Exploring Novel Variants of the Cytochrome P450 Reductase Gene (POR) from the Genome Aggregation Database by Integrating Bioinformatic Tools and Functional Assays
Abstract
1. Introduction
2. Materials and Methods
2.1. Control Variants
2.2. Database Collection
2.3. Single Amino Acid Variant Prediction Tools
- (1)
- PANTHER
- (2)
- PhD-SNP
- (3)
- SIFT
- (4)
- SNAP
- (5)
- MAPP
- (6)
- PolyPhen-1 and PolyPhen-2
- (7)
- MetaSNP
- (8)
- MutPred2
- (9)
- SNPs&Go
- (10)
- PredictSNP2
2.4. Data Integration and Evaluation
2.5. Expression of POR in E. coli
2.6. Purification of Recombinant Human POR from Isolated E. coli Membranes
2.7. Urea Denaturation Assay
2.8. POR Assays with Cytochrome c
2.9. Assay of Cytochrome P450 (CYP) Activity in Reconstituted Liposomes
2.10. Statistical Analysis
3. Results
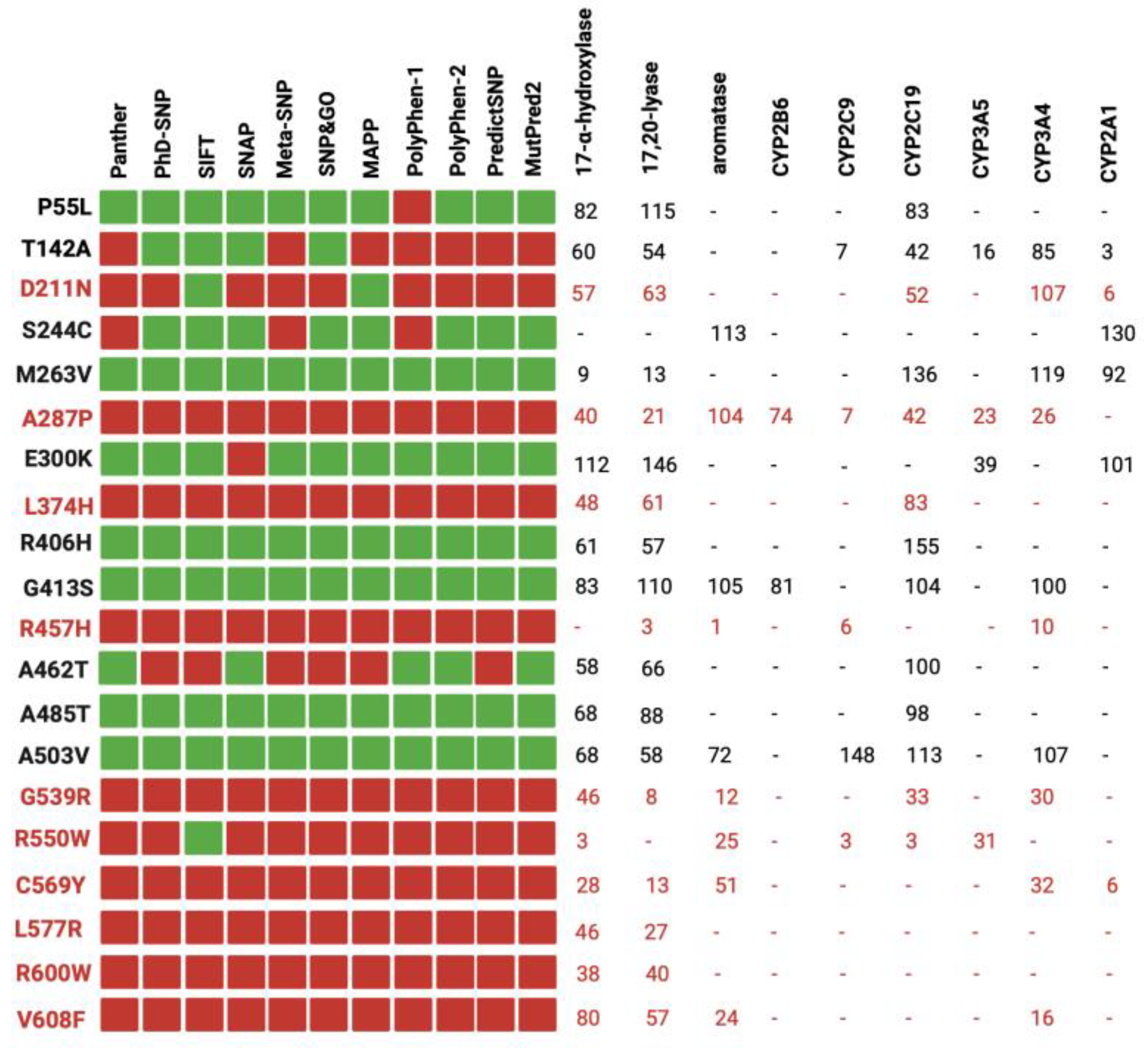
3.1. Performance of the Prediction Tools
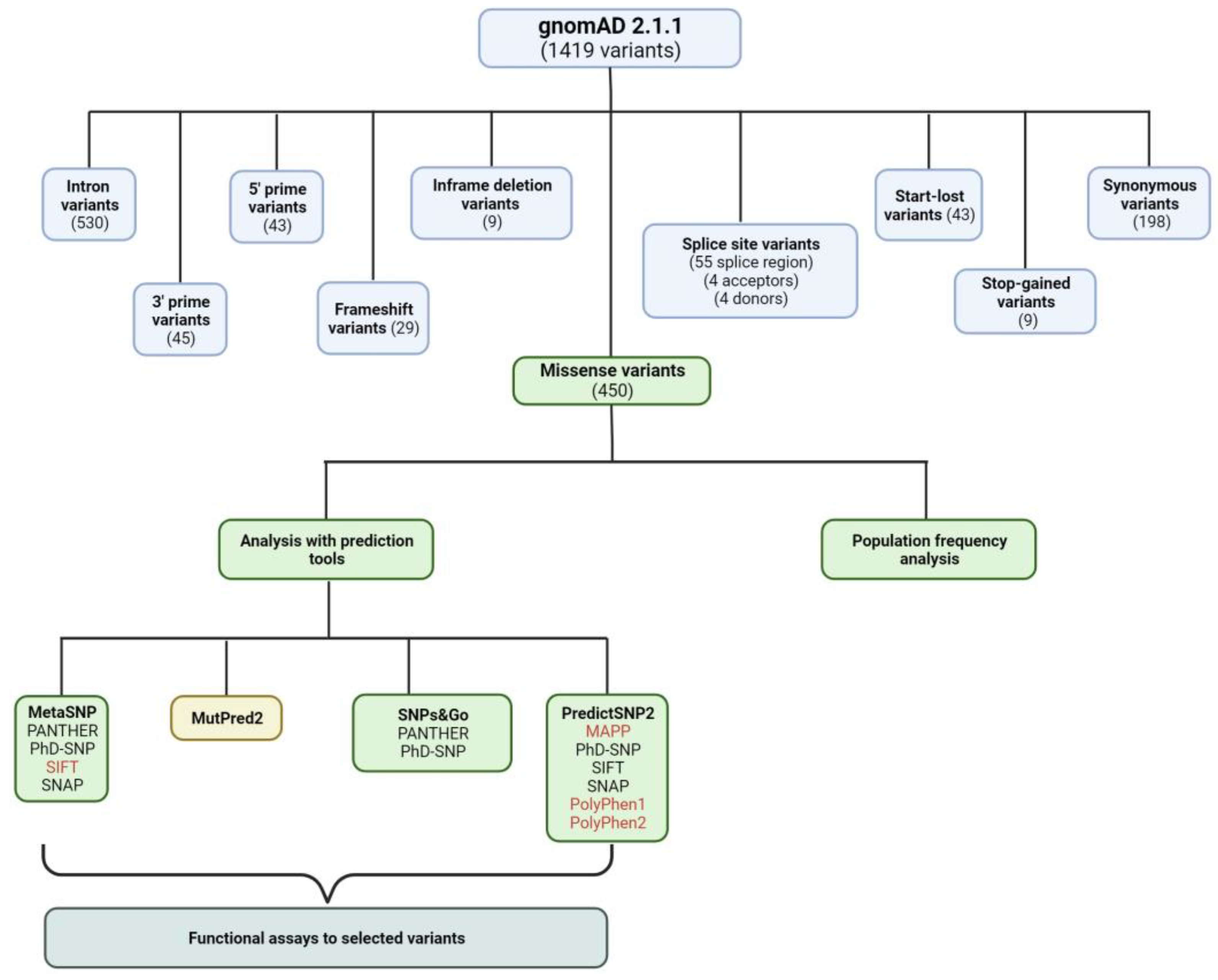
3.2. Curating Variants from gnomAD
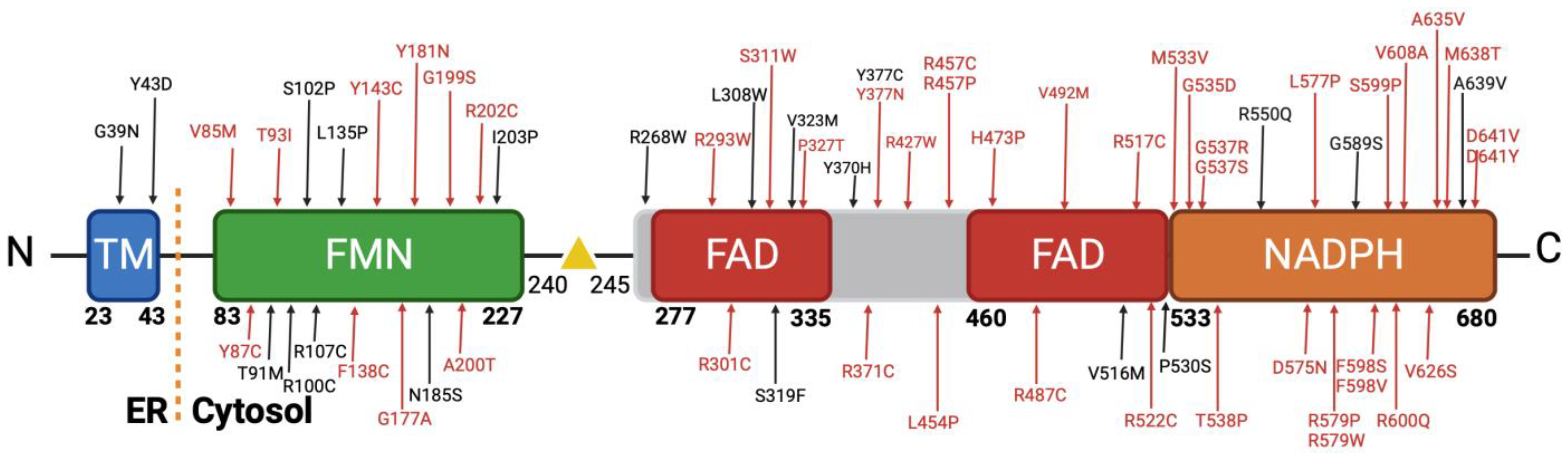
3.3. Analysis of the Missense Variants
3.4. Functional Assay of Selected Variants
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pandey, A.V.; Flück, C.E. NADPH P450 oxidoreductase: Structure, function, and pathology of diseases. Pharmacol. Ther. 2013, 138, 229–254. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Panda, S.P.; Marohnic, C.C.; Martasek, P.; Masters, B.S.; Kim, J.J. Structural basis for human NADPH-cytochrome P450 oxidoreductase deficiency. Proc. Natl. Acad. Sci. USA 2011, 108, 13486–13491. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.V.; Sproll, P. Pharmacogenomics of human P450 oxidoreductase. Front. Pharmacol. 2014, 5, 103. [Google Scholar] [CrossRef] [PubMed]
- Riddick, D.S.; Ding, X.; Wolf, C.R.; Porter, T.D.; Pandey, A.V.; Zhang, Q.Y.; Gu, J.; Finn, R.D.; Ronseaux, S.; McLaughlin, L.A.; et al. NADPH-cytochrome P450 oxidoreductase: Roles in physiology, pharmacology, and toxicology. Drug Metab. Dispos. 2013, 41, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.Y.H.; Coon, M.J. Role of Hemoprotein P-450 in Fatty Acid Omega-Hydroxylation in a Soluble Enzyme System from Liver Microsomes. J. Biol. Chem. 1968, 243, 1331–1332. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.Y.H.; Junk, K.W.; Coon, M.J. Resolution of Cytochrome P-450-Containing Omega-Hydroxylation System of Liver Microsomes into 3 Components. J. Biol. Chem. 1969, 244, 3714–3721. [Google Scholar] [CrossRef]
- Burkhard, F.Z.; Parween, S.; Udhane, S.S.; Flück, C.E.; Pandey, A.V. P450 Oxidoreductase deficiency: Analysis of mutations and polymorphisms. J. Steroid Biochem. Mol. Biol. 2017, 165, 38–50. [Google Scholar] [CrossRef]
- Flück, C.E.; Rojas Velazquez, M.N.; Pandey, A.V. Chapter 12—P450 oxidoreductase deficiency. In Genetic Steroid Disorders, 2nd ed.; New, M.I., Ed.; Academic Press: San Diego, CA, USA, 2023; pp. 239–264. [Google Scholar] [CrossRef]
- Flück, C.E.; Pandey, A.V. Human P450 Oxidoreductase Deficiency. In Encyclopedia of Endocrine Diseases, 2nd ed.; Huhtaniemi, I., Martini, L., Eds.; Academic Press: Oxford, UK, 2019; pp. 431–443. [Google Scholar] [CrossRef]
- Parween, S.; Fernández-Cancio, M.; Benito-Sanz, S.; Camats, N.; Velazquez, M.N.R.; López-Siguero, J.P.; Udhane, S.S.; Kagawa, N.; Flück, C.E.; Audí, L.; et al. Molecular basis of CYP19A1 deficiency in a 46, XX patient with R550W mutation in POR: Expanding the PORD phenotype. J. Clin. Endocrinol. Metab. 2020, 105, e1272–e1290. [Google Scholar] [CrossRef]
- Parween, S.; Roucher-Boulez, F.; Flück, C.E.; Lienhardt-Roussie, A.; Mallet, D.; Morel, Y.; Pandey, A.V. P450 Oxidoreductase Deficiency: Loss of Activity Caused by Protein Instability From a Novel L374H Mutation. J. Clin. Endocrinol. Metab. 2016, 101, 4789–4798. [Google Scholar] [CrossRef][Green Version]
- Parween, S.; DiNardo, G.; Baj, F.; Zhang, C.; Gilardi, G.; Pandey, A.V. Differential effects of variations in human P450 oxidoreductase on the aromatase activity of CYP19A1 polymorphisms R264C and R264H. J. Steroid Biochem. Mol. Biol. 2020, 196, 105507. [Google Scholar] [CrossRef]
- Udhane, S.S.; Parween, S.; Kagawa, N.; Pandey, A.V. Altered CYP19A1 and CYP3A4 Activities Due to Mutations A115V, T142A, Q153R and P284L in the Human P450 Oxidoreductase. Front. Pharmacol. 2017, 8, 580. [Google Scholar] [CrossRef]
- Velazquez, M.N.R.; Parween, S.; Udhane, S.S.; Pandey, A.V. Variability in human drug metabolizing cytochrome P450 CYP2C9, CYP2C19 and CYP3A5 activities caused by genetic variations in cytochrome P450 oxidoreductase. Biochem. Biophys. Res. Commun. 2019, 515, 133–138. [Google Scholar] [CrossRef]
- Rojas Velazquez, M.N.; Noebauer, M.; Pandey, A.V. Loss of Protein Stability and Function Caused by P228L Variation in NADPH-Cytochrome P450 Reductase Linked to Lower Testosterone Levels. Int. J. Mol. Sci. 2022, 23, 10141. [Google Scholar] [CrossRef]
- Flück, C.E.; Tajima, T.; Pandey, A.V.; Arlt, W.; Okuhara, K.; Verge, C.F.; Jabs, E.W.; Mendonça, B.B.; Fujieda, K.; Miller, W.L. Mutant P450 oxidoreductase causes disordered steroidogenesis with and without Antley-Bixler syndrome. Nat. Genet. 2004, 36, 228–230. [Google Scholar] [CrossRef]
- Williamson, L.; Arlt, W.; Shackleton, C.; Kelley, R.I.; Braddock, S.R. Linking Antley-Bixler syndrome and congenital adrenal hyperplasia: A novel case of P450 oxidoreductase deficiency. Am. J. Med. Genet. A 2006, 140A, 1797–1803. [Google Scholar] [CrossRef]
- Krone, N.; Dhir, V.; Ivison, H.E.; Arlt, W. Congenital adrenal hyperplasia and P450 oxidoreductase deficiency. Clin. Endocrinol. 2007, 66, 162–172. [Google Scholar] [CrossRef]
- Shackleton, C.; Marcos, J.; Arlt, W.; Hauffa, B.P. Prenatal diagnosis of p450 oxidoreductase deficiency (ORD): A disorder causing low pregnancy estriol, maternal and fetal virilization, and the Antley-Bixler syndrome. Am. J. Med. Genet. A 2004, 129A, 105–112. [Google Scholar] [CrossRef]
- Miller, W.L. Disorders of androgen synthesis--from cholesterol to dehydroepiandrosterone. Med. Princ. Pract. 2005, 14 (Suppl. S1), 58–68. [Google Scholar] [CrossRef] [PubMed]
- Baronio, F.; Ortolano, R.; Menabo, S.; Cassio, A.; Baldazzi, L.; Di Natale, V.; Tonti, G.; Vestrucci, B.; Balsamo, A. 46,XX DSD due to Androgen Excess in Monogenic Disorders of Steroidogenesis: Genetic, Biochemical, and Clinical Features. Int. J. Mol. Sci. 2019, 20, 4605. [Google Scholar] [CrossRef] [PubMed]
- Yauy, K.; Mau-Them, F.T.; Willems, M.; Coubes, C.; Blanchet, P.; Herlin, C.; Arrada, I.T.; Sanchez, E.; Faure, J.M.; Le Gac, M.P.; et al. B3GAT3-related disorder with craniosynostosis and bone fragility due to a unique mutation. Genet. Med. 2018, 20, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Wang, H.; Chen, L.; Li, H. Advance in clinical research on Antley-Bixler syndrome. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2018, 35, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Tomkova, M.; Marohnic, C.C.; Baxova, A.; Martasek, P. Antley-Bixler syndrome or POR deficiency? Cas. Lek. Cesk 2008, 147, 261–265. [Google Scholar] [PubMed]
- Shackleton, C.; Marcos, J.; Malunowicz, E.M.; Szarras-Czapnik, M.; Jira, P.; Taylor, N.F.; Murphy, N.; Crushell, E.; Gottschalk, M.; Hauffa, B.; et al. Biochemical diagnosis of Antley-Bixler syndrome by steroid analysis. Am. J. Med. Genet. A 2004, 128A, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Rwere, F.; Im, S.; Shen, A.L.; Waskell, L.; Kim, J.P. Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH- Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer. Biochemistry 2018, 57, 945–962. [Google Scholar] [CrossRef] [PubMed]
- Rwere, F.; Xia, C.; Im, S.; Haque, M.M.; Stuehr, D.J.; Waskell, L.; Kim, J.J. Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone. J. Biol. Chem. 2016, 291, 14639–14661. [Google Scholar] [CrossRef]
- Pandey, A.V.; Kempna, P.; Hofer, G.; Mullis, P.E.; Fluck, C.E. Modulation of human CYP19A1 activity by mutant NADPH P450 oxidoreductase. Mol. Endocrinol. 2007, 21, 2579–2595. [Google Scholar] [CrossRef]
- Nicolo, C.; Flück, C.E.; Mullis, P.E.; Pandey, A.V. Restoration of mutant cytochrome P450 reductase activity by external flavin. Mol. Cell Endocrinol. 2010, 321, 245–252. [Google Scholar] [CrossRef]
- Pandey, A.V. Biochemical analysis of mutations in P450 oxidoreductase. Biochem. Soc. Trans. 2006, 34, 1186–1191. [Google Scholar] [CrossRef]
- Parween, S.; Rojas Velazquez, M.N.; Udhane, S.S.; Kagawa, N.; Pandey, A.V. Variability in Loss of Multiple Enzyme Activities Due to the Human Genetic Variation P284T Located in the Flexible Hinge Region of NADPH Cytochrome P450 Oxidoreductase. Front. Pharmacol. 2019, 10, 1187. [Google Scholar] [CrossRef]
- Soneda, S.; Yazawa, T.; Fukami, M.; Adachi, M.; Mizota, M.; Fujieda, K.; Miyamoto, K.; Ogata, T. Proximal promoter of the cytochrome P450 oxidoreductase gene: Identification of microdeletions involving the untranslated exon 1 and critical function of the SP1 binding sites. J. Clin. Endocrinol. Metab. 2011, 96, E1881–E1887. [Google Scholar] [CrossRef]
- Fukami, M.; Nishimura, G.; Homma, K.; Nagai, T.; Hanaki, K.; Uematsu, A.; Ishii, T.; Numakura, C.; Sawada, H.; Nakacho, M.; et al. Cytochrome P450 oxidoreductase deficiency: Identification and characterization of biallelic mutations and genotype-phenotype correlations in 35 Japanese patients. J. Clin. Endocrinol. Metab. 2009, 94, 1723–1731. [Google Scholar] [CrossRef] [PubMed]
- Idkowiak, J.; Malunowicz, E.M.; Dhir, V.; Reisch, N.; Szarras-Czapnik, M.; Holmes, D.M.; Shackleton, C.H.L.; Davies, J.D.; Hughes, I.A.; Krone, N.; et al. Concomitant Mutations in the P450 Oxidoreductase and Androgen Receptor Genes Presenting with 46, XY Disordered Sex Development and Androgenization at Adrenarche. J. Clin. Endocrinol. Metab. 2010, 95, 3418–3427. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Pandey, A.V.; Agrawal, V.; Reardon, W.; Lapunzina, P.D.; Mowat, D.; Jabs, E.W.; Van Vliet, G.; Sack, J.; Flück, C.E.; et al. Diversity and function of mutations in p450 oxidoreductase in patients with Antley-Bixler syndrome and disordered steroidogenesis. Am. J. Hum. Genet. 2005, 76, 729–749. [Google Scholar] [CrossRef] [PubMed]
- Dean, B.; Chrisp, G.L.; Quartararo, M.; Maguire, A.M.; Hameed, S.; King, B.R.; Munns, C.F.; Torpy, D.J.; Falhammar, H.; Rushworth, R.L. P450 Oxidoreductase Deficiency: A Systematic Review and Meta-analysis of Genotypes, Phenotypes, and Their Relationships. J. Clin. Endocrinol. Metab. 2020, 105, e42–e52. [Google Scholar] [CrossRef] [PubMed]
- Gudmundsson, S.; Singer-Berk, M.; Watts, N.A.; Phu, W.; Goodrich, J.K.; Solomonson, M.; Genome Aggregation Database, C.; Rehm, H.L.; MacArthur, D.G.; O’Donnell-Luria, A. Variant interpretation using population databases: Lessons from gnomAD. Hum. Mutat. 2022, 43, 1012–1030. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.P.; Mi, H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef]
- Capriotti, E.; Fariselli, P. PhD-SNPg: A webserver and lightweight tool for scoring single nucleotide variants. Nucleic Acids Res. 2017, 45, W247–W252. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef]
- Hecht, M.; Bromberg, Y.; Rost, B. Better prediction of functional effects for sequence variants. BMC Genom. 2015, 16 (Suppl. S8), S1. [Google Scholar] [CrossRef]
- Stone, E.A.; Sidow, A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005, 15, 978–986. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, E.; Altman, R.B.; Bromberg, Y. Collective judgment predicts disease-associated single nucleotide variants. BMC Genom. 2013, 14 (Suppl. S3), S2. [Google Scholar] [CrossRef]
- Pejaver, V.; Urresti, J.; Lugo-Martinez, J.; Pagel, K.A.; Lin, G.N.; Nam, H.J.; Mort, M.; Cooper, D.N.; Sebat, J.; Iakoucheva, L.M.; et al. Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nat. Commun. 2020, 11, 5918. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, E.; Calabrese, R.; Fariselli, P.; Martelli, P.L.; Altman, R.B.; Casadio, R. WS-SNPs&GO: A web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genom. 2013, 14 (Suppl. S3), S6. [Google Scholar] [CrossRef]
- Bendl, J.; Stourac, J.; Salanda, O.; Pavelka, A.; Wieben, E.D.; Zendulka, J.; Brezovsky, J.; Damborsky, J. PredictSNP: Robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput. Biol. 2014, 10, e1003440. [Google Scholar] [CrossRef] [PubMed]
- Fluck, C.E.; Pandey, A.V. Impact on CYP19A1 activity by mutations in NADPH cytochrome P450 oxidoreductase. J. Steroid Biochem. Mol. Biol. 2017, 165, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Ren, X.; Song, Y.; Su, C.; Fu, J.; Gong, C. Novel phenotypes and genotypes in Antley-Bixler syndrome caused by cytochrome P450 oxidoreductase deficiency: Based on the first cohort of Chinese children. Orphanet J. Rare Dis. 2019, 14, 299. [Google Scholar] [CrossRef]
- Wudy, S.A.; Hartmann, M.F.; Draper, N.; Stewart, P.M.; Arlt, W. A male twin infant with skull deformity and elevated neonatal 17-hydroxyprogesterone: A prismatic case of P450 oxidoreductase deficiency. Endocr. Res. 2004, 30, 957–964. [Google Scholar] [CrossRef]
- Tzetis, M.; Konstantinidou, A.; Sofocleous, C.; Kosma, K.; Mitrakos, A.; Tzannatos, C.; Kitsiou-Tzeli, S. Compound heterozygosity of a paternal submicroscopic deletion and a maternal missense mutation in POR gene: Antley-bixler syndrome phenotype in three sibling fetuses. Birth Defects Res. A Clin. Mol. Teratol. 2016, 106, 536–541. [Google Scholar] [CrossRef]
- Sandee, D.; Morrissey, K.; Agrawal, V.; Tam, H.K.; Kramer, M.A.; Tracy, T.S.; Giacomini, K.M.; Miller, W.L. Effects of genetic variants of human P450 oxidoreductase on catalysis by CYP2D6 in vitro. Pharmacogenetics Genom. 2010, 20, 677–686. [Google Scholar] [CrossRef]
- Miller, W.L. P450 oxidoreductase deficiency: A disorder of steroidogenesis with multiple clinical manifestations. Sci. Signal 2012, 5, pt11. [Google Scholar] [CrossRef] [PubMed]
- Sahakitrungruang, T.; Huang, N.W.; Tee, M.K.; Agrawal, V.; Russell, W.E.; Crock, P.; Murphy, N.; Migeon, C.J.; Miller, W.L. Clinical, Genetic, and Enzymatic Characterization of P450 Oxidoreductase Deficiency in Four Patients. J. Clin. Endocr. Metab. 2009, 94, 4992–5000. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.L. Minireview: Regulation of steroidogenesis by electron transfer. Endocrinology 2005, 146, 2544–2550. [Google Scholar] [CrossRef] [PubMed]
- Guaragna-Filho, G.; Castro, C.C.; Carvalho, R.R.; Coeli, F.B.; Ferraz, L.F.; Petroli, R.J.; Mello, M.P.; Sewaybricker, L.E.; Lemos-Marini, S.H.; D’Souza-Li, L.F.; et al. 46,XX DSD and Antley-Bixler syndrome due to novel mutations in the cytochrome P450 oxidoreductase gene. Arq. Bras. Endocrinol. Metabol. 2012, 56, 578–585. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zou, K.H.; O’Malley, A.J.; Mauri, L. Receiver-operating characteristic analysis for evaluating diagnostic tests and predictive models. Circulation 2007, 115, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Prado, M.J.; Ligabue-Braun, R.; Zaha, A.; Rossetti, M.L.R.; Pandey, A.V. Variant predictions in congenital adrenal hyperplasia caused by mutations in CYP21A2. Front. Pharmacol. 2022, 13, 931089. [Google Scholar] [CrossRef] [PubMed]
- Pio, M.G.; Siffo, S.; Scheps, K.G.; Molina, M.F.; Adrover, E.; Abelleyro, M.M.; Rivolta, C.M.; Targovnik, H.M. Curating the gnomAD database: Report of novel variants in the thyrogobulin gene using in silico bioinformatics algorithms. Mol. Cell Endocrinol. 2021, 534, 111359. [Google Scholar] [CrossRef] [PubMed]
- Turesky, R.J.; Konorev, D.; Fan, X.; Tang, Y.; Yao, L.; Ding, X.; Xie, F.; Zhu, Y.; Zhang, Q.Y. Effect of Cytochrome P450 Reductase Deficiency on 2-Amino-9H-pyrido [2,3-b]indole Metabolism and DNA Adduct Formation in Liver and Extrahepatic Tissues of Mice. Chem. Res. Toxicol. 2015, 28, 2400–2410. [Google Scholar] [CrossRef]
- Flück, C.E.; Mallet, D.; Hofer, G.; Samara-Boustani, D.; Leger, J.; Polak, M.; Morel, Y.; Pandey, A.V. Deletion of P399_E401 in NADPH cytochrome P450 oxidoreductase results in partial mixed oxidase deficiency. Biochem. Biophys. Res. Commun. 2011, 412, 572–577. [Google Scholar] [CrossRef]
- Fluck, C.E. Congenital Adrenal Hyperplasia Owing to 17 alpha-Hydroxylase/17,20 Lyase and P450 Oxidoreductase Deficiencies. In Hormonal and Genetic Basis of Sexual Differentiation Disorders and Hot Topics in Endocrinology; New, M.I., Simpson, J.L., Eds.; Springer: Berlin, Germany, 2011; Volume 707, pp. 3–5. [Google Scholar]
- Xiao, X.; Ma, G.; Li, S.; Wang, M.; Liu, N.; Ma, L.; Zhang, Z.; Chu, H.; Wang, S.L. Functional POR A503V is associated with the risk of bladder cancer in a Chinese population. Sci. Rep. 2015, 5, 11751. [Google Scholar] [CrossRef]
- Camats, N.; Fluck, C.E.; Audi, L. Oligogenic Origin of Differences of Sex Development in Humans. Int. J. Mol. Sci. 2020, 21, 1809. [Google Scholar] [CrossRef] [PubMed]
- Fluck, C.E.; Nicolo, C.; Pandey, A.V. Clinical, structural and functional implications of mutations and polymorphisms in human NADPH P450 oxidoreductase. Fundam. Clin. Pharmacol. 2007, 21, 399–410. [Google Scholar] [CrossRef] [PubMed]
- David, A.; Sternberg, M.J. The Contribution of Missense Mutations in Core and Rim Residues of Protein-Protein Interfaces to Human Disease. J. Mol. Biol. 2015, 427, 2886–2898. [Google Scholar] [CrossRef] [PubMed]
- Petukh, M.; Kucukkal, T.G.; Alexov, E. On human disease-causing amino acid variants: Statistical study of sequence and structural patterns. Hum. Mutat. 2015, 36, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Iijima, S.; Ohishi, A.; Ohzeki, T. Cytochrome P450 oxidoreductase deficiency with Antley-Bixler syndrome: Steroidogenic capacities. J. Pediatr. Endocrinol. Metab. 2009, 22, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Hosoe, J.; Kadowaki, H.; Miya, F.; Aizu, K.; Kawamura, T.; Miyata, I.; Satomura, K.; Ito, T.; Hara, K.; Tanaka, M.; et al. Structural Basis and Genotype-Phenotype Correlations of INSR Mutations Causing Severe Insulin Resistance. Diabetes 2017, 66, 2713–2723. [Google Scholar] [CrossRef]
- Lim, W.A.; Farruggio, D.C.; Sauer, R.T. Structural and energetic consequences of disruptive mutations in a protein core. Biochemistry 1992, 31, 4324–4333. [Google Scholar] [CrossRef]
- de Beer, T.A.; Laskowski, R.A.; Parks, S.L.; Sipos, B.; Goldman, N.; Thornton, J.M. Amino acid changes in disease-associated variants differ radically from variants observed in the 1000 genomes project dataset. PLoS Comput. Biol. 2013, 9, e1003382. [Google Scholar] [CrossRef]
- Vacic, V.; Markwick, P.R.; Oldfield, C.J.; Zhao, X.; Haynes, C.; Uversky, V.N.; Iakoucheva, L.M. Disease-associated mutations disrupt functionally important regions of intrinsic protein disorder. PLoS Comput. Biol. 2012, 8, e1002709. [Google Scholar] [CrossRef]
- Oh, J.; Song, J.S.; Park, J.E.; Jang, S.Y.; Ki, C.S.; Kim, D.K. A Case of Antley-Bixler Syndrome With a Novel Likely Pathogenic Variant (c.529G>C) in the POR Gene. Ann. Lab. Med. 2017, 37, 559–562. [Google Scholar] [CrossRef][Green Version]
- Scott, R.R.; Gomes, L.G.; Huang, N.; Van Vliet, G.; Miller, W.L. Apparent manifesting heterozygosity in P450 oxidoreductase deficiency and its effect on coexisting 21-hydroxylase deficiency. J. Clin. Endocrinol. Metab. 2007, 92, 2318–2322. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Khan, K.K.; He, Y.A.; He, Y.Q.; Halpert, J.R. Topological changes in the CYP3A4 active site probed with phenyldiazene: Effect of interaction with NADPH-cytochrome P450 reductase and cytochrome b5 and of site-directed mutagenesis. Drug Metab. Dispos. 2004, 32, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Si, S.; Wang, Z.; Yang, H.; Han, Z.; Tao, J.; Chen, H.; Wang, K.; Guo, M.; Tan, R.; Wei, J.F.; et al. Impact of single nucleotide polymorphisms on P450 oxidoreductase and peroxisome proliferator-activated receptor alpha on tacrolimus pharmacokinetics in renal transplant recipients. Pharmacogenom. J. 2019, 19, 42–52. [Google Scholar] [CrossRef]
- Goswami, C.; Chattopadhyay, A.; Chuang, E.Y. Rare variants: Data types and analysis strategies. Ann. Transl. Med. 2021, 9, 961. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sridar, C.; Kenaan, C.; Amunugama, H.; Ballou, D.P.; Hollenberg, P.F. Polymorphic variants of cytochrome P450 2B6 (CYP2B6.4-CYP2B6.9) exhibit altered rates of metabolism for bupropion and efavirenz: A charge- reversal mutation in the K139E variant (CYP2B6.8) impairs formation of a functional cytochrome p450-reductase complex. J. Pharmacol. Exp. Ther. 2011, 338, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Tomkova, M.; Marohnic, C.C.; Gurwitz, D.; Seda, O.; Masters, B.S.; Martasek, P. Identification of six novel P450 oxidoreductase missense variants in Ashkenazi and Moroccan Jewish populations. Pharmacogenomics 2012, 13, 543–554. [Google Scholar] [CrossRef]
- Miller, W.L.; Huang, N.; Flück, C.E.; Pandey, A.V. P450 oxidoreductase deficiency. Lancet 2004, 364, 1663. [Google Scholar] [CrossRef]
- Hershkovitz, E.; Parvari, R.; Wudy, S.A.; Hartmann, M.F.; Gomes, L.G.; Loewental, N.; Miller, W.L. Homozygous mutation G539R in the gene for P450 oxidoreductase in a family previously diagnosed as having 17,20-lyase deficiency. J. Clin. Endocrinol. Metab. 2008, 93, 3584–3588. [Google Scholar] [CrossRef]
- Flück, C.E.; Pandey, A.V. Clinical and biochemical consequences of p450 oxidoreductase deficiency. Endocr. Dev. 2011, 20, 63–79. [Google Scholar] [CrossRef]



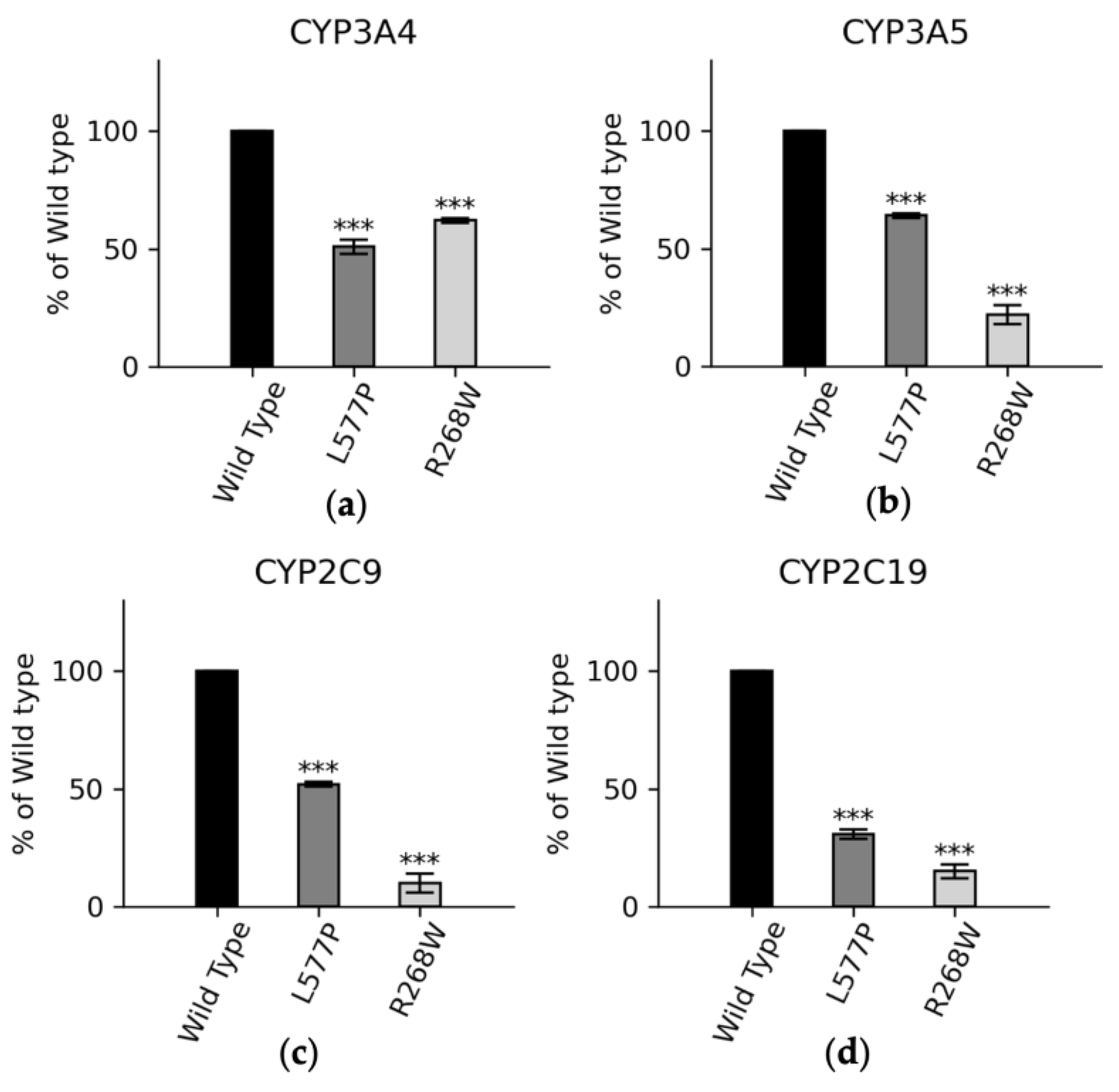
| Wild Type | R268W | L577P | |
|---|---|---|---|
| Vmax (μM/min) | 78 ± 3.4 | 75 ± 1.5 | 55 ± 3.2 |
| KM (μM) | 4.6 ± 0.9 | 3.9 ± 0.3 | 6.4 ± 0.5 |
| Vmax/KM | 17 | 19 | 8.6 |
| % | 100 | 113 | 51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rojas Velazquez, M.N.; Therkelsen, S.; Pandey, A.V. Exploring Novel Variants of the Cytochrome P450 Reductase Gene (POR) from the Genome Aggregation Database by Integrating Bioinformatic Tools and Functional Assays. Biomolecules 2023, 13, 1728. https://doi.org/10.3390/biom13121728
Rojas Velazquez MN, Therkelsen S, Pandey AV. Exploring Novel Variants of the Cytochrome P450 Reductase Gene (POR) from the Genome Aggregation Database by Integrating Bioinformatic Tools and Functional Assays. Biomolecules. 2023; 13(12):1728. https://doi.org/10.3390/biom13121728
Chicago/Turabian StyleRojas Velazquez, Maria Natalia, Søren Therkelsen, and Amit V. Pandey. 2023. "Exploring Novel Variants of the Cytochrome P450 Reductase Gene (POR) from the Genome Aggregation Database by Integrating Bioinformatic Tools and Functional Assays" Biomolecules 13, no. 12: 1728. https://doi.org/10.3390/biom13121728
APA StyleRojas Velazquez, M. N., Therkelsen, S., & Pandey, A. V. (2023). Exploring Novel Variants of the Cytochrome P450 Reductase Gene (POR) from the Genome Aggregation Database by Integrating Bioinformatic Tools and Functional Assays. Biomolecules, 13(12), 1728. https://doi.org/10.3390/biom13121728







