsche, A.; Okeke, M.I. Genomic sequencing and analysis of a novel human cowpox virus with mosaic sequences from north america and old world orthopoxvirus. Front. Microbiol. 2022, 13, 868887. [Google Scholar] [CrossRef]
- Diaz-Cánova, D.; Mavian, C.; Brinkmann, A.; Nitsche, A.; Moens, U.; Okeke, M.I. Genomic Sequencing and phylogenomics of cowpox virus. Viruses 2022, 14, 2134. [Google Scholar] [CrossRef]
- Marennikova, S.S.; Gashnikov, P.V.; Zhukova, O.A.; Riabchikova, E.I.; Strel’tsov, V.V.; Riazankina, O.I.; Chekunova, E.V.; Ianova, N.N.; Shchelkunov, S.N. The biotype and genetic characteristics of an isolate of the cowpox virus causing infection in a child. Zhurnal Mikrobiol. Epidemiol. I Immunobiol. 1996, 4, 6–10. [Google Scholar]
- Pelkonen, P.M.; Tarvainen, K.; Hynninen, A.; Kallio, E.R.; Henttonen, K.; Palva, A.; Vaheri, A.; Vapalahti, O. Cowpox with severe generalized eruption, Finland. Emerg. Infect. Dis. 2003, 9, 1458–1461. [Google Scholar] [CrossRef] [PubMed]
- Popova, A.Y.; Maksyutov, R.A.; Taranov, O.S.; Tregubchak, T.V.; Zaikovskaya, A.V.; Sergeev, A.A.; Vlashchenko, I.V.; Bodnev, S.A.; Ternovoi, V.A.; Alexandrova, N.S.; et al. Cowpox in a human, Russia, 2015. Epidemiol. Infect. 2017, 145, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.; Kurth, A.; Hessler, F.; Kramp, H.; Gokel, M.; Hoffmann, R.; Kuczka, A.; Nitsche, A. Cowpox virus infection in pet rat owners: Not always immediately recognized. Dtsch Arztebl. Int. 2009, 106, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Kalthoff, D.; König, P.; Meyer, H.; Beer, M.; Hoffmann, B. Experimental cowpox virus infection in rats. Vet. Microbiol. 2011, 153, 382–386. [Google Scholar] [CrossRef]
- Ducournau, C.; Ferrier-Rembert, A.; Ferraris, O.; Joffre, A.; Favier, A.L.; Flusin, O.; Van Cauteren, D.; Kecir, K.; Auburtin, B.; Védy, S.; et al. Concomitant human infections with 2 cowpox virus strains in related cases, France, 2011. Emerg. Infect. Dis. 2013, 19, 1996–1999. [Google Scholar] [CrossRef]
- Gavrilova, E.V.; Shcherbakov, D.N.; Maksyutov, R.A.; Shchelkunov, S.N. Development of real-time PCR assay for specific detection of cowpox virus. J. Clin. Virol. 2010, 49, 37–40. [Google Scholar] [CrossRef]
- Mätz-Rensing, K.; Ellerbrok, H.; Ehlers, B.; Pauli, G.; Floto, A.; Alex, M.; Czerny, C.P.; Kaup, F.J. Fatal poxvirus outbreak in a colony of new world monkeys. Vet. Pathol. 2006, 43, 212–218. [Google Scholar] [CrossRef]
- Prkno, A.; Hoffmann, D.; Goerigk, D.; Kaiser, M.; van Maanen, A.C.F.; Jeske, K.; Jenckel, M.; Pfaff, F.; Vahlenkamp, T.W.; Beer, M.; et al. Epidemiological investigations of four cowpox virus outbreaks in alpaca herds, Germany. Viruses 2017, 9, 344. [Google Scholar] [CrossRef]
- Franke, A.; Kershaw, O.; Jenckel, M.; König, L.; Beer, M.; Hoffmann, B.; Hoffmann, D. Fatal cowpox virus infection in an aborted foal. Vector Borne Zoonotic Dis. 2016, 16, 431–433. [Google Scholar] [CrossRef]
- Jungwirth, N.; Puff, C.; Köster, K.; Mischke, R.; Meyer, H.; Stark, A.; Thoma, B.; Zöller, G.; Seehusen, F.; Hewicker-Trautwein, M.; et al. Atypical cowpox virus infection in a series of cats. J. Comp. Pathol. 2018, 158, 71–76. [Google Scholar] [CrossRef]
- Eis-Hübinger, A.M.; Gerritzen, A.; Schneweis, K.E.; Pfeiff, B.; Pullmann, H.; Mayr, A.; Czerny, C.P. Fatal cowpox-like virus infection transmitted by cat. Lancet 1990, 336, 880. [Google Scholar] [CrossRef] [PubMed]
- Esposito, J.J.; Sammons, S.A.; Frace, A.M.; Osborne, J.D.; Olsen-Rasmussen, M.; Zhang, M.; Govil, D.; Damon, I.K.; Kline, R.; Laker, M.; et al. Genome sequence diversity and clues to the evolution of variola (smallpox) virus. Science 2006, 313, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Tryland, M.; Myrmel, H.; Holtet, L.; Haukenes, G.; Traavik, T. Clinical cowpox cases in Norway. Scand. J. Infect. Dis. 1998, 30, 301–303. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, M.; Burck, G.; Meyer, H. Cowpox viruses in Germany: An analysis of 5 cases in 1998. Berl. Munch. Tierarztl. Wochenschr. 1999, 112, 334–338. [Google Scholar] [PubMed]
- Stagegaard, J.; Kurth, A.; Stern, D.; Dabrowski, P.W.; Pocknell, A.; Nitsche, A.; Schrick, L. Seasonal recurrence of cowpox virus outbreaks in captive cheetahs (Acinonyx jubatus). PLoS ONE 2017, 12, e0187089. [Google Scholar] [CrossRef]
- Andreani, J.; Arnault, J.P.; Bou Khalil, J.Y.; Abrahão, J.; Tomei, E.; Vial, E.; Le Bideau, M.; Raoult, D.; La Scola, B. Atypical cowpox virus infection in smallpox-vaccinated patient, France. Emerg. Infect. Dis. 2019, 25, 212–219. [Google Scholar] [CrossRef]
- Kalthoff, D.; Bock, W.I.; Hühn, F.; Beer, M.; Hoffmann, B. Fatal cowpox virus infection in cotton-top tamarins (Saguinus oedipus) in Germany. Vector Borne Zoonotic Dis. 2014, 14, 303–305. [Google Scholar] [CrossRef]
- Okeke, M.I.; Hansen, H.; Traavik, T. A naturally occurring cowpox virus with an ectromelia virus A-type inclusion protein gene displays atypical A-type inclusions. Infect. Genet. Evol. 2012, 12, 160–168. [Google Scholar] [CrossRef]
- Jeske, K.; Weber, S.; Pfaff, F.; Imholt, C.; Jacob, J.; Beer, M.; Ulrich, R.G.; Hoffmann, D. Molecular detection and characterization of the first cowpox virus isolate derived from a bank vole. Viruses 2019, 11, 1075. [Google Scholar] [CrossRef]
- Senkevich, T.G.; Yutin, N.; Wolf, Y.I.; Koonin, E.V.; Moss, B. Ancient gene capture and recent gene loss shape the evolution of orthopoxvirus-host interaction genes. mBio 2021, 12, e0149521. [Google Scholar] [CrossRef]
- Peterson, A.T. Defining viral species: Making taxonomy useful. Virol. J. 2014, 11, 131. [Google Scholar] [CrossRef] [PubMed]
- Baxby, D. Is cowpox misnamed? A review of 10 human cases. Br. Med. J. 1977, 1, 1379–1381. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.; Gaskell, C.J.; Gaskell, R.M.; Baxby, D.; Gruffydd-Jones, T.J. Poxvirus infection in the domestic cat: Some clinical and epidemiological observations. Vet. Rec. 1986, 118, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Wendt, R.; Tittelbach, J.; Schrick, L.; Kellner, N.; Kalbitz, S.; Ruehe, B.; Michel, J.; Schliemann, S.; Elsner, P.; Lübbert, C.; et al. Generalized cowpox virus infection in an immunosuppressed patient. Int. J. Infect. Dis. 2021, 106, 276–278. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Jukes, T.H.; Cantor, C.R. Evolution of Protein Molecules; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Lefkowitz, E.J.; Wang, C.; Upton, C. Poxviruses: Past, present and future. Virus Res. 2006, 117, 105–118. [Google Scholar] [CrossRef]
- Alzhanova, D.; Früh, K. Modulation of the host immune response by cowpox virus. Microbes Infect. 2010, 12, 900–909. [Google Scholar] [CrossRef]
- Haller, S.L.; Peng, C.; McFadden, G.; Rothenburg, S. Poxviruses and the evolution of host range and virulence. Infect. Genet. Evol. 2014, 21, 15–40. [Google Scholar] [CrossRef]
- Bratke, K.A.; McLysaght, A.; Rothenburg, S. A survey of host range genes in poxvirus genomes. Infect. Genet. Evol. 2013, 14, 406–425. [Google Scholar] [CrossRef]
- Patel, D.D.; Pickup, D.J.; Joklik, W.K. Isolation of cowpox virus A-type inclusions and characterization of their major protein component. Virology 1986, 149, 174–189. [Google Scholar] [CrossRef]
- Funahashi, S.; Sato, T.; Shida, H. Cloning and characterization of the gene encoding the major protein of the A-type inclusion body of cowpox virus. J. Gen. Virol. 1988, 69, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Ichihashi, Y.; Matsumoto, S. The relationship between poxvirus and A-type inclusion body during double infection. Virology 1968, 36, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Okeke, M.I.; Adekoya, O.A.; Moens, U.; Tryland, M.; Traavik, T.; Nilssen, Ø. Comparative sequence analysis of A-type inclusion (ATI) and P4c proteins of orthopoxviruses that produce typical and atypical ATI phenotypes. Virus Genes 2009, 39, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Shida, H.; Tanabe, K.; Matsumoto, S. Mechanism of virus occlusion into A-type inclusion during poxvirus infection. Virology 1977, 76, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Kastenmayer, R.J.; Maruri-Avidal, L.; Americo, J.L.; Earl, P.L.; Weisberg, A.S.; Moss, B. Elimination of A-type inclusion formation enhances cowpox virus replication in mice: Implications for orthopoxvirus evolution. Virology 2014, 452, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Drillien, R.; Spehner, D.; Kirn, A. Host range restriction of vaccinia virus in Chinese hamster ovary cells: Relationship to shutoff of protein synthesis. J. Virol. 1978, 28, 843–850. [Google Scholar] [CrossRef]
- Spehner, D.; Gillard, S.; Drillien, R.; Kirn, A. A cowpox virus gene required for multiplication in Chinese hamster ovary cells. J. Virol. 1988, 62, 1297–1304. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Patton, E.E.; Willems, A.R.; Tyers, M. Combinatorial control in ubiquitin-dependent proteolysis: Don’t Skp the F-box hypothesis. Trends Genet. 1998, 14, 236–243. [Google Scholar] [CrossRef]
- Zhang, F.; Meng, X.; Townsend, M.B.; Satheshkumar, P.S.; Xiang, Y. Identification of CP77 as the third orthopoxvirus SAMD9 and SAMD9L inhibitor with unique specificity for a rodent SAMD9L. J. Virol. 2019, 93, e00225-19. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Meng, X.; Zhang, F.; Pathak, P.K.; Chaturvedi, J.; Coronado, J.; Morales, M.; Mao, Y.; Qian, S.B.; Deng, J.; et al. Structure and function of an effector domain in antiviral factors and tumor suppressors SAMD9 and SAMD9L. Proc. Natl. Acad. Sci. USA 2022, 119, e2116550119. [Google Scholar] [CrossRef] [PubMed]
- Conrad, S.J.; Raza, T.; Peterson, E.A.; Liem, J.; Connor, R.; Nounamo, B.; Cannon, M.; Liu, J. Myxoma virus lacking the host range determinant M062 stimulates cGAS-dependent type 1 interferon response and unique transcriptomic changes in human monocytes/macrophages. PLoS Pathog. 2022, 18, e1010316. [Google Scholar] [CrossRef]
- Liu, J.; Wennier, S.; McFadden, G. The immunoregulatory properties of oncolytic myxoma virus and their implications in therapeutics. Microbes Infect. 2010, 12, 1144–1152. [Google Scholar] [CrossRef]
- Früh, K.; Bartee, E.; Gouveia, K.; Mansouri, M. Immune evasion by a novel family of viral PHD/LAP-finger proteins of gamma-2 herpesviruses and poxviruses. Virus Res. 2002, 88, 55–69. [Google Scholar] [CrossRef]
- Dasgupta, A.; Hammarlund, E.; Slifka, M.K.; Früh, K. Cowpox virus evades CTL recognition and inhibits the intracellular transport of MHC class I molecules. J. Immunol. 2007, 178, 1654–1661. [Google Scholar] [CrossRef]
- Alzhanova, D.; Edwards, D.M.; Hammarlund, E.; Scholz, I.G.; Horst, D.; Wagner, M.J.; Upton, C.; Wiertz, E.J.; Slifka, M.K.; Früh, K. Cowpox virus inhibits the transporter associated with antigen processing to evade T cell recognition. Cell Host Microbe 2009, 6, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Luteijn, R.D.; Hoelen, H.; Kruse, E.; van Leeuwen, W.F.; Grootens, J.; Horst, D.; Koorengevel, M.; Drijfhout, J.W.; Kremmer, E.; Früh, K.; et al. Cowpox virus protein CPXV012 eludes CTLs by blocking ATP binding to TAP. J. Immunol. 2014, 193, 1578–1589. [Google Scholar] [CrossRef]
- Tamošiūnaitė, A.; Weber, S.; Schippers, T.; Franke, A.; Xu, Z.; Jenckel, M.; Pfaff, F.; Hoffmann, D.; Newell, M.; Tischer, B.K.; et al. What a difference a gene makes: Identification of virulence factors of cowpox virus. J. Virol. 2020, 94, e01625-19. [Google Scholar] [CrossRef]
- Park, C.; Peng, C.; Rahman, M.J.; Haller, S.L.; Tazi, L.; Brennan, G.; Rothenburg, S. Orthopoxvirus K3 orthologs show virus- and host-specific inhibition of the antiviral protein kinase PKR. PLoS Pathog. 2021, 17, e1009183. [Google Scholar] [CrossRef]
- Wu, S.; Kaufman, R.J. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J. Biol. Chem. 1997, 272, 1291–1296. [Google Scholar] [CrossRef] [PubMed]
- Smee, D.F.; Gowen, B.B.; Wandersee, M.K.; Wong, M.H.; Skirpstunas, R.T.; Baldwin, T.J.; Hoopes, J.D.; Sidwell, R.W. Differential pathogenesis of cowpox virus intranasal infections in mice induced by low and high inoculum volumes and effects of cidofovir treatment. Int. J. Antimicrob. Agents 2008, 31, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Marennikova, S.S. Field and experimental studies of poxvirus infections in rodents. Bull. World Health Organ. 1979, 57, 461–464. [Google Scholar] [PubMed]
- Maiboroda, A.D. Experimental infection of norvegian rats (Rattus norvegicus) with ratpox virus. Acta Virol. 1982, 26, 288–291. [Google Scholar]
- Breithaupt, A.; Kalthoff, D.; Deutskens, F.; König, P.; Hoffmann, B.; Beer, M.; Meyer, H.; Teifke, J.P. Clinical course and pathology in rats (Rattus norvegicus) after experimental cowpox virus infection by percutaneous and intranasal application. Vet. Pathol. 2012, 49, 941–949. [Google Scholar] [CrossRef]
- Franke, A.; Ulrich, R.G.; Weber, S.; Osterrieder, N.; Keller, M.; Hoffmann, D.; Beer, M. Experimental cowpox virus (CPXV) infections of bank voles: Exceptional clinical resistance and variable reservoir competence. Viruses 2017, 9, 391. [Google Scholar] [CrossRef] [PubMed]
- Weber, S.; Jeske, K.; Ulrich, R.G.; Imholt, C.; Jacob, J.; Beer, M.; Hoffmann, D. In vivo characterization of a bank vole-derived cowpox virus isolate in natural hosts and the rat model. Viruses 2020, 12, 237. [Google Scholar] [CrossRef] [PubMed]
- Kramski, M.; Mätz-Rensing, K.; Stahl-Hennig, C.; Kaup, F.J.; Nitsche, A.; Pauli, G.; Ellerbrok, H. A novel highly reproducible and lethal nonhuman primate model for orthopox virus infection. PLoS ONE 2010, 5, e10412. [Google Scholar] [CrossRef]
- Johnson, R.F.; Yellayi, S.; Cann, J.A.; Johnson, A.; Smith, A.L.; Paragas, J.; Jahrling, P.B.; Blaney, J.E. Cowpox virus infection of cynomolgus macaques as a model of hemorrhagic smallpox. Virology 2011, 418, 102–112. [Google Scholar] [CrossRef]
- Smith, A.L.; St Claire, M.; Yellayi, S.; Bollinger, L.; Jahrling, P.B.; Paragas, J.; Blaney, J.E.; Johnson, R.F. Intrabronchial inoculation of cynomolgus macaques with cowpox virus. J. Gen. Virol. 2012, 93, 159–164. [Google Scholar] [CrossRef]
- Duraffour, S.; Mertens, B.; Meyer, H.; van den Oord, J.J.; Mitera, T.; Matthys, P.; Snoeck, R.; Andrei, G. Emergence of cowpox: Study of the virulence of clinical strains and evaluation of antivirals. PLoS ONE 2013, 8, e55808. [Google Scholar] [CrossRef]
- Bourquain, D.; Nitsche, A. Cowpox virus but not vaccinia virus induces secretion of CXCL1, IL-8 and IL-6 and chemotaxis of monocytes in vitro. Virus Res. 2013, 171, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Salinas, L.; Verdugo-Rodriguez, A.; Rodriguez, L.L.; Borca, M.V. The role of interleukin 6 during viral infections. Front. Microbiol. 2019, 10, 1057. [Google Scholar] [CrossRef]
- Haydon, D.T.; Cleaveland, S.; Taylor, L.H.; Laurenson, M.K. Identifying reservoirs of infection: A conceptual and practical challenge. Emerg. Infect. Dis. 2002, 8, 1468–1473. [Google Scholar] [CrossRef] [PubMed]
- Lei, V.; Petty, A.J.; Atwater, A.R.; Wolfe, S.A.; MacLeod, A.S. Skin viral infections: Host Antiviral innate immunity and viral immune evasion. Front. Immunol. 2020, 11, 593901. [Google Scholar] [CrossRef]
- Baxby, D. Medical Microbiology. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- James, W.; Elston, D.; Treat, J.; Rosenbach, M.; Neuhaus, I. Andrews’ Diseases of the Skin: Clinical Dermatology; Elsevier: Edinburgh, UK, 2020; pp. 387–389. [Google Scholar]
- Tack, D.M.; Reynolds, M.G. Zoonotic poxviruses associated with companion animals. Animals 2011, 1, 377–395. [Google Scholar] [CrossRef] [PubMed]
- MacNeill, A.L. Comparative pathology of zoonotic orthopoxviruses. Pathogens 2022, 11, 892. [Google Scholar] [CrossRef]
- Haase, O.; Moser, A.; Rose, C.; Kurth, A.; Zillikens, D.; Schmidt, E. Generalized cowpox infection in a patient with darier disease. Br. J. Dermatol. 2011, 164, 1116–1118. [Google Scholar] [CrossRef]
- Fassbender, P.; Zange, S.; Ibrahim, S.; Zoeller, G.; Herbstreit, F.; Meyer, H. Generalized cowpox virus infection in a patient with HIV, Germany, 2012. Emerg. Infect. Dis. 2016, 22, 553–555. [Google Scholar] [CrossRef]
- Gazzani, P.; Gach, J.E.; Colmenero, I.; Martin, J.; Morton, H.; Brown, K.; Milford, D.V. Fatal disseminated cowpox virus infection in an adolescent renal transplant recipient. Pediatr. Nephrol. 2017, 32, 533–536. [Google Scholar] [CrossRef]
- Crouch, A.C.; Baxby, D.; McCracken, C.M.; Gaskell, R.M.; Bennett, M. Serological evidence for the reservoir hosts of cowpox virus in british wildlife. Epidemiol. Infect. 1995, 115, 185–191. [Google Scholar] [CrossRef]
- Bennett, M.; Crouch, A.J.; Begon, M.; Duffy, B.; Feore, S.; Gaskell, R.M.; Kelly, D.F.; McCracken, C.M.; Vicary, L.; Baxby, D. Cowpox in british voles and mice. J. Comp. Pathol. 1997, 116, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Chantrey, J.; Meyer, H.; Baxby, D.; Begon, M.; Bown, K.J.; Hazel, S.M.; Jones, T.; Montgomery, W.I.; Bennett, M. Cowpox: Reservoir hosts and geographic range. Epidemiol. Infect. 1999, 122, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Begon, M.; Hazel, S.M.; Baxby, D.; Bown, K.; Cavanagh, R.; Chantrey, J.; Jones, T.; Bennett, M. Transmission dynamics of a zoonotic pathogen within and between wildlife host species. Proc. Biol. Sci. 1999, 266, 1939–1945. [Google Scholar] [CrossRef] [PubMed]
- Telfer, S.; Bennett, M.; Bown, K.; Carslake, D.; Cavanagh, R.; Hazel, S.; Jones, T.; Begon, M. Infection with cowpox virus decreases female maturation rates in wild populations of woodland rodents. Oikos 2005, 109, 317–322. [Google Scholar]
- Kaplan, C.; Healing, T.D.; Evans, N.; Healing, L.; Prior, A. Evidence of infection by viruses in small british field rodents. J. Hyg. 1980, 84, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Hazel, S.M.; Bennett, M.; Chantrey, J.; Bown, K.; Cavanagh, R.; Jones, T.R.; Baxby, D.; Begon, M. A longitudinal study of an endemic disease in its wildlife reservoir: Cowpox and wild rodents. Epidemiol. Infect. 2000, 124, 551–562. [Google Scholar] [CrossRef]
- Boulanger, D.; Crouch, A.; Brochier, B.; Bennett, M.; Clément, J.; Gaskell, R.M.; Baxby, D.; Pastoret, P.P. Serological survey for orthopoxvirus infection of wild mammals in areas where a recombinant rabies virus is used to vaccinate foxes. Vet. Rec. 1996, 138, 247–249. [Google Scholar] [CrossRef]
- Sandvik, T.; Tryland, M.; Hansen, H.; Mehl, R.; Moens, U.; Olsvik, O.; Traavik, T. Naturally occurring orthopoxviruses: Potential for recombination with vaccine vectors. J. Clin. Microbiol. 1998, 36, 2542–2547. [Google Scholar] [CrossRef]
- Tryland, M.; Sandvik, T.; Mehl, R.; Bennett, M.; Traavik, T.; Olsvik, O. Serosurvey for orthopoxviruses in rodents and shrews from Norway. J. Wildl. Dis. 1998, 34, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Oldal, M.; Sironen, T.; Henttonen, H.; Vapalahti, O.; Madai, M.; Horváth, G.; Dallos, B.; Kutas, A.; Földes, F.; Kemenesi, G.; et al. Serologic survey of orthopoxvirus infection among rodents in hungary. Vector Borne Zoonotic Dis. 2015, 15, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Forbes, K.M.; Voutilainen, L.; Jääskeläinen, A.; Sironen, T.; Kinnunen, P.M.; Stuart, P.; Vapalahti, O.; Henttonen, H.; Huitu, O. Serological survey of rodent-borne viruses in finnish field voles. Vector Borne Zoonotic Dis. 2014, 14, 278–283. [Google Scholar] [CrossRef]
- Kinnunen, P.M.; Henttonen, H.; Hoffmann, B.; Kallio, E.R.; Korthase, C.; Laakkonen, J.; Niemimaa, J.; Palva, A.; Schlegel, M.; Ali, H.S.; et al. Orthopox virus infections in eurasian wild rodents. Vector Borne Zoonotic Dis. 2011, 11, 1133–1140. [Google Scholar] [CrossRef]
- Laakkonen, J.; Kallio-Kokko, H.; Oktem, M.A.; Blasdell, K.; Plyusnina, A.; Niemimaa, J.; Karataş, A.; Plyusnin, A.; Vaheri, A.; Henttonen, H. Serological survey for viral pathogens in turkish rodents. J. Wildl. Dis. 2006, 42, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Ashford, R.W. What it takes to be a reservoir host. Belg. J. Zool. 1997, 127, 85–90. [Google Scholar]
- Baxby, D.; Bennett, M. Cowpox: A re-evaluation of the risks of human cowpox based on new epidemiological information. Arch. Virol. Suppl. 1997, 13, 1–12. [Google Scholar] [CrossRef]
- Silva, N.I.O.; de Oliveira, J.S.; Kroon, E.G.; Trindade, G.S.; Drumond, B.P. Here, there, and everywhere: The wide host range and geographic distribution of zoonotic orthopoxviruses. Viruses 2020, 13, 43. [Google Scholar] [CrossRef] [PubMed]
- Von Bomhard, W.; Mauldin, E.A.; Breuer, W.; Pfleghaar, S.; Nitsche, A. Localized cowpox infection in a 5-month-old Rottweiler. Vet. Dermatol. 2011, 22, 111–114. [Google Scholar] [CrossRef]
- Kurth, A.; Wibbelt, G.; Gerber, H.P.; Petschaelis, A.; Pauli, G.; Nitsche, A. Rat-to-elephant-to-human transmission of cowpox virus. Emerg. Infect. Dis. 2008, 14, 670–671. [Google Scholar] [CrossRef]
- Martina, B.E.; van Doornum, G.; Dorrestein, G.M.; Niesters, H.G.; Stittelaar, K.J.; Wolters, M.A.; van Bolhuis, H.G.; Osterhaus, A.D. Cowpox virus transmission from rats to monkeys, the Netherlands. Emerg. Infect. Dis. 2006, 12, 1005–1007. [Google Scholar] [CrossRef]
- Fischer, C.P.; Romero, L.M. Chronic captivity stress in wild animals is highly species-specific. Conserv. Physiol. 2019, 7, coz093. [Google Scholar] [CrossRef]
- Hentschke, J.; Meyer, H.; Wittstatt, U.; Ochs, A.; Burkshardt, S.; Aue, A. Kuhpocken bei kanadischen bibern (castor fiver canadensis) und katzenbaren (ailurus fulgens). Tierärztliche Umschau. 1999, 54, 311–317. [Google Scholar]
- Bennett, M.; Gaskell, C.J.; Baxby, D.; Gaskell, R.M.; Kelly, D.F.; Naidoo, J. Feline cowpox virus infection. J. Small Anim. Pract. 1990, 31, 163–173. [Google Scholar]
- Appl, C.; von Bomhard, W.; Hanczaruk, M.; Meyer, H.; Bettenay, S.; Mueller, R. Feline cowpoxvirus infections in Germany: Clinical and epidemiological aspects. Berl. Munch. Tierarztl. Wochenschr. 2013, 126, 55–61. [Google Scholar] [PubMed]
- Scarff, D. Cowpox (orthopox virus) in the cat. Companion Anim. 2012, 17, 36–38. [Google Scholar] [CrossRef]
- Lawn, R. Risk of Cowpox to Small Animal Pracitioners. Vet. Rec. 2010, 166, 631. [Google Scholar] [PubMed]
- Scott, D.W.; Miller, W.H.; Griffin, C.E. Muller & Kirk’s Small Animal Dermatology; Viral, Rickettsial, and Protozoal Skin Diseases; W.B. Saunders: Philadelphia, PA, USA, 2001; pp. 517–542. [Google Scholar]
- McInerney, J.; Papasouliotis, K.; Simpson, K.; English, K.; Cook, S.; Milne, E.; Gunn-Moore, D.A. Pulmonary cowpox in cats: Five cases. J. Feline Med. Surg 2016, 18, 518–525. [Google Scholar] [CrossRef]
- Meyer, H.; Osterrieder, N.; Czerny, C.P. Identification of binding sites for neutralizing monoclonal antibodies on the 14-kDa fusion protein of orthopox viruses. Virology 1994, 200, 778–783. [Google Scholar] [CrossRef] [PubMed]
- Wolfs, T.F.; Wagenaar, J.A.; Niesters, H.G.; Osterhaus, A.D. Rat-to-human transmission of cowpox infection. Emerg. Infect. Dis. 2002, 8, 1495–1496. [Google Scholar] [CrossRef] [PubMed]
- Elsendoorn, A.; Agius, G.; Le Moal, G.; Aajaji, F.; Favier, A.L.; Wierzbicka-Hainault, E.; Béraud, G.; Flusin, O.; Crance, J.M.; Roblot, F. Severe ear chondritis due to cowpox virus transmitted by a pet rat. J. Infect. 2011, 63, 391–393. [Google Scholar] [CrossRef]
- Piskurek, O.; Okada, N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc. Natl. Acad. Sci. USA 2007, 104, 12046–12051. [Google Scholar] [CrossRef]
- Kunasekaran, M.P.; Chen, X.; Costantino, V.; Chughtai, A.A.; MacIntyre, C.R. Evidence for residual immunity to smallpox after vaccination and implications for re-emergence. Mil. Med. 2019, 184, e668–e679. [Google Scholar] [CrossRef] [PubMed]
- Taub, D.D.; Ershler, W.B.; Janowski, M.; Artz, A.; Key, M.L.; McKelvey, J.; Muller, D.; Moss, B.; Ferrucci, L.; Duffey, P.L.; et al. Immunity from smallpox vaccine persists for decades: A longitudinal study. Am. J. Med. 2008, 121, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Ferrier, A.; Frenois-Veyrat, G.; Schvoerer, E.; Henard, S.; Jarjaval, F.; Drouet, I.; Timera, H.; Boutin, L.; Mosca, E.; Peyrefitte, C.; et al. Fatal cowpox virus infection in human fetus, France, 2017. Emerg. Infect. Dis. 2021, 27, 2570–2577. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.G. Fetal infection in chickenpox and alastrim, with histopathologic study of the placenta. Pediatrics 1963, 32, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Fulginiti, V.A.; Papier, A.; Lane, J.M.; Neff, J.M.; Henderson, D.A. Smallpox vaccination: A review, part II. Adverse events. Clin. Infect. Dis. 2003, 37, 251–271. [Google Scholar] [CrossRef]
- Vellozzi, C.; Lane, J.M.; Averhoff, F.; Maurer, T.; Norton, S.; Damon, I.; Casey, C. Generalized vaccinia, progressive vaccinia, and eczema vaccinatum are rare following smallpox (vaccinia) vaccination: United States surveillance, 2003. Clin. Infect. Dis. 2005, 41, 689–697. [Google Scholar] [CrossRef]
- Padval, D.G. Human to human transmission of poxviruses have been observed previously. Dtsch. Arztebl. Int. 2009, 106, 623–624. [Google Scholar] [CrossRef]
- Heskin, J.; Belfield, A.; Milne, C.; Brown, N.; Walters, Y.; Scott, C.; Bracchi, M.; Moore, L.S.; Mughal, N.; Rampling, T.; et al. Transmission of monkeypox virus through sexual contact—A novel route of infection. J. Infect. 2022, 85, 334–363. [Google Scholar] [CrossRef]
- Antinori, A.; Mazzotta, V.; Vita, S.; Carletti, F.; Tacconi, D.; Lapini, L.E.; D’Abramo, A.; Cicalini, S.; Lapa, D.; Pittalis, S.; et al. Epidemiological, clinical and virological characteristics of four cases of monkeypox support transmission through sexual contact, Italy, May 2022. Eurosurveillance 2022, 27, 2200421. [Google Scholar] [CrossRef] [PubMed]
- Bergman, A.; McGee, K.; Farley, J.; Kwong, J.; McNabb, K.; Voss, J. Combating stigma in the era of monkeypox—Is history repeating itself? J. Assoc. Nurses AIDS Care 2022, 33, 668–675. [Google Scholar] [CrossRef]
- Brennan, G.; Stoian, A.M.M.; Yu, H.; Rahman, M.J.; Banerjee, S.; Stroup, J.N.; Park, C.; Tazi, L.; Rothenburg, S. Molecular mechanisms of poxvirus evolution. mBio 2022, e0152622. [Google Scholar] [CrossRef] [PubMed]
- Grange, Z.L.; Goldstein, T.; Johnson, C.K.; Anthony, S.; Gilardi, K.; Daszak, P.; Olival, K.J.; O’Rourke, T.; Murray, S.; Olson, S.H.; et al. Ranking the risk of animal-to-human spillover for newly discovered viruses. Proc. Natl. Acad. Sci. USA 2021, 118, e2002324118. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, R.N.; Kennedy, J.S. ACAM2000: A newly licensed cell culture-based live vaccinia smallpox vaccine. Expert Opin. Investig. Drugs 2008, 17, 555–564. [Google Scholar] [CrossRef]
- Kennedy, J.S.; Greenberg, R.N. IMVAMUNE®: Modified vaccinia Ankara strain as an attenuated smallpox vaccine. Expert Rev. Vaccines 2009, 8, 13–24. [Google Scholar] [CrossRef]
- Diamond, D.; Nirappil, F.; Sun, L.H. Inside America’s Monkeypox Crisis—And the Mistakes That Made It Worse. The Washington Post. 2022. Available online: https://www.washingtonpost.com/health/2022/08/17/monkeypox-biden-vaccine-testing-mistakes/ (accessed on 6 February 2023).
- Bubola, E.; Pronczuk, M. Unequal Access to Monkeypox Shots Gives Europe Pandemic Flashbacks. The New York Times. 2022. Available online: https://www.nytimes.com/2022/08/17/world/europe/monkeypox-vaccine-inequality-europe.html (accessed on 6 February 2023).
- Kozlov, M. Monkeypox in Africa: The science the world ignored. Nature 2022, 607, 17–18. [Google Scholar] [CrossRef] [PubMed]
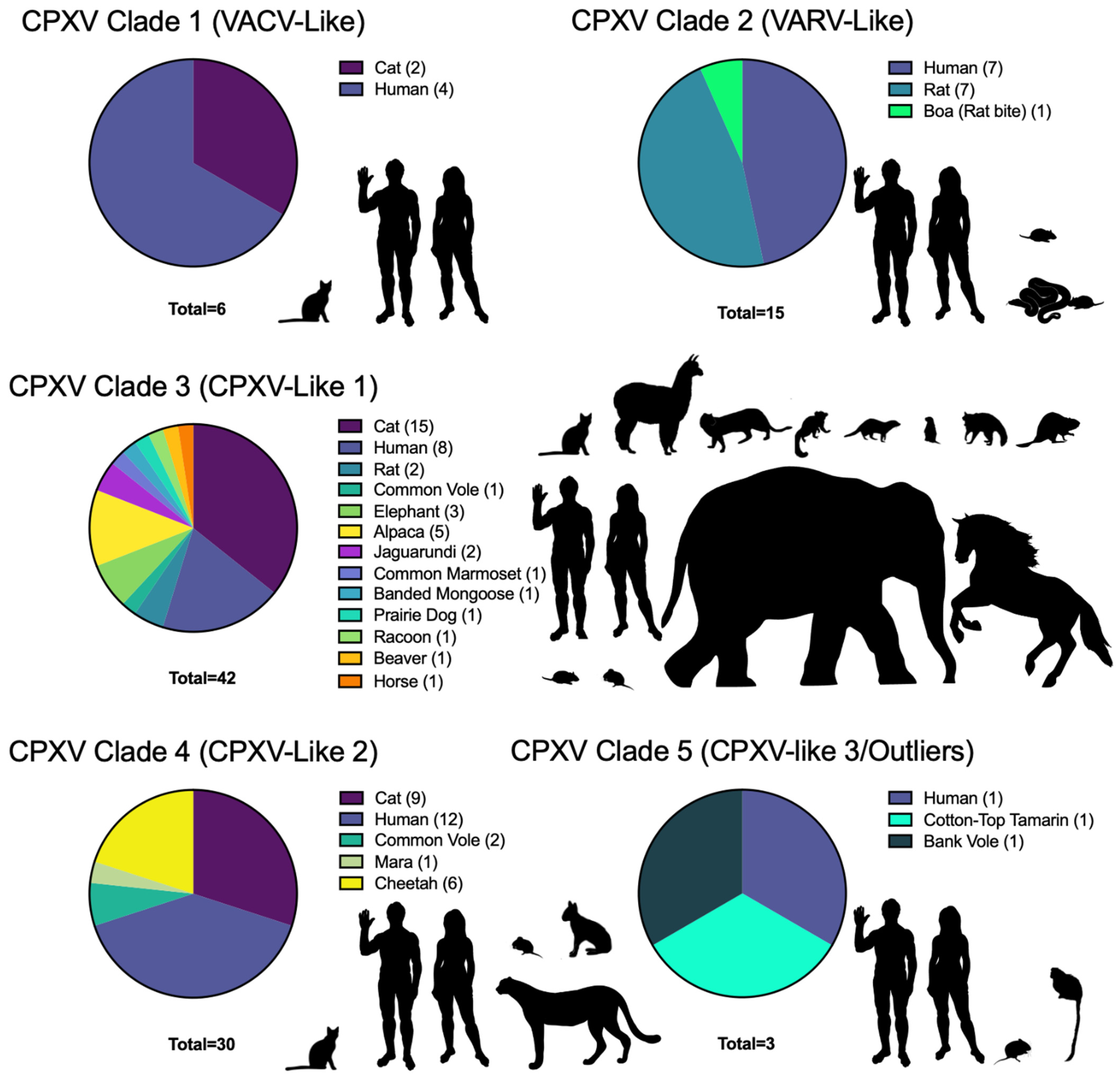
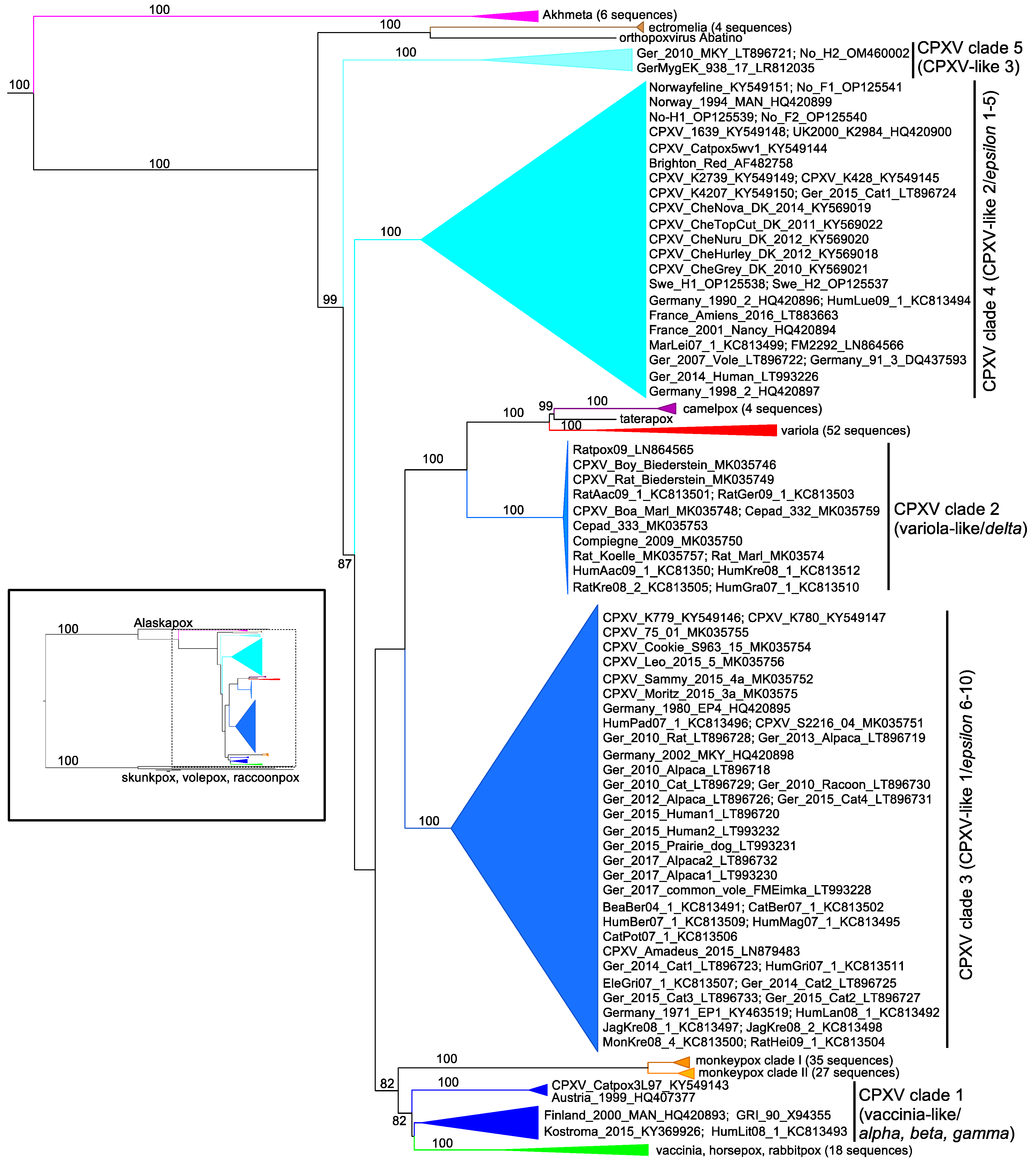
| CPXV Classification | Name | Location | Year | Host | Accession Number | References |
|---|---|---|---|---|---|---|
| CPXV Clade 1 | CPXV_Catpox3L97 | UK | 1977 | Cat | KY549143.2 | [126] |
| CPXV Clade 1 | GRI-90 | Russia, Moscow | 1990 | Human | X94355.2 | [141] |
| CPXV Clade 1 | Austria 1999 | Austria, Texing | 1999 | Cat | HQ407377.1 | [134] |
| CPXV Clade 1 | Finland_2000_MAN | Finland, Tohmajärvi | 2000 | Human | HQ420893.1 | [134,142] |
| CPXV Clade 1 | HumLit08/1 | Lithuania, Vilnius | 2008 | Human | KC813493.1 | [135] |
| CPXV Clade 1 | Kostroma_2015 | Russia, Kostroma | 2015 | Human | KY369926.1 | [143] |
| CPXV Clade 2 | HumAac09/1 | Germany, Aachen | 1990 | Human | KC813508.1 | [135,144] |
| CPXV Clade 2 | HumGra07/1 | Austria, Graz | 2007 | Human | KC813510.1 | [135,144] |
| CPXV Clade 2 | HumKre08/1 | Germany, Krefeld | 2008 | Human | KC813512.1 | [135,144] |
| CPXV Clade 2 | RatKre08/2 | Germany, Krefeld | 2008 | Rat | KC813505.1 | [135,144] |
| CPXV Clade 2 | Ratpox09 | Germany, Munich | 2009 | Rat | LN864565.1 | [136,145] |
| CPXV Clade 2 | RatGer09/1 | Germany, Germering | 2009 | Rat | KC813503.1 | [135,144] |
| CPXV Clade 2 | RatAac09/1 | Germany, Aachen | 2009 | Rat | KC813501.1 | [135,144] |
| CPXV Clade 2 | CPXV/Rat Koelle | Germany, Munich | 2009 | Rat | MK035757.1 | [21] |
| CPXV Clade 2 | CPXV/Compiegne 2009 | France, Compiègne | 2009 | Human | MK035750.1 | [21] |
| CPXV Clade 2 | CPXV/Rat Biederstein | Germany, Munich | 2009 | Rat | MK035749.1 | [21] |
| CPXV Clade 2 | CPXV/Boa Marl | Germany, Marl | 2009 | Boa | MK035748.1 | [21] |
| CPXV Clade 2 | CPXV/Rat Marl | Germany, Marl | 2009 | Rat | MK035747.1 | [21] |
| CPXV Clade 2 | CPXV/Boy Biederstein | Germany, Munich | 2009 | Human | MK035746.1 | [21] |
| CPXV Clade 2 | CPXV/Cepad 332 | France, Épinal | 2011 | Human | MK035759.1 | [21,146] |
| CPXV Clade 2 | CPXV/Cepad 333 | France, Épinal | 2011 | Human | MK035753.1 | [21,146] |
| CPXV Clade 3 | Germany_1971_EP1 | Germany | 1971 | Elephant | KY463519.1 | [147] |
| CPXV Clade 3 | Germany_1980_EP4 | Germany, Hameln | 1980 | Elephant | HQ420895.1 | [87,134] |
| CPXV Clade 3 | CPXV_K780 | UK | 2000 | Cat | KY549147.1 | [126] |
| CPXV Clade 3 | CPXV_K779 | UK, Bristol | 2000 | Cat | KY549146.1 | [126] |
| CPXV Clade 3 | CPXV/75/01 | Germany, Detmold | 2001 | Human | MK035755.1 | [21] |
| CPXV Clade 3 | Germany_2002_MKY | Germany, Göttingen | 2002 | Common Marmoset | HQ420898.1 | [134,148] |
| CPXV Clade 3 | BeaBer04/1 | Germany, Berlin | 2004 | Beaver | KC813491.1 | [135] |
| CPXV Clade 3 | CPXV/S2216/04 | Germany, Hannover | 2005 | Cat | MK035751.1 | [21] |
| CPXV Clade 3 | HumGri07/1 | Germany, Grimmem | 2007 | Human | KC813511.1 | [135] |
| CPXV Clade 3 | HumBer07/1 | Germany, Berlin | 2007 | Human | KC813509.1 | [135] |
| CPXV Clade 3 | EleGri07/1 | Germany, Grimmem | 2007 | Elephant | KC813507.1 | [135] |
| CPXV Clade 3 | CatPot07/1 | Germany, Potsdam | 2007 | Cat | KC813506.1 | [135] |
| CPXV Clade 3 | CatBer07/1 | Germany, Berlin | 2007 | Cat | KC813502.1 | [135] |
| CPXV Clade 3 | HumMag07/1 | Germany, Magdeburg | 2007 | Human | KC813495.1 | [135] |
| CPXV Clade 3 | HumPad07/1 | Germany, Paderborn | 2007 | Human | KC813496.1 | [135] |
| CPXV Clade 3 | MonKre08/4 | Germany, Krefeld | 2008 | Banded Mongoose | KC813500.1 | [18,135] |
| CPXV Clade 3 | JagKre08/2 | Germany, Krefeld | 2008 | Jaguarundi | KC813498.1 | [18,135] |
| CPXV Clade 3 | JagKre08/1 | Germany, Krefeld | 2008 | Jaguarundi | KC813497.1 | [18,135] |
| CPXV Clade 3 | HumLan08/1 | Germany, Landau | 2008 | Human | KC813492.1 | [135] |
| CPXV Clade 3 | RatHei09/1 | Germany, Heidelberg | 2009 | Rat | KC813504.1 | [135] |
| CPXV Clade 3 | Ger/2010/Raccoon | Germany, Ellrich | 2010 | Raccoon | LT896730.1 | [14] |
| CPXV Clade 3 | Ger/2010/Cat | Germany, Nordhausen | 2010 | Cat | LT896729.1 | [14] |
| CPXV Clade 3 | Ger/2010/Alpaca | Germany, Oberwiesenthal | 2010 | Alpaca | LT896718.1 | [14] |
| CPXV Clade 3 | Ger/2010/Rat | Germany, Hannover | 2010 | Rat | LT896728.1 | [14] |
| CPXV Clade 3 | Ger/2012/Alpaca | Germany, Rositz | 2012 | Alpaca | LT896726.1 | [14,149] |
| CPXV Clade 3 | Ger/2013/Alpaca | Germany, Zernitz | 2013 | Alpaca | LT896719.1 | [14,149] |
| CPXV Clade 3 | Ger/2014/Cat1 | Germany, Bleckede | 2014 | Cat | LT896723.1 | [14] |
| CPXV Clade 3 | Ger/2014/Cat2 | Germany, Nordhausen | 2014 | Cat | LT896725.1 | [14] |
| CPXV Clade 3 | Ger/2015/Cat2 | Germany, Rostock | 2015 | Cat | LT896727.1 | [14] |
| CPXV Clade 3 | CPXV Amadeus 2015 | Germany, Berlin | 2015 | Horse | LN879483.1 | [150] |
| CPXV Clade 3 | Ger/2015/Human2 | Germany, Leipzig | 2015 | Human | LT993232.1 | [14] |
| CPXV Clade 3 | Ger/2015/Prairie-dog | Germany, Dresden | 2015 | Prairie Dog | LT993231.1 | [14] |
| CPXV Clade 3 | Ger/2015/Cat3 | Germany, Vogtlandkreis | 2015 | Cat | LT896733.1 | [14] |
| CPXV Clade 3 | Ger/2015/Cat4 | Germany, Hengelbach | 2015 | Cat | LT896731.1 | [14] |
| CPXV Clade 3 | Ger/2015/Human1 | Germany, Leipzig | 2015 | Human | LT896720.1 | [14] |
| CPXV Clade 3 | CPXV/Leo 2015/5 | Germany, Extertal | 2015 | Cat | MK035756.1 | [21] |
| CPXV Clade 3 | CPXV/Cookie S963_15 | Germany, Benthe | 2015 | Cat | MK035754.1 | [21] |
| CPXV Clade 3 | CPXV/Sammy 2015/4a | Germany, Bad Münder | 2015 | Cat | MK035752.1 | [21] |
| CPXV Clade 3 | CPXV/Moritz 2015/3a | Germany, Aerzen | 2015 | Cat | MK035758.1 | [21,151] |
| CPXV Clade 3 | Ger/2017/Alpaca2 | Germany, Merzdorf | 2017 | Alpaca | LT896732.2 | [14,149] |
| CPXV Clade 3 | Ger/2017/Alpaca1 | Germany, Brand-Erbisdorf | 2017 | Alpaca | LT993230.1 | [14,149] |
| CPXV Clade 3 | Ger/2017/common vole FMEimka | Germany, Brand-Erbisdorf | 2017 | Common Vole | LT993228.1 | [149] |
| CPXV Clade 4 | Brighton Red | UK, Brighton | 1937 | Human | AF482758.2 | [17,70,77] |
| CPXV Clade 4 | CPXV_Catpox5wv1 | UK, London | 1972 | Cheetah | KY549144.1 | [126] |
| CPXV Clade 4 | No-Swe-H1 | Sweden | 1990 | Human | OP125538.1 | [118,140] |
| CPXV Clade 4 | Germany_1990_2 | Germany, Bonn | 1990 | Human | HQ420896.1 | [152] |
| CPXV Clade 4 | Germany 91-3 | Germany, Munich | 1991 | Human | DQ437593.1 | [117,153] |
| CPXV Clade 4 | Norway_1994_MAN | Norway, Bergen | 1994 | Human | HQ420899.1 | [134,154] |
| CPXV Clade 4 | UK2000_K2984 | UK, Bristol | 2000 | Cat | HQ420900.1 | [134] |
| CPXV Clade 4 | France_2001_Nancy | France, Nancy | 2001 | Human | HQ420894.1 | [134] |
| CPXV Clade 4 | Ger/2007/Vole | Germany, Rottweil | 2007 | Common Vole | LT896722.1 | [14] |
| CPXV Clade 4 | MarLei07/1 | Germany, Leipzig | 2007 | Mara | KC813499.1 | [135] |
| CPXV Clade 4 | HumLue09/1 | Germany, Lübeck | 2009 | Human | KC813494.1 | [135] |
| CPXV Clade 4 | FM2292 | Germany, Rutesheim | 2011 | Common Vole | LN864566.1 | [136] |
| CPXV Clade 4 | Ger/2014/Human | Germany, Freiburg | 2014 | Human | LT993226.1 | [14] |
| CPXV Clade 4 | Ger/2015/Cat1 | Germany, Vogtlandkreis | 2015 | Cat | LT896724.1 | [14] |
| CPXV Clade 4 | No-Swe-H2 | Sweden | 1990 | Human | OP125537.1 | [118,140] |
| CPXV Clade 4 | Norwayfeline | Norway, Bergen | 1994 | Cat | KY549151.1 | [126] |
| CPXV Clade 4 | No-H1 | Norway | 1994 | Human | OP125539.1 | [118,140] |
| CPXV Clade 4 | No-F1 | Norway | 1994 | Cat | OP125541.1 | [118,140] |
| CPXV Clade 4 | Germany_1998_2 | Germany, Eckental | 1998 | Human | HQ420897.1 | [134,155] |
| CPXV Clade 4 | No-F2 | Norway | 1999 | Cat | OP125540.1 | [140] |
| CPXV Clade 4 | CPXV_K2739 | UK, Bristol | 2000 | Cat | KY549149.1 | [126] |
| CPXV Clade 4 | CPXV_K4207 | UK, Bristol | 2000 | Cat | KY549150.2 | [126] |
| CPXV Clade 4 | CPXV_1639 | UK, Bristol | 2000 | Cat | KY549148.1 | [126] |
| CPXV Clade 4 | CPXV_K428 | UK, Bristol | 2000 | Cat | KY549145.1 | [126] |
| CPXV Clade 4 | CPXV CheGrey_DK_2010 | Denmark, Djursland | 2010 | Cheetah | KY569021.1 | [156] |
| CPXV Clade 4 | CPXV CheTopCut_DK_2011 | Denmark, Djursland | 2011 | Cheetah | KY569022.1 | [156] |
| CPXV Clade 4 | CPXV CheNuru_DK_2012 | Denmark, Djursland | 2012 | Cheetah | KY569020.1 | [156] |
| CPXV Clade 4 | CPXV CheHurley_DK_2012 | Denmark, Djursland | 2012 | Cheetah | KY569018.1 | [156] |
| CPXV Clade 4 | CPXV CheNova_DK_2014 | Denmark, Djursland | 2014 | Cheetah | KY569019.1 | [156] |
| CPXV Clade 4 | France Amiens 2016 | France, Amiens | 2016 | Human | LT883663.1 | [157] |
| CPXV Clade 5 | Ger 2010 MKY | Germany, Bad Liebenstein | 2010 | Cotton-top Tamarin | LT896721.1 | [14,158] |
| CPXV Clade 5 | No-H2 | Norway, Nordland | 2001 | Human | OM460002.1 | [159] |
| CPXV Clade 5 | GerMygEK 938/17 | Germany, Thuringia | 2017 | Bank Vole | LR812035.1 | [160] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bruneau, R.C.; Tazi, L.; Rothenburg, S. Cowpox Viruses: A Zoo Full of Viral Diversity and Lurking Threats. Biomolecules 2023, 13, 325. https://doi.org/10.3390/biom13020325
Bruneau RC, Tazi L, Rothenburg S. Cowpox Viruses: A Zoo Full of Viral Diversity and Lurking Threats. Biomolecules. 2023; 13(2):325. https://doi.org/10.3390/biom13020325
Chicago/Turabian StyleBruneau, Ryan C., Loubna Tazi, and Stefan Rothenburg. 2023. "Cowpox Viruses: A Zoo Full of Viral Diversity and Lurking Threats" Biomolecules 13, no. 2: 325. https://doi.org/10.3390/biom13020325
APA StyleBruneau, R. C., Tazi, L., & Rothenburg, S. (2023). Cowpox Viruses: A Zoo Full of Viral Diversity and Lurking Threats. Biomolecules, 13(2), 325. https://doi.org/10.3390/biom13020325








