Roles of Human Endogenous Retroviruses and Endogenous Virus-Like Elements in Cancer Development and Innate Immunity
Abstract
:1. Introduction
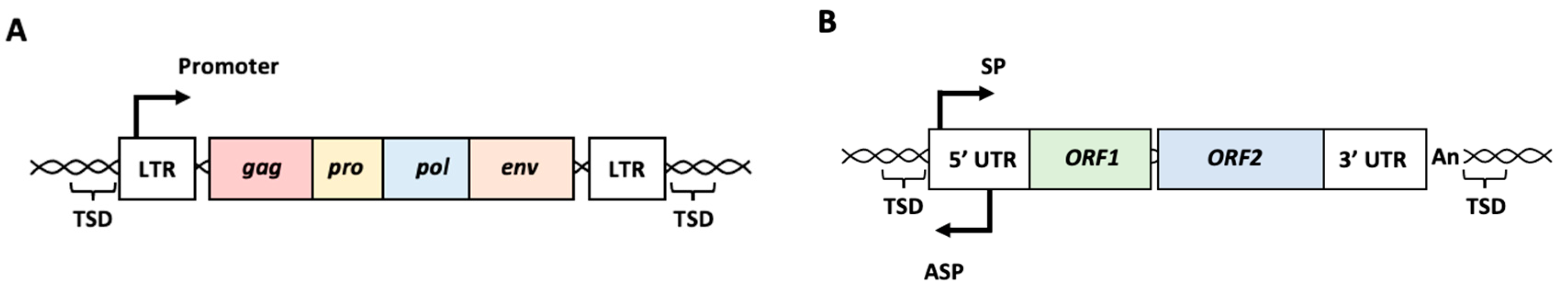
2. Involvement of HERVs and LINEs in Cancers
2.1. HERVs and Cancers
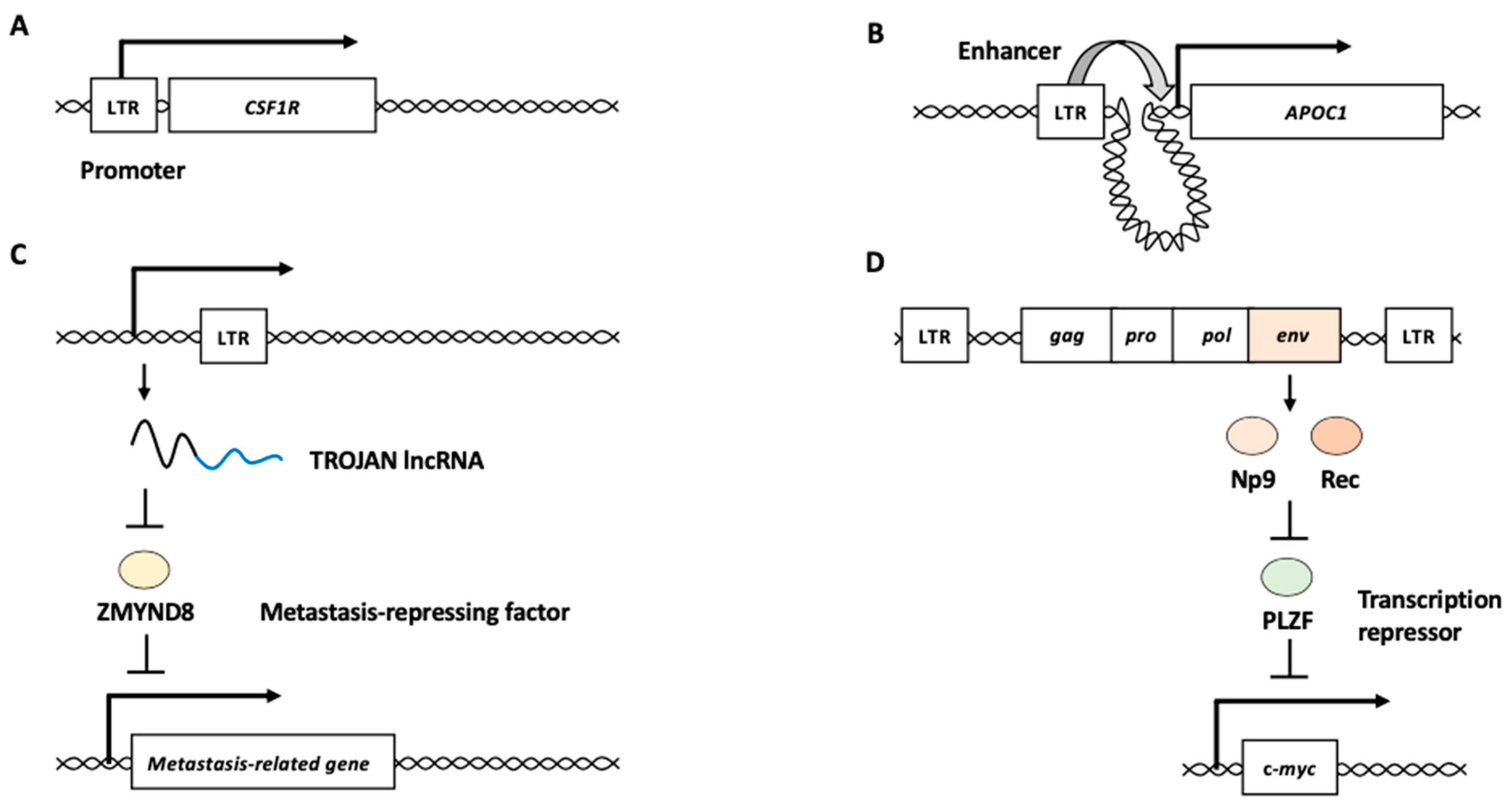
2.1.1. LTRs as Promoter/Enhancer Elements
2.1.2. HERV-Derived RNAs and Proteins
2.2. L1 and Cancers
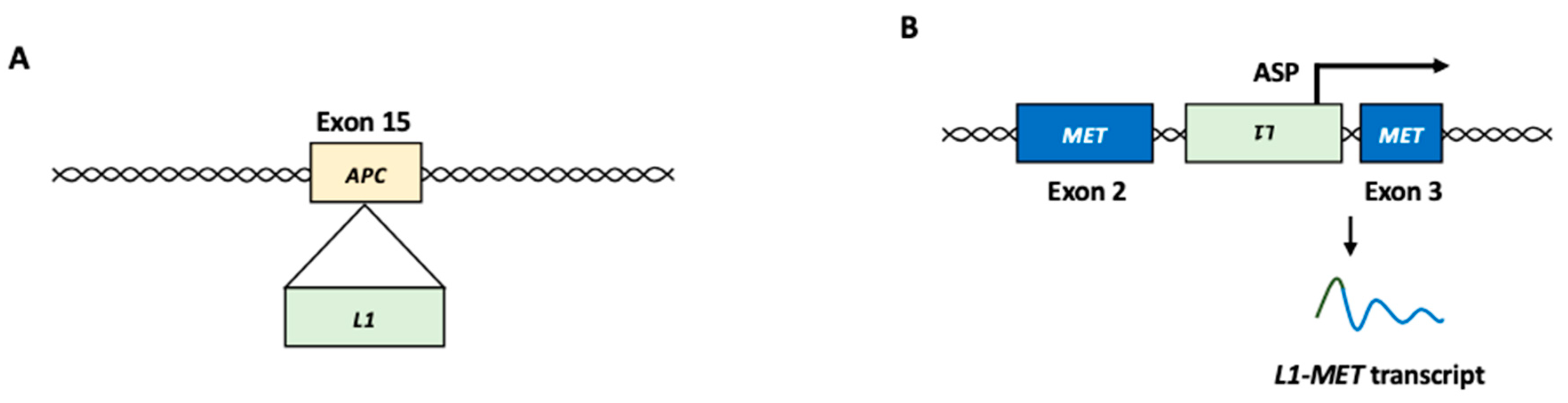
2.2.1. L1 Insertional Mutagenesis
2.2.2. L1 Expression
2.2.3. L1 Chimeric Transcript
3. Involvement of HERVs, LINEs, and nrEVEs in Immunity
3.1. HERVs and Innate Immunity
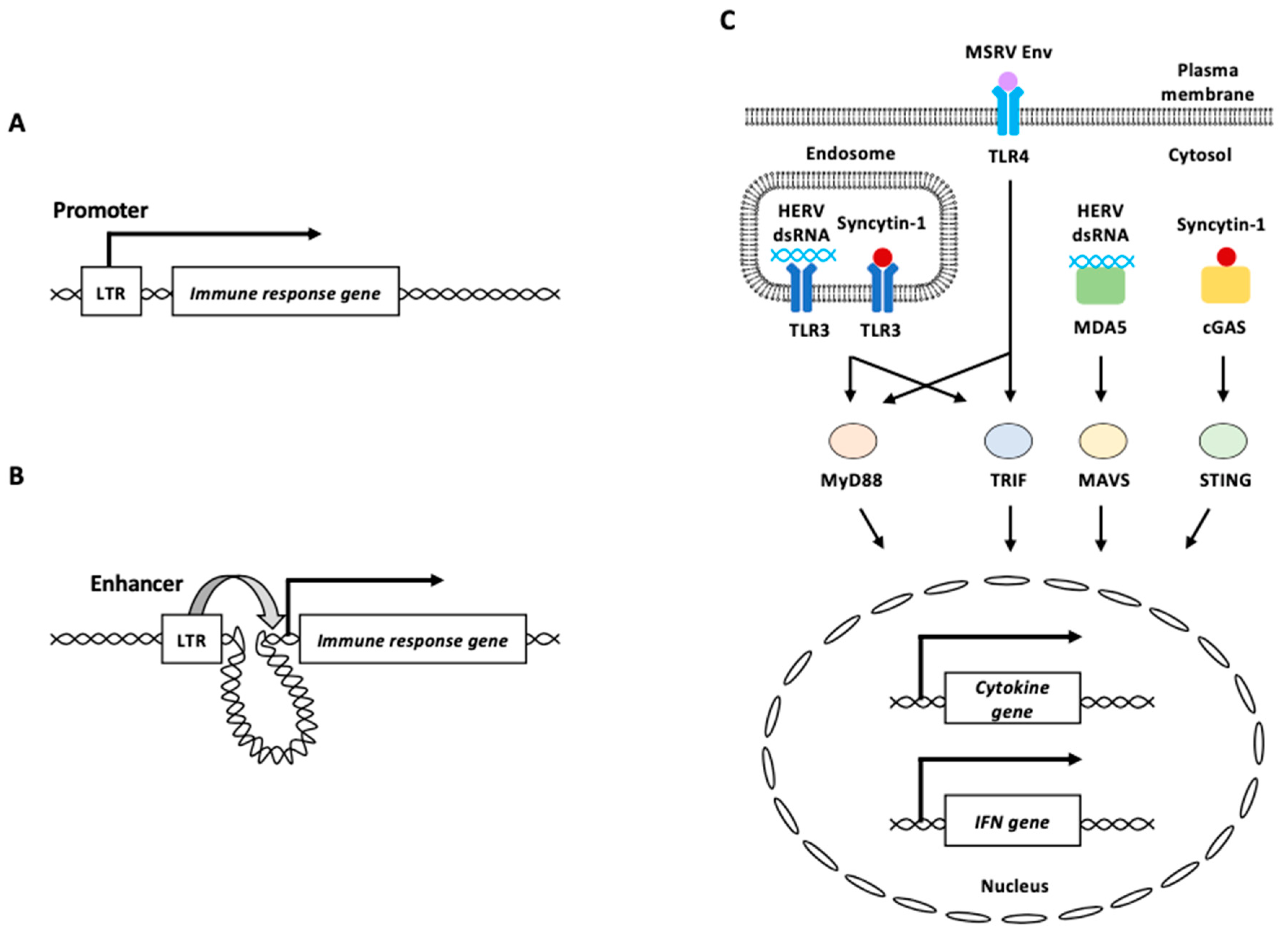
3.1.1. LTRs as Promoter/Enhancer Elements
3.1.2. HERV-Derived cDNAs, RNAs, and Proteins
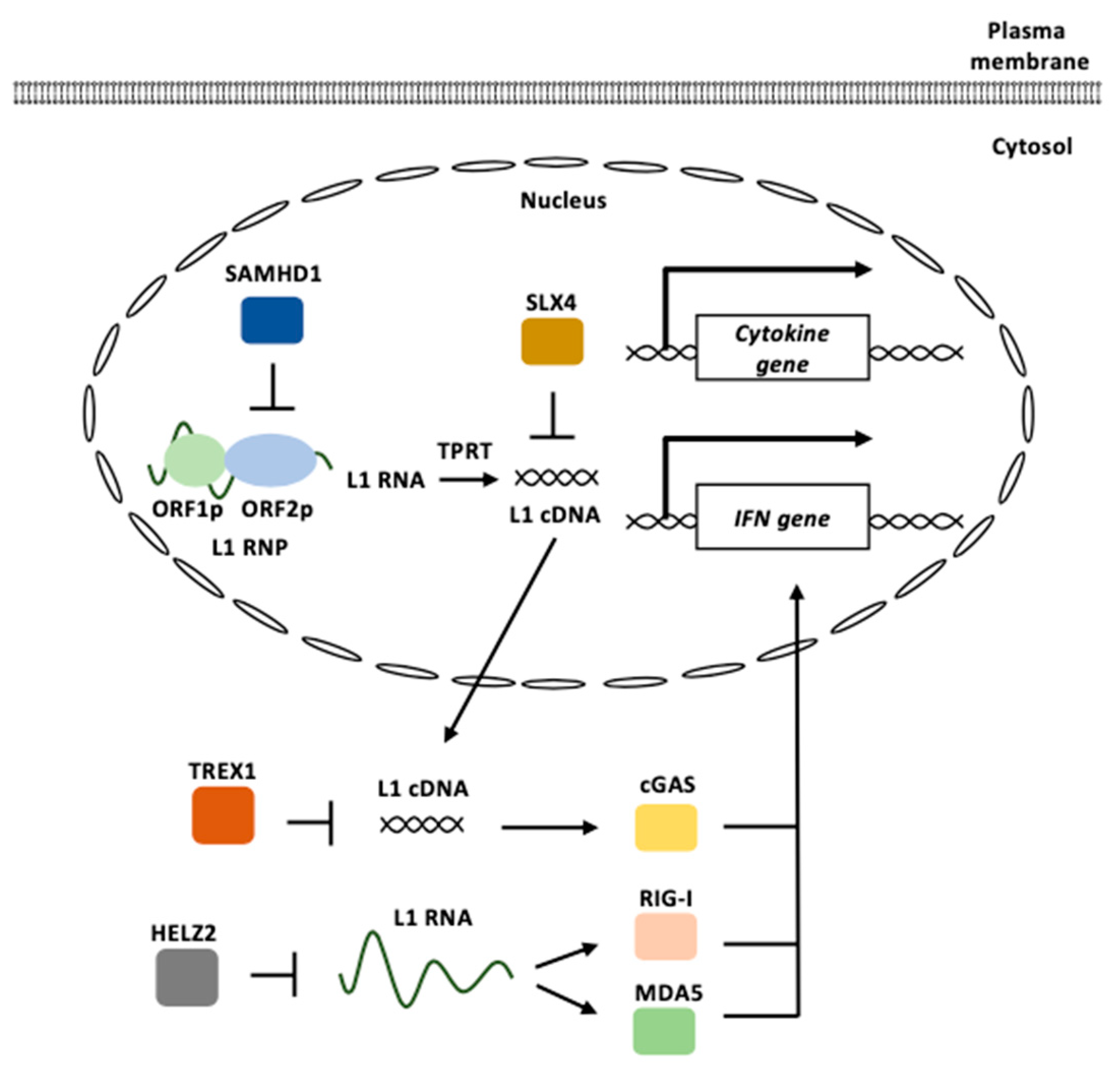
3.2. L1 and Innate Immunity
3.3. Non-Retroviral Endogenous Viral Elements and Innate Immunity
4. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kim, Y.-J.; Lee, J.; Han, K. Transposable Elements: No More “Junk DNA”. Genom. Inform. 2012, 10, 226–233. [Google Scholar] [CrossRef]
- Honda, T.; Tomonaga, K. Endogenous Non-Retroviral RNA Virus Elements Evidence a Novel Type of Antiviral Immunity. Mob. Genet. Elem. 2016, 6, 1548–1554. [Google Scholar] [CrossRef]
- Ogawa, H.; Honda, T. Viral Sequences Are Repurposed for Controlling Antiviral Responses as Non-Retroviral Endogenous Viral Elements. Acta Med. Okayama 2022, 76, 503–510. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; Fitzhugh, W.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Kazazian, H.H.; Moran, J.V. Mobile DNA in Health and Disease. N. Engl. J. Med. 2017, 377, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Chénais, B. Transposable Elements and Human Diseases: Mechanisms and Implication in the Response to Environmental Pollutants. Int. J. Mol. Sci. 2022, 23, 2551. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.; Macia, A.; Muotri, A.R. Transposable Elements, Inflammation, and Neurological Disease. Front. Neurol. 2019, 10, 894. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Lee, X.; Li, X.; Veldman, G.M.; Finnerty, H.; Racie, L.; Lavallie, E.; Tang, X.; Edouard, P.; Howes, S.; et al. Syncytin Is a Captive Retroviral Envelope Protein Involved in placental morphogenesis. Nature 2000, 403, 785–789. [Google Scholar] [CrossRef]
- Sexton, C.E.; Tillett, R.L.; Han, M.V. The Essential but Enigmatic Regulatory Role of HERVH in Pluripotency. Trends Genet. 2022, 38, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Mueller, C.; Aschacher, T.; Wolf, B.; Bergmann, M. A Role of Line-1 in Telomere Regulation. Front. Biosci. Landmark 2018, 23, 1310–1319. [Google Scholar] [CrossRef]
- Stoye, J.P. Endogenous Retroviruses: Still Active after All These Years? Curr. Biol. 2001, 11, R914–R916. [Google Scholar] [CrossRef] [PubMed]
- Jern, P.; Coffin, J.M. Effects of Retroviruses on Host Genome Function. Annu. Rev. Genet. 2008, 42, 709–732. [Google Scholar] [CrossRef]
- Hurst, T.P.; Magiorkinis, G. Epigenetic Control of Human Endogenous Retrovirus Expression: Focus on Regulation of Long-Terminal Repeats (LTRs). Viruses 2017, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Kemp, J.R.; Longworth, M.S. Crossing the LINE toward Genomic Instability: LINE-1 Retrotransposition in Cancer. Front. Chem. 2015, 3, 68. [Google Scholar] [CrossRef] [PubMed]
- Xiao-Jie, L.; Hui-Ying, X.; Qi, X.; Jiang, X.; Shi-Jie, M. LINE-1 in Cancer: Multifaceted Functions and Potential Clinical Implications. Genet. Med. 2016, 18, 431–439. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, Y.; Du, J.; Gao, P.; Zhao, K. The Interplay Among HIV, LINE-1, and the Interferon Signaling System. Front. Immunol. 2021, 12, 732775. [Google Scholar] [CrossRef] [PubMed]
- Warkocki, Z. An Update on Post-Transcriptional Regulation of Retrotransposons. FEBS Lett. 2022, 597, 380–406. [Google Scholar] [CrossRef]
- Beck, C.R.; Collier, P.; Macfarlane, C.; Malig, M.; Kidd, J.M.; Eichler, E.E.; Badge, R.M.; Moran, J.V. LINE-1 Retrotransposition Activity in Human Genomes. Cell 2010, 141, 1159–1170. [Google Scholar] [CrossRef]
- Brouha, B.; Schustak, J.; Badge, R.M.; Lutz-Prigge, S.; Farley, A.H.; Morant, J.V.; Kazazian, H.H. Hot L1s Account for the Bulk of Retrotransposition in the Human Population. Proc. Natl. Acad. Sci. USA 2003, 100, 5280–5285. [Google Scholar] [CrossRef]
- Konkel, M.K.; Batzer, M.A. A Mobile Threat to Genome Stability: The Impact of Non-LTR Retrotransposons upon the Human Genome. Semin. Cancer Biol. 2010, 20, 211–221. [Google Scholar] [CrossRef]
- Bhat, A.; Ghatage, T.; Bhan, S.; Lahane, G.P.; Dhar, A.; Kumar, R.; Pandita, R.K.; Bhat, K.M.; Ramos, K.S.; Pandita, T.K. Role of Transposable Elements in Genome Stability: Implications for Health and Disease. Int. J. Mol. Sci. 2022, 23, 7802. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, K.; Shinkai, Y. SETDB1-Mediated Silencing of Retroelements. Viruses 2020, 12, 596. [Google Scholar] [CrossRef]
- Zhang, Q.; Pan, J.; Cong, Y.; Mao, J. Transcriptional Regulation of Endogenous Retroviruses and Their Misregulation in Human Diseases. Int. J. Mol. Sci. 2022, 23, 10112. [Google Scholar] [CrossRef]
- Grandi, N.; Tramontano, E. HERV Envelope Proteins: Physiological Role and Pathogenic Potential in Cancer and Autoimmunity. Front. Microbiol. 2018, 9, 462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liang, J.Q.; Zheng, S. Expressional Activation and Functional Roles of Human Endogenous Retroviruses in Cancers. Rev. Med. Virol. 2019, 29, e2025. [Google Scholar] [CrossRef]
- Del Re, B.; Giorgi, G. Long INterspersed Element-1 Mobility as a Sensor of Environmental Stresses. Environ. Mol. Mutagen. 2020, 61, 465–493. [Google Scholar] [CrossRef] [PubMed]
- Katoh, H.; Asai, H.; Takemoto, K.; Tomizawa, R.; Honda, C.; Watanabe, M.; Honda, T. Cap Analysis of Gene Expression Clarifies Transcriptomic Divergence Within Monozygotic Twin Pairs. Twin Res. Human. Genet. 2023, 26, 269–276. [Google Scholar] [CrossRef]
- Gonzalez-Cao, M.; Iduma, P.; Karachaliou, N.; Santarpia, M.; Blanco, J.; Rosell, R. Human Endogenous Retroviruses and Cancer. Cancer Biol. Med. 2016, 13, 483–488. [Google Scholar]
- Yu, H.L.; Zhao, Z.K.; Zhu, F. The Role of Human Endogenous Retroviral Long Terminal Repeat Sequences in Human Cancer (Review). Int. J. Mol. Med. 2013, 32, 755–762. [Google Scholar] [CrossRef]
- Lavia, P.; Sciamanna, I.; Spadafora, C. An Epigenetic LINE-1-Based Mechanism in Cancer. Int. J. Mol. Sci. 2022, 23, 14610. [Google Scholar] [CrossRef]
- Kitsou, K.; Lagiou, P.; Magiorkinis, G. Human Endogenous Retroviruses in Cancer: Oncogenesis Mechanisms and Clinical Implications. J. Med. Virol. 2023, 95, e28350. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.C.; Devine, S.E. The Role of Somatic L1 Retrotransposition in Human Cancers. Viruses 2017, 9, 131. [Google Scholar] [CrossRef] [PubMed]
- Jansz, N.; Faulkner, G.J. Endogenous Retroviruses in the Origins and Treatment of Cancer. Genome Biol. 2021, 22, 147. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Nishikawa, Y.; Nishimura, K.; Teng, D.; Takemoto, K.; Ueda, K. Effects of Activation of the LINE-1 Antisense Promoter on the Growth of Cultured Cells. Sci. Rep. 2020, 10, 22136. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Zhang, R.; Zhang, X.; Sun, Y.; Liu, P.; Francoeur, N.; Han, L.; Lam, W.Y.; Yi, Z.; Sebra, R.; et al. LINE-1 Promotes Tumorigenicity and Exacerbates Tumor Progression via Stimulating Metabolism Reprogramming in Non-Small Cell Lung Cancer. Mol. Cancer 2022, 21, 147. [Google Scholar] [CrossRef]
- Swergold, G.D. Identification, Characterization, and Cell Specificity of a Human LINE-1 Promoter. Mol. Cell. Biol. 1990, 10, 6718–6729. [Google Scholar] [CrossRef] [PubMed]
- Moran, J.V.; DeBerardinis, R.J.; Kazazian, H.H. Exon Shuffling by L1 Retrotransposition. Science 1999, 283, 1530–1534. [Google Scholar] [CrossRef]
- Goodier, J.L.; Ostertag, E.M.; Kazazian, H.H. Transduction of 3′-Flanking Sequences Is Common in L1 Retrotransposition. Hum. Mol. Genet. 2000, 9, 653–657. [Google Scholar] [CrossRef]
- Speek, M. Antisense Promoter of Human L1 Retrotransposon Drives Transcription of Adjacent Cellular Genes. Mol. Cell. Biol. 2001, 21, 1973–1985. [Google Scholar] [CrossRef]
- Stacey, K.J.; Sagulenko, V. A Clear Link between Endogenous Retroviral LTR Activity and Hodgkin’s Lymphoma. Cell Res. 2010, 20, 869–871. [Google Scholar] [CrossRef]
- Lamprecht, B.; Walter, K.; Kreher, S.; Kumar, R.; Hummel, M.; Lenze, D.; Köchert, K.; Bouhlel, M.A.; Richter, J.; Soler, E.; et al. Derepression of an Endogenous Long Terminal Repeat Activates the CSF1R Proto-Oncogene in Human Lymphoma. Nat. Med. 2010, 16, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Deniz, Ö.; Ahmed, M.; Todd, C.D.; Rio-Machin, A.; Dawson, M.A.; Branco, M.R. Endogenous Retroviruses Are a Source of Enhancers with Oncogenic Potential in Acute Myeloid Leukaemia. Nat. Commun. 2020, 11, 3506. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.Z.; Liu, Y.R.; Xu, X.E.; Jin, X.; Hu, X.; Yu, K.D.; Shao, Z.M. Transcriptome Analysis of Triple-Negative Breast Cancer Reveals an Integrated MRNA-LncRNA Signature with Predictive and Prognostic Value. Cancer Res. 2016, 76, 2105–2114. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Xu, X.E.; Jiang, Y.Z.; Liu, Y.R.; Sun, W.; Guo, Y.J.; Ren, Y.X.; Zuo, W.J.; Hu, X.; Huang, S.L.; et al. The Endogenous Retrovirus-Derived Long Noncoding RNA TROJAN Promotes Triple-Negative Breast Cancer Progression via ZMYND8 Degradation. Sci. Adv. 2019, 5, eaat9820. [Google Scholar] [CrossRef]
- Manasanch, E.E.; Orlowski, R.Z. Proteasome Inhibitors in Cancer Therapy. Nat. Rev. Clin. Oncol. 2017, 14, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Lower, R.; Lower, J.; Kurth, R. The Viruses in All of Us: Characteristics and Biological Significance of Human Endogenous Retrovirus Sequences. Proc. Natl. Acad. Sci. USA 1996, 93, 5177–5184. [Google Scholar] [CrossRef]
- Xue, B.; Sechi, L.A.; Kelvin, D.J. Human Endogenous Retrovirus K (HML-2) in Health and Disease. Front. Microbiol. 2020, 11, 1690. [Google Scholar] [CrossRef]
- Ma, W.; Hong, Z.; Liu, H.; Chen, X.; Ding, L.; Liu, Z.; Zhou, F.; Yuan, Y. Human Endogenous Retroviruses-k (HML-2) Expression Is Correlated with Prognosis and Progress of Hepatocellular Carcinoma. BioMed Res. Int. 2016, 2016, 8201642. [Google Scholar] [CrossRef]
- Li, M.; Radvanyi, L.; Yin, B.; Li, J.; Chivukula, R.; Lin, K.; Lu, Y.; Shen, J.J.; Chang, D.Z.; Li, D.; et al. Downregulation of Human Endogenous Retrovirus Type K (HERV-K) Viral Env RNA in Pancreatic Cancer Cells Decreases Cell Proliferation and Tumor Growth. Clin. Cancer Res. 2017, 23, 5892–5911. [Google Scholar] [CrossRef]
- Curty, G.; Marston, J.L.; De Mulder Rougvie, M.; Leal, F.E.; Nixon, D.F.; Soares, M.A. Human Endogenous Retrovirus K in Cancer: A Potential Biomarker and Immunotherapeutic Target. Viruses 2020, 12, 726. [Google Scholar] [CrossRef]
- Armbruester, V.; Sauter, M.; Krautkraemer, E.; Meese, E.; Kleiman, A.; Best, B.; Roemer, K.; Mueller-Lantzsch, N. A Novel Gene from the Human Endogenous Retrovirus K Expressed in Transformed Cells. Clin. Cancer Res. 2002, 8, 1800–1807. [Google Scholar]
- Denne, M.; Sauter, M.; Armbruester, V.; Licht, J.D.; Roemer, K.; Mueller-Lantzsch, N. Physical and Functional Interactions of Human Endogenous Retrovirus Proteins Np9 and Rec with the Promyelocytic Leukemia Zinc Finger Protein. J. Virol. 2007, 81, 5607–5616. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Chen, T.; Meng, Z.; Gan, Y.; Wang, X.; Xu, F.; Gu, Y.; Xu, X.; Tang, J.; Zhou, H.; et al. The Viral Oncogene Np9 Acts as a Critical Molecular Switch for Co-Activating β-Catenin, ERK, Akt and Notch1 and Promoting the Growth of Human Leukemia Stem/Progenitor Cells. Leukemia 2013, 27, 1469–1478. [Google Scholar] [CrossRef]
- Bergallo, M.; Montanari, P.; Mareschi, K.; Merlino, C.; Berger, M.; Bini, I.; Daprà, V.; Galliano, I.; Fagioli, F. Expression of the Pol Gene of Human Endogenous Retroviruses HERV-K and -W in Leukemia Patients. Arch. Virol. 2017, 162, 3639–3644. [Google Scholar] [CrossRef] [PubMed]
- Grandi, N.; Cadeddu, M.; Blomberg, J.; Tramontano, E. Contribution of Type W Human Endogenous Retroviruses to the Human Genome: Characterization of HERV-W Proviral Insertions and Processed Pseudogenes. Retrovirology 2016, 13, 67. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Chen, P.T.; Chang, G.D.; Huang, C.J.; Chen, H. Functional Characterization of the Placental Fusogenic Membrane Protein Syncytin. Biol. Reprod. 2004, 71, 1956–1962. [Google Scholar] [CrossRef]
- Wang, Q.; Shi, Y.; Bian, Q.; Zhang, N.; Wang, M.; Wang, J.; Li, X.; Lai, L.; Zhao, Z.; Yu, H. Molecular Mechanisms of Syncytin-1 in Tumors and Placental Development Related Diseases. Discov. Oncol. 2023, 14, 104. [Google Scholar] [CrossRef]
- Strick, R.; Ackermann, S.; Langbein, M.; Swiatek, J.; Schubert, S.W.; Hashemolhosseini, S.; Koscheck, T.; Fasching, P.A.; Schild, R.L.; Beckmann, M.W.; et al. Proliferation and Cell-Cell Fusion of Endometrial Carcinoma Are Induced by the Human Endogenous Retroviral Syncytin-1 and Regulated by TGF-β. J. Mol. Med. 2007, 85, 23–38. [Google Scholar] [CrossRef]
- Liu, C.; Xu, J.; Wen, F.; Yang, F.; Li, X.; Geng, D.; Li, L.; Chen, J.; Zheng, J. Upregulation of Syncytin-1 Promotes Invasion and Metastasis by Activating Epithelial-Mesenchymal Transition-Related Pathway in Endometrial Carcinoma. Onco Targets Ther. 2019, 12, 31–40. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, L.; Liu, Y.; Zhou, P.; Yan, Q.; Yu, H.; Chen, X.; Zhu, F. Implication of Human Endogenous Retrovirus W Family Envelope in Hepatocellular Carcinoma Promotes MEK/ERK-Mediated Metastatic Invasiveness and Doxorubicin Resistance. Cell Death Discov. 2021, 7, 177. [Google Scholar] [CrossRef]
- Yu, H.; Liu, T.; Zhao, Z.; Chen, Y.; Zeng, J.; Liu, S.; Zhu, F. Mutations in 3′-Long Terminal Repeat of HERV-W Family in Chromosome 7 Upregulate Syncytin-1 Expression in Urothelial Cell Carcinoma of the Bladder through Interacting with c-Myb. Oncogene 2014, 33, 3947–3958. [Google Scholar] [CrossRef]
- Tristem, M. Identification and Characterization of Novel Human Endogenous Retrovirus Families by Phylogenetic Screening of the Human Genome Mapping Project Database. J. Virol. 2000, 74, 3715–3730. [Google Scholar] [CrossRef]
- Yi, J.M.; Kim, H.M.; Kim, H.S. Human Endogenous Retrovirus HERV-H Family in Human Tissues and Cancer Cells: Expression, Identification, and Phylogeny. Cancer Lett. 2006, 231, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Wentzensen, N.; Coy, J.F.; Knaebel, H.P.; Linnebacher, M.; Wilz, B.; Gebert, J.; Von Knebel Doeberitz, M. Expression of an Endogenous Retroviral Sequence from the HERV-H Group in Gastrointestinal Cancers. Int. J. Cancer 2007, 121, 1417–1423. [Google Scholar] [CrossRef] [PubMed]
- Alves, P.M.S.; Lévy, N.; Stevenson, B.J.; Bouzourene, H.; Theiler, G.; Bricard, G.; Viatte, S.; Ayyoub, M.; Vuilleumier, H.; Givel, J.-C.R.; et al. Identification of Tumor-Associated Antigens by Large-Scale Analysis of Genes Expressed in Human Colorectal Cancer. Cancer Immun. 2008, 8, 11. [Google Scholar]
- Honda, T.; Rahman, M.A. Profiling of LINE-1-Related Genes in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2019, 20, 645. [Google Scholar] [CrossRef] [PubMed]
- Honda, T. Links between Human LINE-1 Retrotransposons and Hepatitis Virus-Related Hepatocellular Carcinoma. Front. Chem. 2016, 4, 21. [Google Scholar] [CrossRef]
- Ding, D.; Lou, X.; Hua, D.; Yu, W.; Li, L.; Wang, J.; Gao, F.; Zhao, N.; Ren, G.; Li, L.; et al. Recurrent Targeted Genes of Hepatitis B Virus in the Liver Cancer Genomes Identified by a Next-Generation Sequencing-Based Approach. PLoS Genet. 2012, 8, e1003065. [Google Scholar] [CrossRef]
- Lau, C.C.; Sun, T.; Ching, A.K.K.; He, M.; Li, J.W.; Wong, A.M.; Co, N.N.; Chan, A.W.H.; Li, P.S.; Lung, R.W.M.; et al. Viral-Human Chimeric Transcript Predisposes Risk to Liver Cancer Development and Progression. Cancer Cell 2014, 25, 335–349. [Google Scholar] [CrossRef]
- Cohen, S.B.; Graham, M.E.; Lovrecz, G.O.; Bache, N.; Robinson, P.J.; Reddel, R.R. Protein Composition of Catalytically Active Human Telomerase from Immortal Cells. Science 2007, 315, 1850–1853. [Google Scholar] [CrossRef]
- Nault, J.C.; Datta, S.; Imbeaud, S.; Franconi, A.; Mallet, M.; Couchy, G.; Letouzé, E.; Pilati, C.; Verret, B.; Blanc, J.F.; et al. Recurrent AAV2-Related Insertional Mutagenesis in Human Hepatocellular Carcinomas. Nat. Genet. 2015, 47, 1187–1193. [Google Scholar] [CrossRef] [PubMed]
- Miki, Y.; Nishisho, I.; Horii, A.; Miyoshi, Y.; Utsunomiya, J.; Kinzler, K.W.; Vogelstein, B.; Nakamura, Y. Disruption of the APC Gene by a Retrotransposal Insertion of L1 Sequence in a Colon Cancer. Cancer Res. 1992, 52, 643–645. [Google Scholar] [PubMed]
- Scott, E.C.; Gardner, E.J.; Masood, A.; Chuang, N.T.; Vertino, P.M.; Devine, S.E. A Hot L1 Retrotransposon Evades Somatic Repression and Initiates Human Colorectal Cancer. Genome Res. 2016, 26, 745–755. [Google Scholar] [CrossRef]
- Fodde, R. The APC Gene in Colorectal Cancer. Eur. J. Cancer 2002, 38, 867–871. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Upton, K.R.; Muñoz-Lopez, M.; Gerhardt, D.J.; Fisher, M.E.; Nguyen, T.; Brennan, P.M.; Baillie, J.K.; Collino, A.; Ghisletti, S.; et al. Endogenous Retrotransposition Activates Oncogenic Pathways in Hepatocellular Carcinoma. Cell 2013, 153, 101–111. [Google Scholar] [CrossRef]
- Fukuyama, R.; Niculaita, R.; Ng, K.P.; Obusez, E.; Sanchez, J.; Kalady, M.; Aung, P.P.; Casey, G.; Sizemore, N. Mutated in Colorectal Cancer, a Putative Tumor Suppressor for Serrated Colorectal Cancer, Selectively Represses β-Catenin-Dependent Transcription. Oncogene 2008, 27, 6044–6055. [Google Scholar] [CrossRef]
- Nakayama, R.; Ueno, Y.; Ueda, K.; Honda, T. Latent Infection with Kaposi’s Sarcoma-Associated Herpesvirus Enhances Retrotransposition of Long Interspersed Element-1. Oncogene 2019, 38, 4340–4351. [Google Scholar] [CrossRef]
- Nishikawa, Y.; Nakayama, R.; Obika, S.; Ohsaki, E.; Ueda, K.; Honda, T. Inhibition of LINE-1 Retrotransposition by Capsaicin. Int. J. Mol. Sci. 2018, 19, 3243. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Nohara, S.; Nishikawa, Y.; Suzuki, Y.; Kawamura, Y.; Miura, K.; Tomonaga, K.; Ueda, K.; Honda, T. Characterization of an Active LINE-1 in the Naked Mole-Rat Genome. Sci. Rep. 2021, 11, 5725. [Google Scholar] [CrossRef]
- Rodić, N.; Sharma, R.; Sharma, R.; Zampella, J.; Dai, L.; Taylor, M.S.; Hruban, R.H.; Iacobuzio-Donahue, C.A.; Maitra, A.; Torbenson, M.S.; et al. Long Interspersed Element-1 Protein Expression Is a Hallmark of Many Human Cancers. Am. J. Pathol. 2014, 184, 1280–1286. [Google Scholar] [CrossRef]
- Sciamanna, I.; Landriscina, M.; Pittoggi, C.; Quirino, M.; Mearelli, C.; Beraldi, R.; Mattei, E.; Serafino, A.; Cassano, A.; Sinibaldi-Vallebona, P.; et al. Inhibition of Endogenous Reverse Transcriptase Antagonizes Human Tumor Growth. Oncogene 2005, 24, 3923–3931. [Google Scholar] [CrossRef]
- Aschacher, T.; Wolf, B.; Enzmann, F.; Kienzl, P.; Messner, B.; Sampl, S.; Svoboda, M.; Mechtcheriakova, D.; Holzmann, K.; Bergmann, M. LINE-1 Induces HTERT and Ensures Telomere Maintenance in Tumour Cell Lines. Oncogene 2016, 35, 94–104. [Google Scholar] [CrossRef]
- Cervantes-Ayalc, A.; Ruiz Esparza-Garrido, R.; Velázquez-Flores, M.Á. Long Interspersed Nuclear Elements 1 (LINE1): The Chimeric Transcript L1-MET and Its Involvement in Cancer. Cancer Genet. 2020, 241, 1–11. [Google Scholar] [CrossRef]
- Hur, K.; Cejas, P.; Feliu, J.; Moreno-Rubio, J.; Burgos, E.; Boland, C.R.; Goel, A. Hypomethylation of Long Interspersed Nuclear Element-1 (LINE-1) Leads to Activation of Protooncogenes in Human Colorectal Cancer Metastasis. Gut 2014, 63, 635–646. [Google Scholar] [CrossRef]
- Zhu, C.; Utsunomiya, T.; Ikemoto, T.; Yamada, S.; Morine, Y.; Imura, S.; Arakawa, Y.; Takasu, C.; Ishikawa, D.; Imoto, I.; et al. Hypomethylation of Long Interspersed Nuclear Element-1 (LINE-1) Is Associated with Poor Prognosis via Activation of c-MET in Hepatocellular Carcinoma. Ann. Surg. Oncol. 2014, 21, 729–735. [Google Scholar] [CrossRef]
- Birchmeier, C.; Birchmeier, W.; Gherardi, E.; Vande Woude, G.F. Met, Metastasis, Motility and More. Nat. Rev. Mol. Cell Biol. 2003, 4, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Solyom, S.; Ewing, A.D.; Rahrmann, E.P.; Doucet, T.; Nelson, H.H.; Burns, M.B.; Harris, R.S.; Sigmon, D.F.; Casella, A.; Erlanger, B.; et al. Extensive Somatic L1 Retrotransposition in Colorectal Tumors. Genome Res. 2012, 22, 2328–2338. [Google Scholar] [CrossRef] [PubMed]
- Tubio, J.M.C.; Li, Y.; Ju, Y.S.; Martincorena, I.; Cooke, S.L.; Tojo, M.; Gundem, G.; Pipinikas, C.P.; Zamora, J.; Raine, K.; et al. Extensive Transduction of Nonrepetitive DNA Mediated by L1 Retrotransposition in Cancer Genomes. Science 2014, 345, 1251343. [Google Scholar] [CrossRef] [PubMed]
- Russ, E.; Iordanskiy, S. Endogenous Retroviruses as Modulators of Innate Immunity. Pathogens 2023, 12, 162. [Google Scholar] [CrossRef] [PubMed]
- Grandi, N.; Tramontano, E. Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses. Front. Immunol. 2018, 9, 2039. [Google Scholar] [CrossRef] [PubMed]
- Grandi, N.; Tramontano, E. Type W Human Endogenous Retrovirus (HERV-W) Integrations and Their Mobilization by L1 Machinery: Contribution to the Human Transcriptome and Impact on the Host Physiopathology. Viruses 2017, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Helmy, M.; Selvarajoo, K. Systems Biology to Understand and Regulate Human Retroviral Proinflammatory Response. Front. Immunol. 2021, 12, 736349. [Google Scholar] [CrossRef]
- Suntsova, M.; Garazha, A.; Ivanova, A.; Kaminsky, D.; Zhavoronkov, A.; Buzdin, A. Molecular Functions of Human Endogenous Retroviruses in Health and Disease. Cell. Mol. Life Sci. 2015, 72, 3653–3675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, R.; Yu, J. New Understanding of the Relevant Role of LINE-1 Retrotransposition in Human Disease and Immune Modulation. Front. Cell Dev. Biol. 2020, 8, 657. [Google Scholar] [CrossRef]
- Mathavarajah, S.; Dellaire, G. LINE-1: An Emerging Initiator of CGAS-STING Signalling and Inflammation That Is Dysregulated in Disease. Biochem. Cell Biol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V. The Trinity of CGAS, TLR9, and ALRs Guardians of the Cellular Galaxy Against Host-Derived Self-DNA. Front. Immunol. 2021, 11, 624597. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Du, J.; Peng, Y.; Li, P.; Wang, S.; Wang, Y.; Hou, J.; Kang, J.; Zheng, W.; Hua, S.; et al. LINE1 Contributes to Autoimmunity through Both RIG-I- and MDA5-Mediated RNA Sensing Pathways. J. Autoimmun. 2018, 90, 105–115. [Google Scholar] [CrossRef]
- Carty, M.; Guy, C.; Bowie, A.G. Detection of Viral Infections by Innate Immunity. Biochem. Pharmacol. 2021, 183, 114316. [Google Scholar] [CrossRef]
- Webb, L.G.; Fernandez-Sesma, A. RNA Viruses and the CGAS-STING Pathway: Reframing Our Understanding of Innate Immune Sensing. Curr. Opin. Virol. 2022, 53, 101206. [Google Scholar] [CrossRef]
- Behzadi, P.; García-Perdomo, H.A.; Karpiński, T.M. Toll-Like Receptors: General Molecular and Structural Biology. J. Immunol. Res. 2021, 2021, 9914854. [Google Scholar] [CrossRef]
- Reikine, S.; Nguyen, J.B.; Modis, Y. Pattern Recognition and Signaling Mechanisms of RIG-I and MDA5. Front. Immunol. 2014, 5, 342. [Google Scholar] [CrossRef]
- Said, E.A.; Tremblay, N.; Al-Balushi, M.S.; Al-Jabri, A.A.; Lamarre, D. Viruses Seen by Our Cells: The Role of Viral RNA Sensors. J. Immunol. Res. 2018, 2018, 9480497. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Chiang, C.; Gack, M.U. Endogenous Nucleic Acid Recognition by RIG-I-Like Receptors and CGAS. J. Interferon Cytokine Res. 2019, 39, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhao, H.; Shen, Y.; Chen, Q. A Variety of Nucleic Acid Species Are Sensed by CGAS, Implications for Its Diverse Functions. Front. Immunol. 2022, 13, 826880. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Sun, L.; Chen, X.; Du, F.; Shi, H.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP Is an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Science 2013, 339, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Crosse, K.M.; Monson, E.A.; Beard, M.R.; Helbig, K.J. Interferon-Stimulated Genes as Enhancers of Antiviral Innate Immune Signaling. J. Innate Immun. 2018, 10, 85–93. [Google Scholar] [CrossRef]
- Michalska, A.; Blaszczyk, K.; Wesoly, J.; Bluyssen, H.A.R. A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses. Front. Immunol. 2018, 9, 1135. [Google Scholar] [CrossRef]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef]
- Manghera, M.; Ferguson-Parry, J.; Lin, R.; Douville, R.N. NF-κB and IRF1 Induce Endogenous Retrovirus K Expression via Interferon-Stimulated Response Elements in Its 5′ Long Terminal Repeat. J. Virol. 2016, 90, 9338–9349. [Google Scholar] [CrossRef]
- Durnaoglu, S.; Lee, S.K.; Ahnn, J. Human Endogenous Retroviruses as Gene Expression Regulators: Insights from Animal Models into Human Diseases. Mol. Cells 2021, 44, 861–878. [Google Scholar] [CrossRef]
- Honda, T.; Takemoto, K.; Ueda, K. Identification of a Retroelement-Containing Human Transcript Induced in the Nucleus by Vaccination. Int. J. Mol. Sci. 2019, 20, 2875. [Google Scholar] [CrossRef]
- Ito, J.; Sugimoto, R.; Nakaoka, H.; Yamada, S.; Kimura, T.; Hayano, T.; Inoue, I. Systematic Identification and Characterization of Regulatory Elements Derived from Human Endogenous Retroviruses. PLoS Genet. 2017, 13, e1006883. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory Evolution of Innate Immunity through Co-Option of Endogenous Retroviruses. Science 2016, 351, 1083–1087. [Google Scholar] [CrossRef] [PubMed]
- Domansky, A.N.; Kopantzev, E.P.; Snezhkov, E.V.; Lebedev, Y.B.; Leib-Mosch, C.; Sverdlov, E.D. Solitary HERV-K LTRs Possess Bi-Directional Promoter Activity and Contain a Negative Regulatory Element in the U5 Region. FEBS Lett. 2000, 472, 191–195. [Google Scholar] [CrossRef]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via DsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef]
- Mikhalkevich, N.; O’Carroll, I.P.; Tkavc, R.; Lund, K.; Sukumar, G.; Dalgard, C.L.; Johnson, K.R.; Li, W.; Wang, T.; Nath, A.; et al. Response of Human Macrophages to Gamma Radiation Is Mediated via Expression of Endogenous Retroviruses. PLoS Pathog. 2021, 17, e1009305. [Google Scholar] [CrossRef] [PubMed]
- Perron, H.; Garson, J.A.; Bedin, F.; Beseme, F.; Paranhos-Baccala, G.; Komurian-Pradel, F.; Mallet, F.; Tuke, P.W.; Voisset, C.; Blond, J.L.; et al. Molecular Identification of a Novel Retrovirus Repeatedly Isolated from Patients with Multiple Sclerosis. The Collaborative Research Group on Multiple Sclerosis. Proc. Natl. Acad. Sci. USA 1997, 94, 7583–7588. [Google Scholar] [CrossRef] [PubMed]
- Rolland, A.; Jouvin-Marche, E.; Viret, C.; Faure, M.; Perron, H.; Marche, P.N. The Envelope Protein of a Human Endogenous Retrovirus-W Family Activates Innate Immunity through CD14/TLR4 and Promotes Th1-Like Responses. J. Immunol. 2006, 176, 7636–7644. [Google Scholar] [CrossRef] [PubMed]
- Duperray, A.; Barbe, D.; Raguenez, G.; Weksler, B.B.; Romero, I.A.; Couraud, P.O.; Perron, H.; Marche, P.N. Inflammatory Response of Endothelial Cells to a Human Endogenous Retrovirus Associated with Multiple Sclerosis Is Mediated by TLR4. Int. Immunol. 2015, 27, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, Z.; Wang, P.; Li, S.; Zeng, J.; Tu, X.; Yan, Q.; Xiao, Z.; Pan, M.; Zhu, F. Syncytin-1, an Endogenous Retroviral Protein, Triggers the Activation of CRP via TLR3 Signal Cascade in Glial Cells. Brain Behav. Immun. 2018, 67, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Wu, X.; Zhou, P.; Zhou, Y.; Li, X.; Liu, Z.; Tan, H.; Yao, W.; Xia, Y.; Zhu, F. HERV-W Envelope Triggers Abnormal Dopaminergic Neuron Process through DRD2/PP2A/AKT1/GSK3 for Schizophrenia Risk. Viruses 2022, 14, 145. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yan, Q.; Liu, L.; Xue, X.; Yao, W.; Li, X.; Li, W.; Ding, S.; Xia, Y.; Zhang, D.; et al. Domesticated HERV-W Env Contributes to the Activation of the Small Conductance Ca2+-Activated K+ Type 2 Channels via Decreased 5-HT4 Receptor in Recent-Onset Schizophrenia. Virol. Sin. 2023, 38, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, X.; Li, W.; Yan, Q.; Zhou, P.; Xia, Y.; Yao, W.; Zhu, F. HERV-W ENV Induces Innate Immune Activation and Neuronal Apoptosis via Linc01930/CGAS Axis in Recent-Onset Schizophrenia. Int. J. Mol. Sci. 2023, 24, 3000. [Google Scholar] [CrossRef]
- Perron, H.; Mekaoui, L.; Bernard, C.; Veas, F.; Stefas, I.; Leboyer, M. Endogenous Retrovirus Type W GAG and Envelope Protein Antigenemia in Serum of Schizophrenic Patients. Biol. Psychiatry 2008, 64, 1019–1023. [Google Scholar] [CrossRef]
- Rangel, S.C.; da Silva, M.D.; da Silva, A.L.; dos Santos, J.d.M.B.; Neves, L.M.; Pedrosa, A.; Rodrigues, F.M.; Trettel, C.d.S.; Furtado, G.E.; de Barros, M.P.; et al. Human Endogenous Retroviruses and the Inflammatory Response: A Vicious Circle Associated with Health and Illness. Front. Immunol. 2022, 13, 1057791. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Z.; Wu, Z.; Ren, J.; Fan, Y.; Sun, L.; Cao, G.; Niu, Y.; Zhang, B.; Ji, Q.; et al. Resurrection of Endogenous Retroviruses during Aging Reinforces Senescence. Cell 2023, 186, 287–304.e26. [Google Scholar] [CrossRef]
- Morozov, V.A.; Dao Thi, V.L.; Denner, J. The Transmembrane Protein of the Human Endogenous Retrovirus—K (HERV-K) Modulates Cytokine Release and Gene Expression. PLoS ONE 2013, 8, e70399. [Google Scholar] [CrossRef]
- Laska, M.J.; Troldborg, A.; Hauge, E.M.; Bahrami, S.; Stengaard-Pedersen, K. Human Endogenous Retroviral Genetic Element With Immunosuppressive Activity in Both Human Autoimmune Diseases and Experimental Arthritis. Arthritis Rheumatol. 2017, 69, 398–409. [Google Scholar] [CrossRef]
- Lokossou, A.G.; Toudic, C.; Nguyen, P.T.; Elisseeff, X.; Vargas, A.; Rassart, É.; Lafond, J.; Leduc, L.; Bourgault, S.; Gilbert, C.; et al. Endogenous Retrovirus-Encoded Syncytin-2 Contributes to Exosome-Mediated Immunosuppression of T Cells. Biol. Reprod. 2020, 102, 185–198. [Google Scholar] [CrossRef]
- Mazur, D.J.; Perrino, F.W. Identification and Expression of the TREX1 and TREX2 CDNA Sequences Encoding Mammalian 3′→5′ Exonucleases. J. Biol. Chem. 1999, 274, 19655–19660. [Google Scholar] [CrossRef] [PubMed]
- Stetson, D.B.; Ko, J.S.; Heidmann, T.; Medzhitov, R. Trex1 Prevents Cell-Intrinsic Initiation of Autoimmunity. Cell 2008, 134, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.A.; Tejwani, L.; Trujillo, C.A.; Negraes, P.D.; Herai, R.H.; Mesci, P.; Macia, A.; Crow, Y.J.; Muotri, A.R. Modeling of TREX1-Dependent Autoimmune Disease Using Human Stem Cells Highlights L1 Accumulation as a Source of Neuroinflammation. Cell Stem Cell 2017, 21, 319–331.e8. [Google Scholar] [CrossRef] [PubMed]
- Crow, Y.J.; Hayward, B.E.; Parmar, R.; Robins, P.; Leitch, A.; Ali, M.; Black, D.N.; Van Bokhoven, H.; Brunner, H.G.; Hamel, B.C.; et al. Mutations in the Gene Encoding the 3′-5′ DNA Exonuclease TREX1 Cause Aicardi-Goutières Syndrome at the AGS1 Locus. Nat. Genet. 2006, 38, 917–920. [Google Scholar] [CrossRef] [PubMed]
- Rice, G.I.; Bond, J.; Asipu, A.; Brunette, R.L.; Manfield, I.W.; Carr, I.M.; Fuller, J.C.; Jackson, R.M.; Lamb, T.; Briggs, T.A.; et al. Mutations Involved in Aicardi-Goutières Syndrome Implicate SAMHD1 as Regulator of the Innate Immune Response. Nat. Genet. 2009, 41, 829–832. [Google Scholar] [CrossRef] [PubMed]
- Thiele, H.; Du Moulin, M.; Barczyk, K.; George, C.; Schwindt, W.; Nürnberg, G.; Frosch, M.; Kurlemann, G.; Roth, J.; Nürnberg, P.; et al. Cerebral Arterial Stenoses and Stroke: Novel Features of Aicardi-Goutières Syndrome Caused by the Arg164X Mutation in SAMHD1 Are Associated with Altered Cytokine Expression. Hum. Mutat. 2010, 31, E1836–E1850. [Google Scholar] [CrossRef]
- Xin, B.; Jones, S.; Puffenberger, E.G.; Hinze, C.; Bright, A.; Tan, H.; Zhou, A.; Wu, G.; Jilda, V.A.; Agamanolis, D.; et al. Homozygous Mutation in SAMHD1 Gene Causes Cerebral Vasculopathy and Early Onset Stroke. Proc. Natl. Acad. Sci. USA 2011, 108, 5372–5377. [Google Scholar] [CrossRef] [PubMed]
- Goldstone, D.C.; Ennis-Adeniran, V.; Hedden, J.J.; Groom, H.C.T.; Rice, G.I.; Christodoulou, E.; Walker, P.A.; Kelly, G.; Haire, L.F.; Yap, M.W.; et al. HIV-1 Restriction Factor SAMHD1 Is a Deoxynucleoside Triphosphate Triphosphohydrolase. Nature 2011, 480, 379–382. [Google Scholar] [CrossRef]
- Lahouassa, H.; Daddacha, W.; Hofmann, H.; Ayinde, D.; Logue, E.C.; Dragin, L.; Bloch, N.; Maudet, C.; Bertrand, M.; Gramberg, T.; et al. SAMHD1 Restricts the Replication of Human Immunodeficiency Virus Type 1 by Depleting the Intracellular Pool of Deoxynucleoside Triphosphates. Nat. Immunol. 2012, 13, 223–228. [Google Scholar] [CrossRef]
- Zhao, K.; Du, J.; Han, X.; Goodier, J.L.; Li, P.; Zhou, X.; Wei, W.; Evans, S.L.; Li, L.; Zhang, W.; et al. Modulation of LINE-1 and Alu/SVA Retrotransposition by Aicardi-Goutières Syndrome-Related SAMHD1. Cell Rep. 2013, 4, 1108–1115. [Google Scholar] [CrossRef]
- Svendsen, J.M.; Smogorzewska, A.; Sowa, M.E.; O’Connell, B.C.; Gygi, S.P.; Elledge, S.J.; Harper, J.W. Mammalian BTBD12/SLX4 Assembles a Holliday Junction Resolvase and Is Required for DNA Repair. Cell 2009, 138, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Cybulski, K.E.; Howlett, N.G. FANCP/SLX4: A Swiss Army Knife of DNA Interstrand Crosslink Repair. Cell Cycle 2011, 10, 1757–1763. [Google Scholar] [CrossRef] [PubMed]
- Brégnard, C.; Guerra, J.; Déjardin, S.; Passalacqua, F.; Benkirane, M.; Laguette, N. Upregulated LINE-1 Activity in the Fanconi Anemia Cancer Susceptibility Syndrome Leads to Spontaneous Pro-Inflammatory Cytokine Production. EBioMedicine 2016, 8, 184–194. [Google Scholar] [CrossRef]
- Nepal, M.; Che, R.; Zhang, J.; Ma, C.; Fei, P. Fanconi Anemia Signaling and Cancer. Trends Cancer 2017, 3, 840–856. [Google Scholar] [CrossRef] [PubMed]
- Luqman-Fatah, A.; Watanabe, Y.; Uno, K.; Ishikawa, F.; Moran, J.V.; Miyoshi, T. The Interferon Stimulated Gene-Encoded Protein HELZ2 Inhibits Human LINE-1 Retrotransposition and LINE-1 RNA-Mediated Type I Interferon Induction. Nat. Commun. 2023, 14, 203. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Carbone, C.J.; Katlinskaya, Y.V.; Zheng, H.; Zheng, K.; Luo, M.; Wang, P.J.; Greenberg, R.A.; Fuchs, S.Y. Type I Interferon Controls Propagation of Long Interspersed Element-1. J. Biol. Chem. 2015, 290, 10191–10199. [Google Scholar] [CrossRef] [PubMed]
- Horie, M.; Honda, T.; Suzuki, Y.; Kobayashi, Y.; Daito, T.; Oshida, T.; Ikuta, K.; Jern, P.; Gojobori, T.; Coffin, J.M.; et al. Endogenous Non-Retroviral RNA Virus Elements in Mammalian Genomes. Nature 2010, 463, 84–87. [Google Scholar] [CrossRef]
- Sofuku, K.; Parrish, N.F.; Honda, T.; Tomonaga, K. Transcription Profiling Demonstrates Epigenetic Control of Non-Retroviral RNA Virus-Derived Elements in the Human Genome. Cell Rep. 2015, 12, 1548–1554. [Google Scholar] [CrossRef]
- Sofuku, K.; Honda, T. Influence of Endogenous Viral Sequences on Gene Expression. In Gene Expression and Regulation in Mammalian Cells—Transcription from General Aspects; IntechOpen: Rijeka, Croatia, 2018. [Google Scholar]
- Parrish, N.F.; Fujino, K.; Shiromoto, Y.; Iwasaki, Y.W.; Ha, H.; Xing, J.; Makino, A.; Kuramochi-Miyagawa, S.; Nakano, T.; Siomi, H.; et al. PiRNAs Derived from Ancient Viral Processed Pseudogenes as Transgenerational Sequence-Specific Immune Memory in Mammals. RNA 2015, 21, 1691–1703. [Google Scholar] [CrossRef]
- Fujino, K.; Horie, M.; Honda, T.; Merriman, D.K.; Tomonaga, K. Inhibition of Borna Disease Virus Replication by an Endogenous Bornavirus-like Element in the Ground Squirrel Genome. Proc. Natl. Acad. Sci. USA 2014, 111, 13175–13180. [Google Scholar] [CrossRef]
- Belyi, V.A.; Levine, A.J.; Skalka, A.M. Unexpected Inheritance: Multiple Integrations of Ancient Bornavirus and Ebolavirus/Marburgvirus Sequences in Vertebrate Genomes. PLoS Pathog. 2010, 6, e1001030. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.J.; Leach, R.W.; Bruenn, J. Filoviruses Are Ancient and Integrated into Mammalian Genomes. BMC Evol. Biol. 2010, 10, 193. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.J.; Dittmar, K.; Ballinger, M.J.; Bruenn, J.A. Evolutionary Maintenance of Filovirus-like Genes in Bat Genomes. BMC Evol. Biol. 2011, 11, 336. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.J.; Ballinger, M.J.; Zhan, J.J.; Hanzly, L.E.; Bruenn, J.A. Evidence That Ebolaviruses and Cuevaviruses Have Been Diverging from Marburgviruses since the Miocene. PeerJ 2014, 2, e556. [Google Scholar] [CrossRef]
- Kondoh, T.; Manzoor, R.; Nao, N.; Maruyama, J.; Furuyama, W.; Miyamoto, H.; Shigeno, A.; Kuroda, M.; Matsuno, K.; Fujikura, D.; et al. Putative Endogenous Filovirus VP35-like Protein Potentially Functions as an IFN Antagonist but Not a Polymerase Cofactor. PLoS ONE 2017, 12, e0186450. [Google Scholar] [CrossRef]
- Cárdenas, W.B.; Loo, Y.-M.; Gale, M.; Hartman, A.L.; Kimberlin, C.R.; Martínez-Sobrido, L.; Saphire, E.O.; Basler, C.F. Ebola Virus VP35 Protein Binds Double-Stranded RNA and Inhibits Alpha/Beta Interferon Production Induced by RIG-I Signaling. J. Virol. 2006, 80, 5168–5178. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Katoh, H.; Honda, T. Roles of Human Endogenous Retroviruses and Endogenous Virus-Like Elements in Cancer Development and Innate Immunity. Biomolecules 2023, 13, 1706. https://doi.org/10.3390/biom13121706
Katoh H, Honda T. Roles of Human Endogenous Retroviruses and Endogenous Virus-Like Elements in Cancer Development and Innate Immunity. Biomolecules. 2023; 13(12):1706. https://doi.org/10.3390/biom13121706
Chicago/Turabian StyleKatoh, Hirokazu, and Tomoyuki Honda. 2023. "Roles of Human Endogenous Retroviruses and Endogenous Virus-Like Elements in Cancer Development and Innate Immunity" Biomolecules 13, no. 12: 1706. https://doi.org/10.3390/biom13121706
APA StyleKatoh, H., & Honda, T. (2023). Roles of Human Endogenous Retroviruses and Endogenous Virus-Like Elements in Cancer Development and Innate Immunity. Biomolecules, 13(12), 1706. https://doi.org/10.3390/biom13121706



