An Interpretable Classification Model Using Gluten-Specific TCR Sequences Shows Diagnostic Potential in Coeliac Disease
Abstract
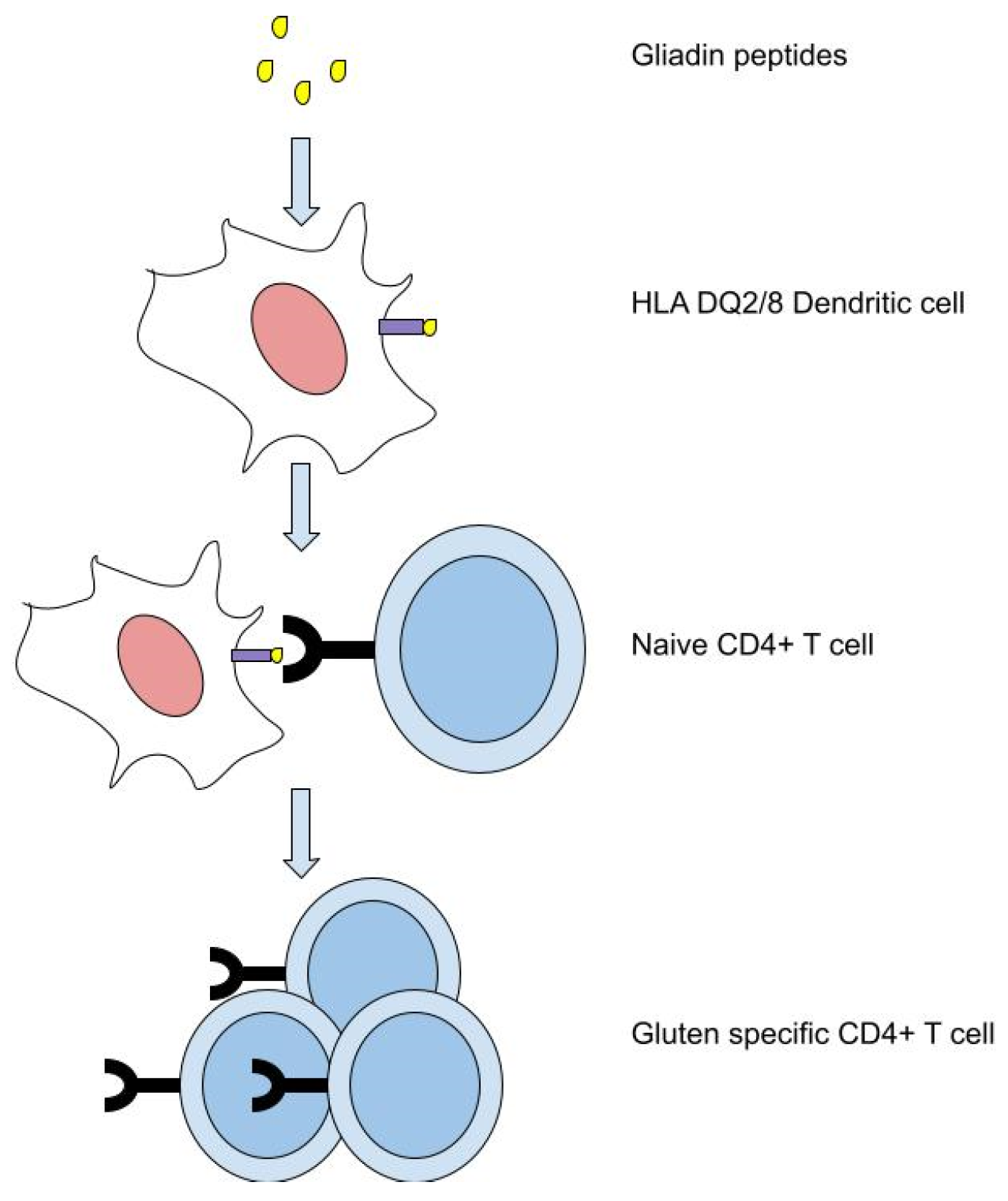
:1. Introduction
2. Materials and Methods
2.1. Identification of Published Coeliac-Specific TCR Sequences
2.2. Cohort 1: Training Dataset
2.2.1. Study Subjects
2.2.2. Processing Intestinal Biopsies
2.2.3. Cell Sorting and Sequencing
2.3. Cohort 2: Testing Dataset
2.4. Data Processing
2.5. Evaluation Measures
2.6. Training, Cross-Validation, and Testing Classification Model with Feature Selection
3. Results
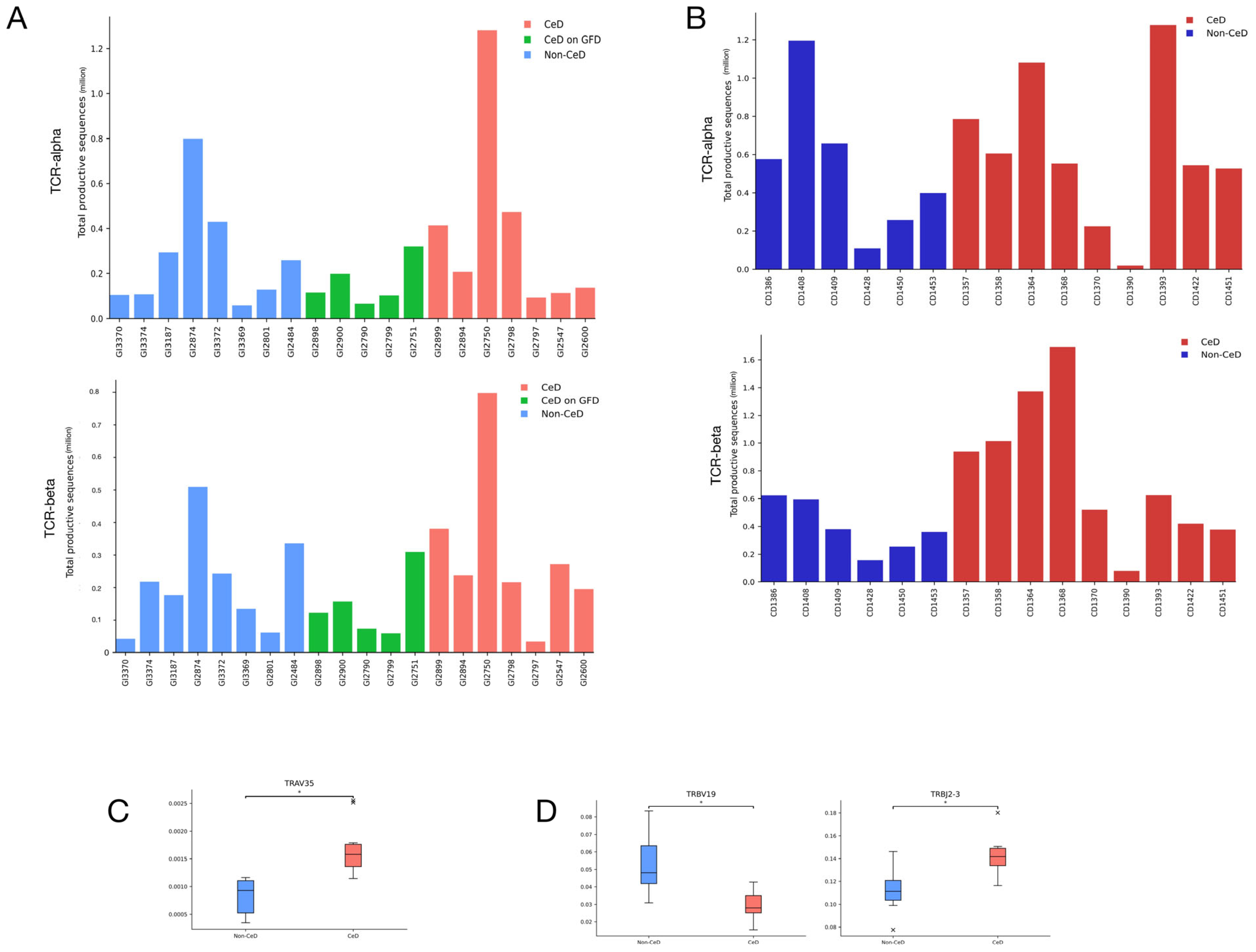
3.1. CeD Patients Cannot Be Identified through Repertoire Characteristics Alone
3.2. Published Gluten-Specific TCRs Are Predominantly TCR-α and TCR-β
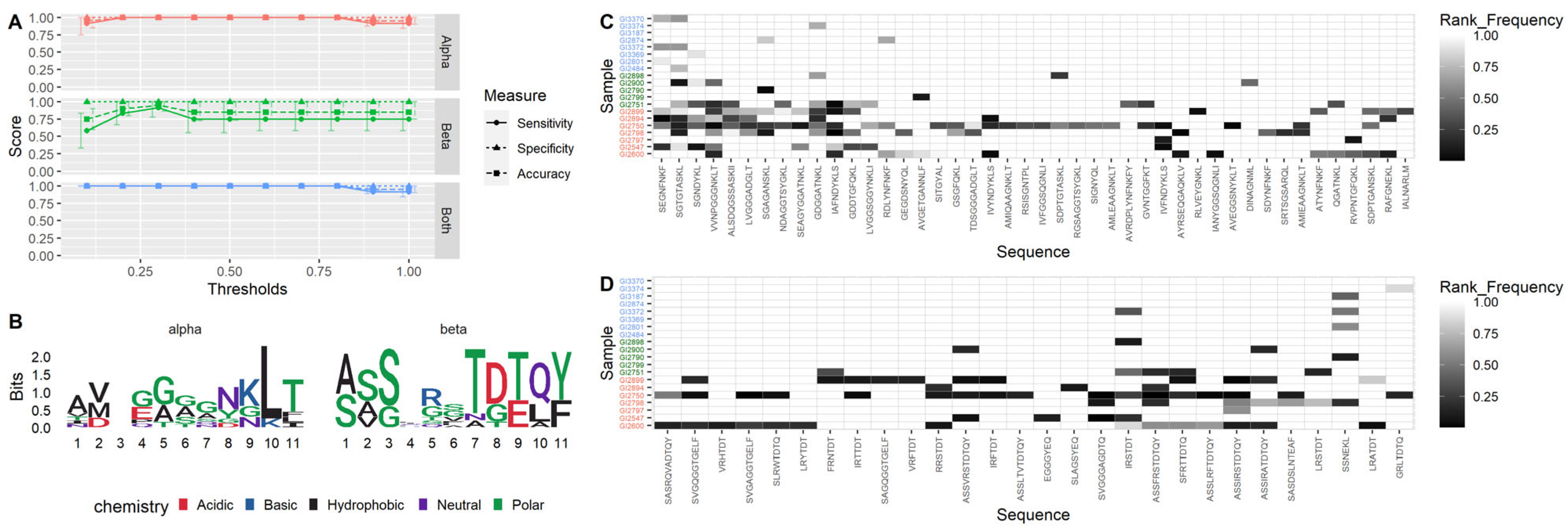
3.3. TCR-α Alone Provides 100% Accuracy on Training Dataset
3.4. CeD Predicted with 100% Accuracy for Patients on a Gluten-Free Diet
3.5. Cross-Validation Shows Model Robustness
3.6. Using TCR-α Alone Provides 80% Testing Accuracy
3.7. Coeliac-Predictive TCR Sequences with Highest Diagnostic Potential Identified
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Caio, G.; Volta, U.; Sapone, A.; Leffler, D.A.; De Giorgio, R.; Catassi, C.; Fasano, A. Celiac Disease: A Comprehensive Current Review. BMC Med. 2019, 17, 142. [Google Scholar] [CrossRef]
- Dieli-Crimi, R.; Cénit, M.C.; Núñez, C. The Genetics of Celiac Disease: A Comprehensive Review of Clinical Implications. J. Autoimmun. 2015, 64, 26–41. [Google Scholar] [CrossRef]
- Abadie, V.; Sollid, L.M.; Barreiro, L.B.; Jabri, B. Integration of Genetic and Immunological Insights into a Model of Celiac Disease Pathogenesis. Annu. Rev. Immunol. 2011, 29, 493–525. [Google Scholar] [CrossRef]
- Krangel, M.S. Mechanics of T Cell Receptor Gene Rearrangement. Curr. Opin. Immunol. 2009, 21, 133–139. [Google Scholar] [CrossRef]
- Kockelbergh, H.; Evans, S.; Deng, T.; Clyne, E.; Kyriakidou, A.; Economou, A.; Luu Hoang, K.N.; Woodmansey, S.; Foers, A.; Fowler, A.; et al. Utility of Bulk T-Cell Receptor Repertoire Sequencing Analysis in Understanding Immune Responses to COVID-19. Diagnostics 2022, 12, 1222. [Google Scholar] [CrossRef]
- Wange, R.L.; Samelson, L.E. Complex Complexes: Signaling at the TCR. Immunity 1996, 5, 197–205. [Google Scholar] [CrossRef]
- Hennecke, J.; Wiley, D.C. T Cell Receptor–MHC Interactions up Close. Cell 2001, 104, 1–4. [Google Scholar] [CrossRef]
- Chien, Y.; Jores, R.; Crowley, M.P. Recognition by γ/δ T Cells. Annu. Rev. Immunol. 1996, 14, 511–532. [Google Scholar] [CrossRef]
- Quarsten, H.; McAdam, S.N.; Jensen, T.; Arentz-Hansen, H.; Molberg, Ø.; Lundin, K.E.A.; Sollid, L.M. Staining of Celiac Disease-Relevant T Cells by Peptide-DQ2 Multimers. J. Immunol. 2001, 167, 4861–4868. [Google Scholar] [CrossRef]
- Arentz-Hansen, H.; Körner, R.; Molberg, Ø.; Quarsten, H.; Vader, W.; Kooy, Y.M.C.; Lundin, K.E.A.; Koning, F.; Roepstorff, P.; Sollid, L.M.; et al. The Intestinal T Cell Response to α-Gliadin in Adult Celiac Disease Is Focused on a Single Deamidated Glutamine Targeted by Tissue Transglutaminase. J. Exp. Med. 2000, 191, 603–612. [Google Scholar] [CrossRef]
- Bonner, W.A.; Hulett, H.R.; Sweet, R.G.; Herzenberg, L.A. Fluorescence Activated Cell Sorting. Rev. Sci. Instrum. 1972, 43, 404–409. [Google Scholar] [CrossRef]
- Smithson, G.; Siegelman, J.; Oki, T.; Maxwell, J.R.; Leffler, D.A. The Evolving Landscape of Biomarkers in Celiac Disease: Leading the Way to Clinical Development. Front. Immunol. 2021, 12, 665756. [Google Scholar] [CrossRef]
- Elhanati, Y.; Sethna, Z.; Callan, C.G.; Mora, T.; Walczak, A.M. Predicting the Spectrum of TCR Repertoire Sharing with a Data-driven Model of Recombination. Immunol. Rev. 2018, 284, 167–179. [Google Scholar] [CrossRef]
- Li, H.; Ye, C.; Ji, G.; Han, J. Determinants of Public T Cell Responses. Cell Res. 2012, 22, 33–42. [Google Scholar] [CrossRef]
- Glanville, J.; Huang, H.; Nau, A.; Hatton, O.; Wagar, L.E.; Rubelt, F.; Ji, X.; Han, A.; Krams, S.M.; Pettus, C.; et al. Identifying Specificity Groups in the T Cell Receptor Repertoire. Nature 2017, 547, 94–98. [Google Scholar] [CrossRef]
- Al-Toma, A.; Volta, U.; Auricchio, R.; Castillejo, G.; Sanders, D.S.; Cellier, C.; Mulder, C.J.; Lundin, K.E.A. European Society for the Study of Coeliac Disease (ESsCD) Guideline for Coeliac Disease and Other Gluten-related Disorders. United Eur. Gastroenterol. J. 2019, 7, 583–613. [Google Scholar] [CrossRef]
- Leffler, D.; Schuppan, D.; Pallav, K.; Najarian, R.; Goldsmith, J.D.; Hansen, J.; Kabbani, T.; Dennis, M.; Kelly, C.P. Kinetics of the Histological, Serological and Symptomatic Responses to Gluten Challenge in Adults with Coeliac Disease. Gut 2013, 62, 996–1004. [Google Scholar] [CrossRef]
- Villanacci, V.; Lorenzi, L.; Donato, F.; Auricchio, R.; Dziechciarz, P.; Gyimesi, J.; Koletzko, S.; Mišak, Z.; Laguna, V.M.; Polanco, I.; et al. Histopathological Evaluation of Duodenal Biopsy in the PreventCD Project: An Observational Interobserver Agreement Study. APMIS 2018, 126, 208–214. [Google Scholar] [CrossRef]
- Weile, B.; Hansen, B.F.; Hagerstrand, I.; Hansen, J.P.H.; Krasilnikoff, P.A. Interobserver Variation in Diagnosing Coeliac Disease. A Joint Study by Danish and Swedish PathologistsNote. APMIS 2000, 108, 380–384. [Google Scholar] [CrossRef]
- Arguelles-Grande, C.; Tennyson, C.A.; Lewis, S.K.; Green, P.H.R.; Bhagat, G. Variability in Small Bowel Histopathology Reporting between Different Pathology Practice Settings: Impact on the Diagnosis of Coeliac Disease. J. Clin. Pathol. 2012, 65, 242–247. [Google Scholar] [CrossRef]
- Foers, A.D.; Shoukat, M.S.; Welsh, O.E.; Donovan, K.; Petry, R.; Evans, S.C.; FitzPatrick, M.E.; Collins, N.; Klenerman, P.; Fowler, A.; et al. Classification of Intestinal T-cell Receptor Repertoires Using Machine Learning Methods Can Identify Patients with Coeliac Disease Regardless of Dietary Gluten Status. J. Pathol. 2021, 253, 279–291. [Google Scholar] [CrossRef]
- FitzPatrick, M.E.B.; Provine, N.M.; Garner, L.C.; Powell, K.; Amini, A.; Irwin, S.L.; Ferry, H.; Ambrose, T.; Friend, P.; Vrakas, G.; et al. Human Intestinal Tissue-Resident Memory T Cells Comprise Transcriptionally and Functionally Distinct Subsets. Cell Rep. 2021, 34, 108661. [Google Scholar] [CrossRef]
- Quigley, M.F.; Greenaway, H.Y.; Venturi, V.; Lindsay, R.; Quinn, K.M.; Seder, R.A.; Douek, D.C.; Davenport, M.P.; Price, D.A. Convergent Recombination Shapes the Clonotypic Landscape of the Naïve T-Cell Repertoire. Proc. Natl. Acad. Sci. USA 2010, 107, 19414–19419. [Google Scholar] [CrossRef]
- Barnitz, R.A.; Imam, S.; Yates, K.; Haining, W.N. Isolation of RNA and the Synthesis and Amplification of CDNA from Antigen-Specific T Cells for Genome-Wide Expression Analysis. In Immune Homeostasis; Snow, A.L., Lenardo, M.J., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 979, pp. 161–173. ISBN 978-1-62703-289-6. [Google Scholar]
- Yao, Y.; Zia, A.; Neumann, R.S.; Pavlovic, M.; Balaban, G.; Lundin, K.E.A.; Sandve, G.K.; Qiao, S.-W. T Cell Receptor Repertoire as a Potential Diagnostic Marker for Celiac Disease. Clin. Immunol. 2021, 222, 108621. [Google Scholar] [CrossRef]
- Risnes, L.F.; Christophersen, A.; Dahal-Koirala, S.; Neumann, R.S.; Sandve, G.K.; Sarna, V.K.; Lundin, K.E.A.; Qiao, S.-W.; Sollid, L.M. Disease-Driving CD4+ T Cell Clonotypes Persist for Decades in Celiac Disease. J. Clin. Investig. 2018, 128, 2642–2650. [Google Scholar] [CrossRef]
- Bolotin, D.A.; Poslavsky, S.; Mitrophanov, I.; Shugay, M.; Mamedov, I.Z.; Putintseva, E.V.; Chudakov, D.M. MiXCR: Software for Comprehensive Adaptive Immunity Profiling. Nat. Methods 2015, 12, 380–381. [Google Scholar] [CrossRef]
- Van Rossum, G.; Drake, F.L. (Eds.) The Python Language Reference; Release 3.0.1 [Repr.]; Python Documentation Manual; Python Software Foundation: Hampton, NH, USA, 2010; ISBN 978-1-4414-1269-0. [Google Scholar]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 56–61. [Google Scholar]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array Programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M. Seaborn: Statistical Data Visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Dunn, O.J. Multiple Comparisons among Means. J. Am. Stat. Assoc. 1961, 56, 52–64. [Google Scholar] [CrossRef]
- Sollid, L.M.; Tye-Din, J.A.; Qiao, S.-W.; Anderson, R.P.; Gianfrani, C.; Koning, F. Update 2020: Nomenclature and Listing of Celiac Disease–Relevant Gluten Epitopes Recognized by CD4+ T Cells. Immunogenetics 2020, 72, 85–88. [Google Scholar] [CrossRef]
- Christophersen, A.; Dahal-Koirala, S.; Chlubnová, M.; Jahnsen, J.; Lundin, K.E.A.; Sollid, L.M. Phenotype-Based Isolation of Antigen-Specific CD4 + T Cells in Autoimmunity: A Study of Celiac Disease. Adv. Sci. 2022, 9, 2104766. [Google Scholar] [CrossRef]
- Han, A.; Newell, E.W.; Glanville, J.; Fernandez-Becker, N.; Khosla, C.; Chien, Y.; Davis, M.M. Dietary Gluten Triggers Concomitant Activation of CD4 + and CD8 + Aβ T Cells and Γδ T Cells in Celiac Disease. Proc. Natl. Acad. Sci. USA 2013, 110, 13073–13078. [Google Scholar] [CrossRef]
- Broughton, S.E.; Petersen, J.; Theodossis, A.; Scally, S.W.; Loh, K.L.; Thompson, A.; van Bergen, J.; Kooy-Winkelaar, Y.; Henderson, K.N.; Beddoe, T.; et al. Biased T Cell Receptor Usage Directed against Human Leukocyte Antigen DQ8-Restricted Gliadin Peptides Is Associated with Celiac Disease. Immunity 2012, 37, 611–621. [Google Scholar] [CrossRef]
- Dahal-Koirala, S.; Neumann, R.S.; Jahnsen, J.; Lundin, K.E.A.; Sollid, L.M. On the Immune Response to Barley in Celiac Disease: Biased and Public T-cell Receptor Usage to a Barley Unique and Immunodominant Gluten Epitope. Eur. J. Immunol. 2020, 50, 256–269. [Google Scholar] [CrossRef]
- Dahal-Koirala, S.; Risnes, L.F.; Christophersen, A.; Sarna, V.K.; Lundin, K.E.; Sollid, L.M.; Qiao, S.W. TCR Sequencing of Single Cells Reactive to DQ2.5-Glia-A2 and DQ2.5-Glia-Ω2 Reveals Clonal Expansion and Epitope-Specific V-Gene Usage. Mucosal Immunol. 2016, 9, 587–596. [Google Scholar] [CrossRef]
- Petersen, J.; van Bergen, J.; Loh, K.L.; Kooy-Winkelaar, Y.; Beringer, D.X.; Thompson, A.; Bakker, S.F.; Mulder, C.J.J.; Ladell, K.; McLaren, J.E.; et al. Determinants of Gliadin-Specific T Cell Selection in Celiac Disease. J. Immunol. 2015, 194, 6112–6122. [Google Scholar] [CrossRef]
- Hardy, M.Y.; Russell, A.K.; Pizzey, C.; Jones, C.M.; Watson, K.A.; La Gruta, N.L.; Cameron, D.J.; Tye-Din, J.A. Characterisation of Clinical and Immune Reactivity to Barley and Rye Ingestion in Children with Coeliac Disease. Gut 2020, 69, 830–840. [Google Scholar] [CrossRef]
- Gunnarsen, K.S.; Høydahl, L.S.; Risnes, L.F.; Dahal-Koirala, S.; Neumann, R.S.; Bergseng, E.; Frigstad, T.; Frick, R.; du Pré, M.F.; Dalhus, B.; et al. A TCRα Framework–Centered Codon Shapes a Biased T Cell Repertoire through Direct MHC and CDR3β Interactions. JCI Insight 2017, 2, e95193. [Google Scholar] [CrossRef]
- Cook, L.; Munier, C.M.L.; Seddiki, N.; Hardy, M.Y.; Anderson, R.P.; Zaunders, J.; Tye-Din, J.A.; Kelleher, A.D.; Bockel, D. Circulating Gluten-specific, but Not CMV-specific, CD39 + Regulatory T Cells Have an Oligoclonal TCR Repertoire. Clin. Transl. Immunol. 2020, 9, e1096. [Google Scholar] [CrossRef]
- Ting, Y.T.; Dahal-Koirala, S.; Kim, H.S.K.; Qiao, S.-W.; Neumann, R.S.; Lundin, K.E.A.; Petersen, J.; Reid, H.H.; Sollid, L.M.; Rossjohn, J. A Molecular Basis for the T Cell Response in HLA-DQ2.2 Mediated Celiac Disease. Proc. Natl. Acad. Sci. USA 2020, 117, 3063–3073. [Google Scholar] [CrossRef]
- Qiao, S.-W.; Christophersen, A.; Lundin, K.E.A.; Sollid, L.M. Biased Usage and Preferred Pairing of α- and β-Chains of TCRs Specific for an Immunodominant Gluten Epitope in Coeliac Disease. Int. Immunol. 2014, 26, 13–19. [Google Scholar] [CrossRef]
- Qiao, S.-W.; Ráki, M.; Gunnarsen, K.S.; Løset, G.-Å.; Lundin, K.E.A.; Sandlie, I.; Sollid, L.M. Posttranslational Modification of Gluten Shapes TCR Usage in Celiac Disease. J. Immunol. 2011, 187, 3064–3071. [Google Scholar] [CrossRef]
- Dahal-Koirala, S.; Ciacchi, L.; Petersen, J.; Risnes, L.F.; Neumann, R.S.; Christophersen, A.; Lundin, K.E.A.; Reid, H.H.; Qiao, S.-W.; Rossjohn, J.; et al. Discriminative T-Cell Receptor Recognition of Highly Homologous HLA-DQ2–Bound Gluten Epitopes. J. Biol. Chem. 2019, 294, 941–952. [Google Scholar] [CrossRef]
- Hardy, M.Y.; Girardin, A.; Pizzey, C.; Cameron, D.J.; Watson, K.A.; Picascia, S.; Auricchio, R.; Greco, L.; Gianfrani, C.; La Gruta, N.L.; et al. Consistency in Polyclonal T-Cell Responses to Gluten Between Children and Adults With Celiac Disease. Gastroenterology 2015, 149, 1541–1552.e2. [Google Scholar] [CrossRef]
- Petersen, J.; Montserrat, V.; Mujico, J.R.; Loh, K.L.; Beringer, D.X.; van Lummel, M.; Thompson, A.; Mearin, M.L.; Schweizer, J.; Kooy-Winkelaar, Y.; et al. T-Cell Receptor Recognition of HLA-DQ2–Gliadin Complexes Associated with Celiac Disease. Nat. Struct. Mol. Biol. 2014, 21, 480–488. [Google Scholar] [CrossRef]
- Christophersen, A.; Ráki, M.; Bergseng, E.; Lundin, K.E.; Jahnsen, J.; Sollid, L.M.; Qiao, S.-W. Tetramer-visualized Gluten-specific CD4+ T Cells in Blood as a Potential Diagnostic Marker for Coeliac Disease without Oral Gluten Challenge. United Eur. Gastroenterol. J. 2014, 2, 268–278. [Google Scholar] [CrossRef]
- Sarna, V.K.; Lundin, K.E.A.; Mørkrid, L.; Qiao, S.-W.; Sollid, L.M.; Christophersen, A. HLA-DQ–Gluten Tetramer Blood Test Accurately Identifies Patients With and Without Celiac Disease in Absence of Gluten Consumption. Gastroenterology 2018, 154, 886–896.e6. [Google Scholar] [CrossRef]
- Anderson, R.P.; Goel, G.; Hardy, M.Y.; Russell, A.K.; Wang, S.; Szymczak, E.; Zhang, R.; Goldstein, K.E.; Neff, K.; Truitt, K.E.; et al. Whole Blood Interleukin-2 Release Test to Detect and Characterize Rare Circulating Gluten-Specific T Cell Responses in Coeliac Disease. Clin. Exp. Immunol. 2021, 204, 321–334. [Google Scholar] [CrossRef]
- Dahal-Koirala, S.; Risnes, L.F.; Neumann, R.S.; Christophersen, A.; Lundin, K.E.A.; Sandve, G.K.; Qiao, S.-W.; Sollid, L.M. Comprehensive Analysis of CDR3 Sequences in Gluten-Specific T-Cell Receptors Reveals a Dominant R-Motif and Several New Minor Motifs. Front. Immunol. 2021, 12, 639672. [Google Scholar] [CrossRef]

| TCR-α | TCR-β | |||
|---|---|---|---|---|
| Cohort 1: Training | Cohort 2: Testing | Cohort 1: Training | Cohort 2: Testing | |
| Number of samples | 20 (12 CeD, 8 controls) | 15 (9 CeD, 6 controls) | 20 (12 CeD, 8 controls) | 15 (9 CeD, 6 controls) |
| Mean number of productive sequences per sample (min–max) | 284,888 (57,629–1,280,631) | 587,482 (19,649–1,276,321) | 228,422 (33,421–797,688) | 627,303 (787,765–1,693,681) |
| Mean number of clones per sample (min–max) | 5203 (1404–16,376) | 1104 (217–3747) | 4076 (830–12,048) | 2231 (362–8613) |
| Mean number of CD4+ T cells sorted per sample (min–max) | 25,603 (6054–104,187) | 8000 (estimated) | 25,603 (6054–104,187) | 8000 (estimated) |
| Publication | Chains (Number of Sequences) | Paired | T-Cell Gluten Epitopes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DQ8-glia-α1 | DQ8-glia-γ1b | DQ2.2-glut-L1 | DQ2.5-glia-α1a | DQ2.5-glia-α2 | DQ2.5-glia-ω1 | DQ2.5-glia-ω2 | DQ2.5-glia-γ2 | DQ2.5-hor-3 | |||
| Broughton [37] | TCR-α (9) TCR-β (9) | Yes |  | ||||||||
| Dahal-Koirala [38] | TCR-α (11) TCR-β (11) | Yes |  | ||||||||
| Dahal-Koirala [39] | TCR-α (10) TCR-β (10) | Yes |  |  | |||||||
| Petersen [40] | TCR-α (18) TCR-β (18) | Yes |  | ||||||||
| Risnes [26] | TCR-α (14) TCR-β (8) | No |  |  |  |  |  |  | |||
| Hardy [41] | TCR-α (211) TCR-β (216) | No |  |  |  | ||||||
| Gunnarsen [42] | TCR-α (9) TCR-β (9) | Yes |  |  |  |  | |||||
| Cook [43] | TCR-β (26) | No |  |  | |||||||
| Ting [44] | TCR-α (23) TCR-β (23) | Yes |  | ||||||||
| Qiao [45] | TCR-α (29) TCR-β (29) | Yes |  | ||||||||
| Qiao [46] | TCR-β (6) | No |  |  |  | ||||||
| Dahal-Koirala [47] | TCR-α (7) TCR-β (7) | Yes |  |  | |||||||
| Hardy [48] | TCR-β (21) | No |  |  | |||||||
| Petersen [49] | TCR-α (20) TCR-β (20) | Yes |  |  | |||||||
| Han [36] | TCR-β (437) TCR-δ (194) | No |  |  | |||||||
| Christophersen [35] | TCR-α (10) TCR-β (7) | Yes |  |  |  |  |  | ||||
| Total TCR-α: 357 Total TCR-β: 799 | |||||||||||
| Loci | Sensitivity | Specificity | Balanced Accuracy | Optimal Threshold Values |
|---|---|---|---|---|
| TCR-α | 1.0 | 0.75 | 0.875 | 0.2–0.8 |
| TCR-β | 0.92 | 0.25 | 0.585 | 0.3 |
| TCR-α and TCR-β | 1.0 | 0.75 | 0.875 | 0.1–0.8 |
| Rank | Sequence | Training: Number of CeD Samples | Validation: Number of Cross-Validation Folds | Testing: Number of CeD Samples |
|---|---|---|---|---|
| 1 | AYRSEQGAQKLV | 2 | 20 | 3 |
| 2 | GDGGATNKL | 6 | 19 | 2 |
| 3 | RDLYNFNKF | 2 | 19 | 2 |
| 4 | GDDTGFQKL | 4 | 20 | 1 |
| 5 | IVFNDYKLS | 3 | 20 | 1 |
| 6= | AVGETGANNLF | 2 | 19 | 1 |
| 6= | SITGYAL | 2 | 19 | 1 |
| 8= | DINAGNML | 1 | 19 | 1 |
| 8= | RGSAGGTSYGKL | 1 | 19 | 1 |
| 8= | AVEGGSNYKLT | 1 | 19 | 1 |
| Rank | Sequence | Training: Number of CeD Samples | Validation: Number of Cross-Validation Folds | Testing: Number of CeD Samples |
|---|---|---|---|---|
| 1 | IRSTDT | 4 | 20 | 3 |
| 2 | VRFTDT | 1 | 19 | 3 |
| 3 | SFRTTDTQ | 4 | 20 | 1 |
| 4 | ASSIRATDTQY | 3 | 20 | 1 |
| 5= | IRTTDT | 2 | 20 | 1 |
| 5= | LRSTDT | 2 | 19 | 1 |
| 5= | LRATDT | 2 | 19 | 1 |
| 5= | SASDSLNTEAF | 2 | 19 | 1 |
| 5= | SLRWTDTQ | 2 | 20 | 1 |
| 10 | ASSLTVTDTQY | 1 | 19 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fowler, A.; FitzPatrick, M.; Shanmugarasa, A.; Ibrahim, A.S.F.; Kockelbergh, H.; Yang, H.-C.; Williams-Walker, A.; Luu Hoang, K.N.; Evans, S.; Provine, N.; et al. An Interpretable Classification Model Using Gluten-Specific TCR Sequences Shows Diagnostic Potential in Coeliac Disease. Biomolecules 2023, 13, 1707. https://doi.org/10.3390/biom13121707
Fowler A, FitzPatrick M, Shanmugarasa A, Ibrahim ASF, Kockelbergh H, Yang H-C, Williams-Walker A, Luu Hoang KN, Evans S, Provine N, et al. An Interpretable Classification Model Using Gluten-Specific TCR Sequences Shows Diagnostic Potential in Coeliac Disease. Biomolecules. 2023; 13(12):1707. https://doi.org/10.3390/biom13121707
Chicago/Turabian StyleFowler, Anna, Michael FitzPatrick, Aberami Shanmugarasa, Amro Sayed Fadel Ibrahim, Hannah Kockelbergh, Han-Chieh Yang, Amelia Williams-Walker, Kim Ngan Luu Hoang, Shelley Evans, Nicholas Provine, and et al. 2023. "An Interpretable Classification Model Using Gluten-Specific TCR Sequences Shows Diagnostic Potential in Coeliac Disease" Biomolecules 13, no. 12: 1707. https://doi.org/10.3390/biom13121707
APA StyleFowler, A., FitzPatrick, M., Shanmugarasa, A., Ibrahim, A. S. F., Kockelbergh, H., Yang, H.-C., Williams-Walker, A., Luu Hoang, K. N., Evans, S., Provine, N., Klenerman, P., & Soilleux, E. J. (2023). An Interpretable Classification Model Using Gluten-Specific TCR Sequences Shows Diagnostic Potential in Coeliac Disease. Biomolecules, 13(12), 1707. https://doi.org/10.3390/biom13121707







