Progressive Exaptation of Endogenous Retroviruses in Placental Evolution in Cattle
Abstract
1. Introduction
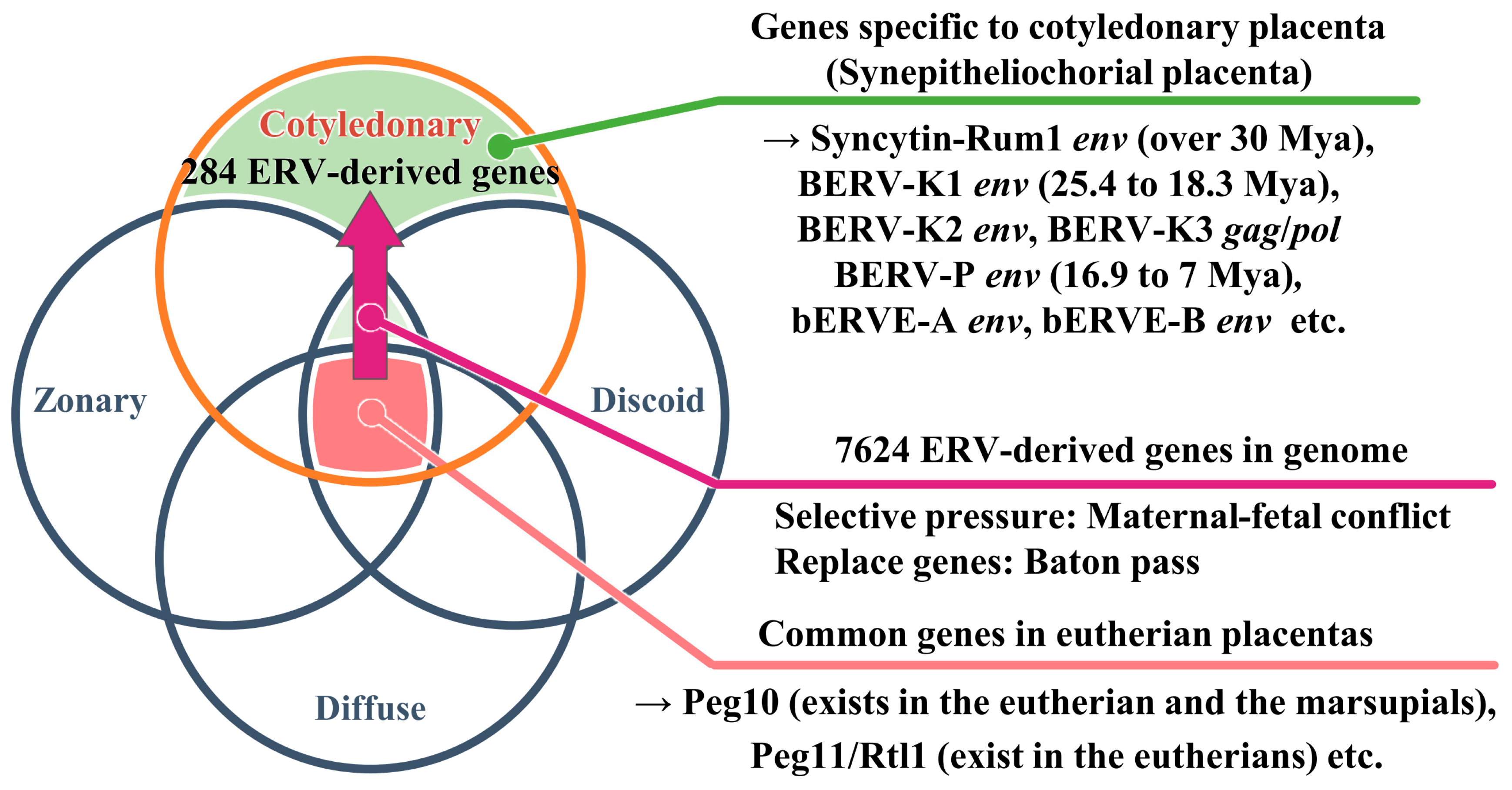
2. Diversity and Classification of Placenta in Mammals
3. The Principle of Genomic Rewiring during Waves of Retroviral Infection
4. Peg10, an LTR-Type Retrotransposon Commonly Expressed in Marsupial and Eutherian Placentas
5. The env Genes, Syncytin and Syncytin-like, Essential for Placentation in Mammals
6. Morphology of the Bovine Placenta
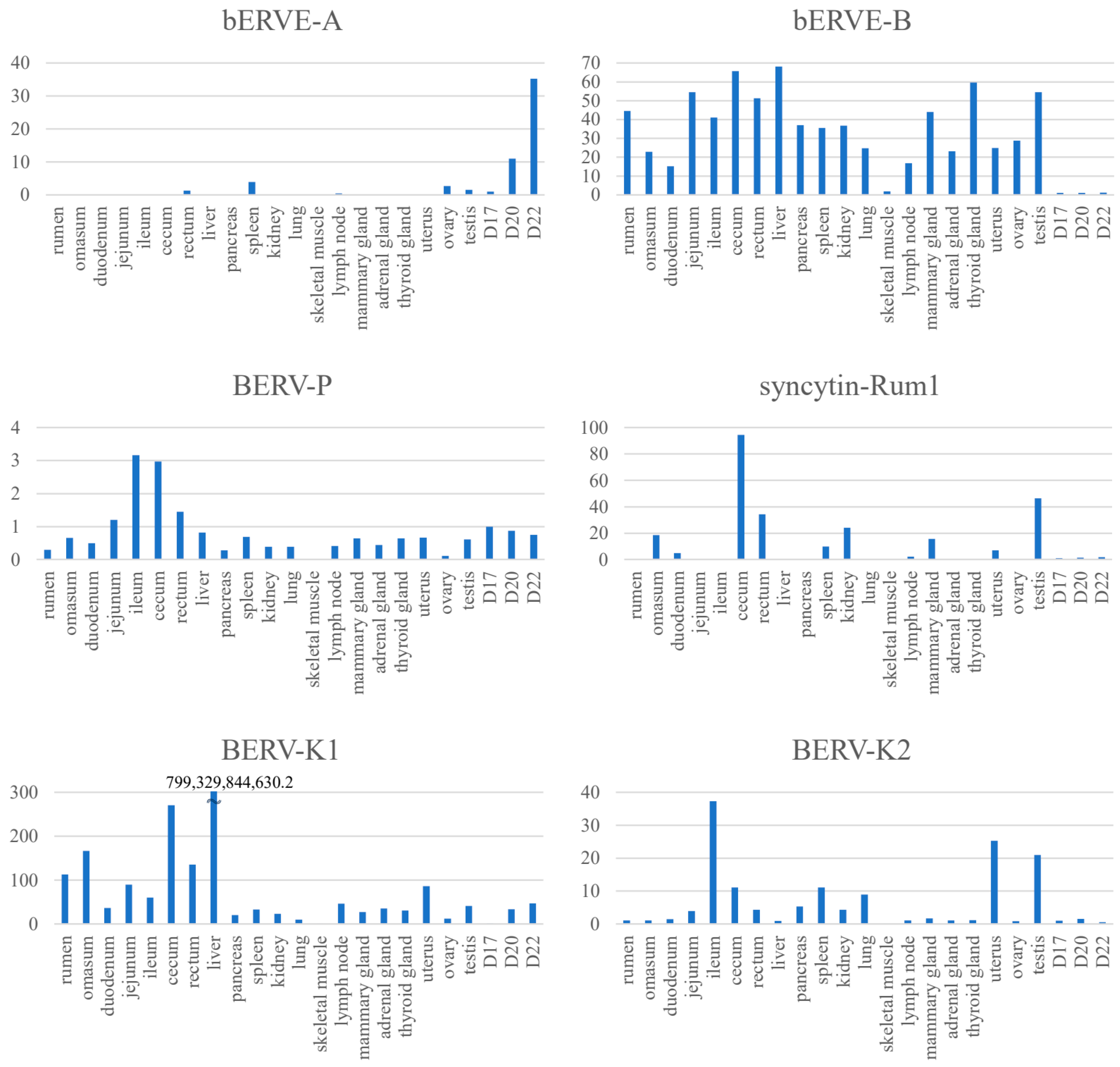
7. Expression of env-Derived Genes from Endogenous Retroviruses in Bovine Placenta
8. Regulatory Mechanisms Involved in BERV Gene Expression in the Bovine Placenta
9. Expression of gag-Derived Genes from Endogenous Retroviruses in Cattle Placenta
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shine, R. Life-history evolution in reptiles. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 23–46. [Google Scholar] [CrossRef]
- Van Dyke, J.U.; Brandley, M.C.; Thompson, M.B. The evolution of viviparity: Molecular and genomic data from squamate reptiles advance understanding of live birth in amniotes. Reproduction 2013, 147, R15–R26. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, D.G. Evolution of vertebrate viviparity and specializations for fetal nutrition: A quantitative and qualitative analysis. J. Morphol. 2015, 276, 961–990. [Google Scholar] [CrossRef]
- Hughes, R.L.; Hall, L.S. Early development and embryology of the platypus. Philos. Trans. R. Soc. B Biol. Sci. 1998, 353, 1101–1114. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.X.; Yuan, C.X.; Meng, Q.J.; Ji, Q. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature 2011, 476, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Bi, S.; Zheng, X.; Wang, X.; Cignetti, N.E.; Yang, S.; Wible, J.R. An Early Cretaceous eutherian and the placental-marsupial dichotomy. Nature 2018, 558, 390–395. [Google Scholar] [CrossRef]
- Roberts, R.M.; Green, J.A.; Schulz, L.C. The evolution of the placenta. Reproduction 2016, 152, R179–R189. [Google Scholar] [CrossRef] [PubMed]
- Foster, C.S.P.; Van Dyke, J.U.; Thompson, M.B.; Smith, N.M.A.; Simpfendorfer, C.A.; Murphy, C.R.; Whittington, C.M. Different Genes are Recruited During Convergent Evolution of Pregnancy and the Placenta. Mol. Biol. Evol. 2022, 39, msac077. [Google Scholar] [CrossRef]
- Mika, K.; Marinić, M.; Singh, M.; Muter, J.; Brosens, J.J.; Lynch, V.J. Evolutionary transcriptomics implicates new genes and pathways in human pregnancy and adverse pregnancy outcomes. Elife 2021, 10, e69584. [Google Scholar] [CrossRef]
- Carter, A.M.; Enders, A.C. Comparative aspects of trophoblast development and placentation. Reprod. Biol. Endocrinol. 2004, 2, 46. [Google Scholar] [CrossRef]
- Armstrong, D.L.; McGowen, M.R.; Weckle, A.; Pantham, P.; Caravas, J.; Agnew, D.; Benirschke, K.; Savage-Rumbaugh, S.; Nevo, E.; Kim, C.J.; et al. The core transcriptome of mammalian placentas and the divergence of expression with placental shape. Placenta 2017, 57, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Haig, D. Genetic conflicts in human pregnancy. Q. Rev. Biol. 1993, 68, 495–532. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Bai, H.; Sakurai, T.; Nakaya, Y.; Konno, T.; Miyazawa, T.; Gojobori, T.; Imakawa, K. Dynamic evolution of endogenous retrovirus-derived genes expressed in bovine conceptuses during the period of placentation. Genome Biol. Evol. 2013, 5, 296–306. [Google Scholar] [CrossRef]
- Sakurai, T.; Nakagawa, S.; Bai, H.; Bai, R.; Kusama, K.; Ideta, A.; Aoyagi, Y.; Kaneko, K.; Iga, K.; Yasuda, J.; et al. Novel endogenous retrovirus-derived transcript expressed in the bovine placenta is regulated by WNT signaling. Biochem. J. 2017, 474, 3499–3512. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, K.; Hattori, M.; Yada, T.; Gojobori, T.; Sakaki, Y.; Okada, N. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003, 4, R74. [Google Scholar] [CrossRef] [PubMed]
- Kazazian, H.H., Jr. Mobile elements: Drivers of genome evolution. Science 2004, 303, 1626–1632. [Google Scholar] [CrossRef]
- Bovine Genome Sequencing and Analysis Consortium; Elsik, C.G.; Tellam, R.L.; Worley, K.C.; Gibbs, R.A.; Muzny, D.M.; Weinstock, G.M.; Adelson, D.L.; Eichler, E.E.; Elnitski, L.; et al. The genome sequence of taurine cattle: A window to ruminant biology and evolution. Science 2009, 324, 522–528. [Google Scholar]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef]
- Zhao, P.; Peng, C.; Fang, L.; Wang, Z.; Liu, G.E. Taming transposable elements in livestock and poultry: A review of their roles and applications. Genet. Sel. Evol. 2023, 55, 50. [Google Scholar] [CrossRef]
- Xiong, Y.; Eickbush, T.H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990, 9, 3353–3362. [Google Scholar] [CrossRef]
- Weiss, R.A. The discovery of endogenous retroviruses. Retrovirology 2006, 3, 67. [Google Scholar] [CrossRef] [PubMed]
- Hoyt, S.J.; Storer, J.M.; Hartley, G.A.; Grady, P.G.S.; Gershman, A.; de Lima, L.G.; Limouse, C.; Halabian, R.; Wojenski, L.; Rodriguez, M.; et al. From telomere to telomere: The transcriptional and epigenetic state of human repeat elements. Science 2022, 376, eabk3112. [Google Scholar] [CrossRef] [PubMed]
- Jern, P.; Coffin, J.M. Effects of retroviruses on host genome function. Annu. Rev. Genet. 2008, 42, 709–732. [Google Scholar] [CrossRef] [PubMed]
- Adelson, D.L.; Raison, J.M.; Edgar, R.C. Characterization and distribution of retrotransposons and simple sequence repeats in the bovine genome. Proc. Natl. Acad. Sci. USA 2009, 106, 12855–12860. [Google Scholar] [CrossRef]
- Garcia-Etxebarria, K.; Jugo, B.M. Genomic environment and digital expression of bovine endogenous retroviruses. Gene 2014, 548, 14–21. [Google Scholar] [CrossRef]
- Gifford, R.; Tristem, M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 2003, 26, 291–315. [Google Scholar] [CrossRef]
- de Parseval, N.; Heidmann, T. Human endogenous retroviruses: From infectious elements to human genes. Cytogenet. Genome Res. 2005, 110, 318–332. [Google Scholar] [CrossRef]
- Vargiu, L.; Rodriguez-Tomé, P.; Sperber, G.O.; Cadeddu, M.; Grandi, N.; Blikstad, V.; Tramontano, E.; Blomberg, J. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology 2016, 13, 7. [Google Scholar] [CrossRef]
- Dupressoir, A.; Vernochet, C.; Harper, F.; Guégan, J.; Dessen, P.; Pierron, G.; Heidmann, T. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc. Natl. Acad. Sci. USA 2011, 108, E1164–E1173. [Google Scholar] [CrossRef]
- Kaneko-Ishino, T.; Ishino, F. The role of genes domesticated from LTR retrotransposons and retroviruses in mammals. Front. Microbiol. 2012, 3, 262. [Google Scholar] [CrossRef]
- Ono, R.; Nakamura, K.; Inoue, K.; Naruse, M.; Usami, T.; Wakisaka-Saito, N.; Hino, T.; Suzuki-Migishima, R.; Ogonuki, N.; Miki, H.; et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 2006, 38, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.A.; Mungall, A.J.; Matthews, L.; Ryder, E.; Gray, D.J.; Pask, A.J.; Shaw, G.; Graves, J.A.; Rogers, J.; SAVOIR consortium; et al. The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 2008, 6, e135. [Google Scholar] [CrossRef] [PubMed]
- Sekita, Y.; Wagatsuma, H.; Nakamura, K.; Ono, R.; Kagami, M.; Wakisaka, N.; Hino, T.; Suzuki-Migishima, R.; Kohda, T.; Ogura, A.; et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. 2008, 40, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Blond, J.L.; Lavillette, D.; Cheynet, V.; Bouton, O.; Oriol, G.; Chapel-Fernandes, S.; Mandrand, B.; Mallet, F.; Cosset, F.L. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 2000, 74, 3321–3329. [Google Scholar] [CrossRef]
- Mi, S.; Lee, X.; Li, X.; Veldman, G.M.; Finnerty, H.; Racie, L.; LaVallie, E.; Tang, X.Y.; Edouard, P.; Howes, S.; et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000, 403, 785–789. [Google Scholar] [CrossRef]
- Blaise, S.; de Parseval, N.; Bénit, L.; Heidmann, T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 13013–13018. [Google Scholar] [CrossRef]
- Esnault, C.; Priet, S.; Ribet, D.; Vernochet, C.; Bruls, T.; Lavialle, C.; Weissenbach, J.; Heidmann, T. A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proc. Natl. Acad. Sci. USA 2008, 105, 17532–17537. [Google Scholar] [CrossRef]
- Lokossou, A.G.; Toudic, C.; Barbeau, B. Implication of human endogenous retrovirus envelope proteins in placental functions. Viruses 2014, 6, 4609–4627. [Google Scholar] [CrossRef]
- Dupressoir, A.; Marceau, G.; Vernochet, C.; Bénit, L.; Kanellopoulos, C.; Sapin, V.; Heidmann, T. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. USA 2005, 102, 725–730. [Google Scholar] [CrossRef]
- Dupressoir, A.; Vernochet, C.; Bawa, O.; Harper, F.; Pierron, G.; Opolon, P.; Heidmann, T. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl. Acad. Sci. USA 2009, 106, 12127–12132. [Google Scholar] [CrossRef]
- Heidmann, O.; Vernochet, C.; Dupressoir, A.; Heidmann, T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: A new “syncytin” in a third order of mammals. Retrovirology 2009, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.; Heidmann, O.; Bernard-Stoecklin, S.; Reynaud, K.; Véron, G.; Mulot, B.; Dupressoir, A.; Heidmann, T. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc. Natl. Acad. Sci. USA 2012, 109, E432–E441. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, K.A.; Palmarini, M.; Adelson, D.L.; Spencer, T.E. Sheep endogenous betaretroviruses (enJSRVs) and the hyaluronidase 2 (HYAL2) receptor in the ovine uterus and conceptus. Biol. Reprod. 2005, 73, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, K.A.; Palmarini, M.; Varela, M.; Burghardt, R.C.; Hayashi, K.; Farmer, J.L.; Spencer, T.E. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc. Natl. Acad. Sci. USA 2006, 103, 14390–14395. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, K.A.; Palmarini, M.; Spencer, T.E. Ovine endogenous betaretroviruses (enJSRVs) and placental morphogenesis. Placenta 2006, 27 (Suppl. A), S135–S140. [Google Scholar] [CrossRef]
- Murcia, P.R.; Arnaud, F.; Palmarini, M. The transdominant endogenous retrovirus enJS56A1 associates with and blocks intracellular trafficking of Jaagsiekte sheep retrovirus Gag. J. Virol. 2007, 81, 1762–1772. [Google Scholar] [CrossRef]
- Armezzani, A.; Arnaud, F.; Caporale, M.; di Meo, G.; Iannuzzi, L.; Murgia, C.; Palmarini, M. The signal peptide of a recently integrated endogenous sheep betaretrovirus envelope plays a major role in eluding gag-mediated late restriction. J. Virol. 2011, 85, 7118–7128. [Google Scholar] [CrossRef]
- Morozov, V.A.; Morozov, A.V.; Lagaye, S. Endogenous JSRV-like proviruses in domestic cattle: Analysis of sequences and transcripts. Virology 2007, 367, 59–70. [Google Scholar] [CrossRef][Green Version]
- Baba, K.; Nakaya, Y.; Shojima, T.; Muroi, Y.; Kizaki, K.; Hashizume, K.; Imakawa, K.; Miyazawa, T. Identification of novel endogenous betaretroviruses which are transcribed in the bovine placenta. J. Virol. 2011, 85, 1237–1245. [Google Scholar] [CrossRef]
- Koshi, K.; Suzuki, Y.; Nakaya, Y.; Imai, K.; Hosoe, M.; Takahashi, T.; Kizaki, K.; Miyazawa, T.; Hashizume, K. Bovine trophoblastic cell differentiation and binucleation involves enhanced endogenous retrovirus element expression. Reprod. Biol. Endocrinol. 2012, 10, 41. [Google Scholar] [CrossRef]
- Cornelis, G.; Heidmann, O.; Degrelle, S.A.; Vernochet, C.; Lavialle, C.; Letzelter, C.; Bernard-Stoecklin, S.; Hassanin, A.; Mulot, B.; Guillomot, M.; et al. Captured retroviral envelope syncytin gene associated with the unique placental structure of higher ruminants. Proc. Natl. Acad. Sci. USA 2013, 110, E828–E837. [Google Scholar] [CrossRef] [PubMed]
- Nakaya, Y.; Koshi, K.; Nakagawa, S.; Hashizume, K.; Miyazawa, T. Fematrin-1 is involved in fetomaternal cell-to-cell fusion in Bovinae placenta and has contributed to diversity of ruminant placentation. J. Virol. 2013, 87, 10563–10572. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.; Vernochet, C.; Carradec, Q.; Souquere, S.; Mulot, B.; Catzeflis, F.; Nilsson, M.A.; Menzies, B.R.; Renfree, M.B.; Pierron, G.; et al. Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc. Natl. Acad. Sci. USA 2015, 112, E487–E496. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W.; Spencer, T.E.; Johnson, G.A. Interferons and uterine receptivity. Semin. Reprod. Med. 2009, 27, 90–102. [Google Scholar] [CrossRef]
- Green, J.A.; Geisert, R.D.; Johnson, G.A.; Spencer, T.E. Implantation and Placentation in Ruminants. Adv. Anat. Embryol. Cell Biol. 2021, 234, 129–154. [Google Scholar]
- Wooding, F.B. Current topic: The synepitheliochorial placenta of ruminants: Binucleate cell fusions and hormone production. Placenta 1992, 13, 101–113. [Google Scholar] [CrossRef]
- Klisch, K.; Pfarrer, C.; Schuler, G.; Hoffmann, B.; Leiser, R. Tripolar acytokinetic mitosis and formation of feto-maternal syncytia in the bovine placentome: Different modes of the generation of multinuclear cells. Anat. Embryol. 1999, 200, 229–237. [Google Scholar] [CrossRef]
- Wooding, F.B.P. The ruminant placental trophoblast binucleate cell: An evolutionary breakthrough. Biol. Reprod. 2022, 107, 705–716. [Google Scholar] [CrossRef]
- Wooding, F.B. Frequency and localization of binucleate cells in the placentomes of ruminants. Placenta 1983, 4, 527–539. [Google Scholar]
- Wooding, F.B.; Wathes, D.C. Binucleate cell migration in the bovine placentome. J. Reprod. Fertil. 1980, 59, 425–430. [Google Scholar] [CrossRef]
- Seo, H.; Bazer, F.W.; Burghardt, R.C.; Johnson, G.A. Immunohistochemical Examination of Trophoblast Syncytialization during Early Placentation in Sheep. Int. J. Mol. Sci. 2019, 20, 4530. [Google Scholar] [CrossRef] [PubMed]
- Yamada, A.; Ohtsuki, K.; Shiga, N.; Green, J.A.; Matsuno, Y.; Imakawa, K. Epithelial-mesenchymal transition and bi- and multi-nucleated trophoblast cell formation in ovine conceptuses during the peri-implantation period. J. Reprod. Dev. 2022, 68, 110–117. [Google Scholar] [CrossRef]
- Imakawa, K.; Nakagawa, S.; Miyazawa, T. Baton pass hypothesis: Successive incorporation of unconserved endogenous retroviral genes for placentation during mammalian evolution. Genes Cells 2015, 20, 771–788. [Google Scholar] [CrossRef]
- Nakaya, Y.; Miyazawa, T. Dysfunction of bovine endogenous retrovirus K2 envelope glycoprotein is related to unsuccessful intracellular trafficking. J. Virol. 2014, 88, 6896–6905. [Google Scholar] [CrossRef] [PubMed]
- Koshi, K.; Nakaya, Y.; Kizaki, K.; Ishiguro-Oonuma, T.; Miyazawa, T.; Spencer, T.E.; Hashizume, K. Induction of ovine trophoblast cell fusion by fematrin-1 in vitro. Anim. Sci. J. 2016, 87, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Khazaee, E.; Farzaneh, N.; Mirshokraei, P.; Tabatabaeizadeh, S.E.; Dehghani, H. Expression of endogenous retroviruses in pre-implantation stages of bovine embryo. Reprod. Domest. Anim. 2018, 53, 1405–1414. [Google Scholar] [CrossRef]
- Garcia-Etxebarria, K.; Jugo, B.M. Genome-wide detection and characterization of endogenous retroviruses in Bos taurus. J. Virol. 2010, 84, 10852–10862. [Google Scholar] [CrossRef]
- Xiao, R.; Park, K.; Lee, H.; Kim, J.; Park, C. Identification and classification of endogenous retroviruses in cattle. J. Virol. 2008, 82, 582–587. [Google Scholar] [CrossRef]
- Garcia-Etxebarria, K.; Jugo, B.M. Evolutionary history of bovine endogenous retroviruses in the Bovidae family. BMC Evol. Biol. 2013, 13, 256. [Google Scholar] [CrossRef]
- Johnson, W.E. Endogenous Retroviruses in the Genomics Era. Annu. Rev. Virol. 2015, 2, 135–159. [Google Scholar] [CrossRef]
- Xiao, R.; Park, K.; Oh, Y.; Kim, J.; Park, C. Structural characterization of the genome of BERV gamma4, the most abundant endogenous retrovirus family in cattle. Mol. Cells 2008, 26, 404–408. [Google Scholar] [PubMed]
- Torresi, C.; Casciari, C.; Giammarioli, M.; Feliziani, F.; De Mia, G.M. Characterization of a novel full-length bovine endogenous retrovirus, BERV-β1. Arch. Virol. 2015, 160, 3105–3114. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Kim, J.; Choi, H.; Park, K.; Lee, H.; Park, C. Characterization of the bovine endogenous retrovirus beta3 genome. Mol. Cells 2008, 25, 142–147. [Google Scholar] [PubMed]
- Wang, X.; Liu, S. Endogenous Jaagsiekte sheep retrovirus envelope protein promotes sheep trophoblast cell fusion by activating PKA/MEK/ERK1/2 signaling. Theriogenology 2022, 193, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, J.; Sugimoto, M.; Bernstein, H.; Jinno, Y.; Schust, D. A novel human endogenous retroviral protein inhibits cell-cell fusion. Sci. Rep. 2013, 3, 1462. [Google Scholar] [CrossRef]
- Kitao, K.; Tanikaga, T.; Miyazawa, T. Identification of a post-transcriptional regulatory element in the human endogenous retroviral syncytin-1. J. Gen. Virol. 2019, 100, 662–668. [Google Scholar] [CrossRef]
- Kitao, K.; Nakagawa, S.; Miyazawa, T. An ancient retroviral RNA element hidden in mammalian genomes and its involvement in co-opted retroviral gene regulation. Retrovirology 2021, 18, 36. [Google Scholar] [CrossRef]
- Kovalskaya, E.; Buzdin, A.; Gogvadze, E.; Vinogradova, T.; Sverdlov, E. Functional human endogenous retroviral LTR transcription start sites are located between the R and U5 regions. Virology 2006, 346, 373–378. [Google Scholar] [CrossRef]
- Criscione, S.W.; Zhang, Y.; Thompson, W.; Sedivy, J.M.; Neretti, N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genom. 2014, 15, 583. [Google Scholar] [CrossRef]
- Thompson, P.J.; Macfarlan, T.S.; Lorincz, M.C. Long Terminal Repeats: From Parasitic Elements to Building Blocks of the Transcriptional Regulatory Repertoire. Mol. Cell 2016, 62, 766–776. [Google Scholar] [CrossRef]
- Manghera, M.; Douville, R.N. Endogenous retrovirus-K promoter: A landing strip for inflammatory transcription factors? Retrovirology 2013, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Akopov, S.B.; Nikolaev, L.G.; Khil, P.P.; Lebedev, Y.B.; Sverdlov, E.D. Long terminal repeats of human endogenous retrovirus K family (HERV-K) specifically bind host cell nuclear proteins. FEBS Lett. 1998, 421, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.F.; Coffin, J.M. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: Implications for human and viral evolution. Proc. Natl. Acad. Sci. USA 2004, 10S1, 1668–1672. [Google Scholar] [CrossRef] [PubMed]
- Belshaw, R.; Watson, J.; Katzourakis, A.; Howe, A.; Woolven-Allen, J.; Burt, A.; Tristem, M. Rate of recombinational deletion among human endogenous retroviruses. J. Virol. 2007, 81, 9437–9442. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.J.; Lock, W.M.; Mager, D.L. Endogenous retroviral LTRs as promoters for human genes: A critical assessment. Gene 2009, 448, 105–114. [Google Scholar] [CrossRef]
- Dunn, C.A.; Romanish, M.T.; Gutierrez, L.E.; van de Lagemaat, L.N.; Mager, D.L. Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene 2006, 366, 335–342. [Google Scholar] [CrossRef]
- Romanish, M.T.; Lock, W.M.; van de Lagemaat, L.N.; Dunn, C.A.; Mager, D.L. Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet. 2007, 3, e10. [Google Scholar] [CrossRef]
- Reiss, D.; Zhang, Y.; Mager, D.L. Widely variable endogenous retroviral methylation levels in human placenta. Nucleic Acids Res. 2007, 35, 4743–4754. [Google Scholar] [CrossRef]
- Matousková, M.; Blazková, J.; Pajer, P.; Pavlícek, A.; Hejnar, J. CpG methylation suppresses transcriptional activity of human syncytin-1 in non-placental tissues. Exp. Cell Res. 2006, 312, 1011–1020. [Google Scholar] [CrossRef]
- Davenport, K.M.; Ortega, M.S.; Liu, H.; O’Neil, E.V.; Kelleher, A.M.; Warren, W.C.; Spencer, T.E. Single-nuclei RNA sequencing (snRNA-seq) uncovers trophoblast cell types and lineages in the mature bovine placenta. Proc. Natl. Acad. Sci. USA 2023, 120, e2221526120. [Google Scholar] [CrossRef]
- Wang, Y.; Ming, H.; Yu, L.; Li, J.; Zhu, L.; Sun, H.X.; Pinzon-Arteaga, C.A.; Wu, J.; Jiang, Z. Establishment of bovine trophoblast stem cells. Cell Rep. 2023, 42, 112439. [Google Scholar] [CrossRef] [PubMed]
| Mode of Distribution of Villi | Degree of Syncytialization | Representative Species | Type of Fused Cells |
|---|---|---|---|
| Cotyledonary | Synepitheliochorial | Cow, Sheep | Fetomaternal hybrid |
| Diffuse | Epitheliochorial | Horse, Pig | None |
| Zonary | Endotheliochorial | Dog, Cat | Syncytiotrophoblast |
| Discoid | Hemochorial | Mouse, Human | Syncytiotrophoblast |
| Appearance in Host Genome | Species | Expressed Tissues (Localization) #1 | Function | References | |
|---|---|---|---|---|---|
| Syncytin-Rum1 env | Over 30 Mya | Most ruminants(except for the family Tragulidae) | Placenta (BNCs) | Fusogenic activity | [51] |
| BERV-K1 env | 25.4 to 18.3 Mya | Bovinae subfamily | Placenta (BNCs), embryonic blastomeres (two- to 16-cell stage) | Fusogenic activity | [52,63] |
| BERV-K2 env | No data | No data | Endometrium, embryonic blastomeres (2- to 16-cell stage) | No data | [49,63] |
| BERV-P env | 16.9 to 7.0 Mya | genus Bos | Conceptus during periattachment (Trophectoderm) | No data | [13] |
| bERVE-A env | No data | Bos taurus | Placenta (BNCs) | No data | [50] |
| bERVE-B env | No data | No data | Endometrium, placenta | No data | [50] |
| BERV-K3 gag/pol | No data | No data | Conceptus after attachment (Trophectoderm), placenta | No data | [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakurai, T.; Kusama, K.; Imakawa, K. Progressive Exaptation of Endogenous Retroviruses in Placental Evolution in Cattle. Biomolecules 2023, 13, 1680. https://doi.org/10.3390/biom13121680
Sakurai T, Kusama K, Imakawa K. Progressive Exaptation of Endogenous Retroviruses in Placental Evolution in Cattle. Biomolecules. 2023; 13(12):1680. https://doi.org/10.3390/biom13121680
Chicago/Turabian StyleSakurai, Toshihiro, Kazuya Kusama, and Kazuhiko Imakawa. 2023. "Progressive Exaptation of Endogenous Retroviruses in Placental Evolution in Cattle" Biomolecules 13, no. 12: 1680. https://doi.org/10.3390/biom13121680
APA StyleSakurai, T., Kusama, K., & Imakawa, K. (2023). Progressive Exaptation of Endogenous Retroviruses in Placental Evolution in Cattle. Biomolecules, 13(12), 1680. https://doi.org/10.3390/biom13121680






