Molecular Insight into Ligand Binding and Transport by the Lentil Lipid Transfer Protein Lc-LTP2: The Role of Basic Amino Acid Residues at Opposite Entrances to the Hydrophobic Cavity
Abstract
:1. Introduction
2. Results
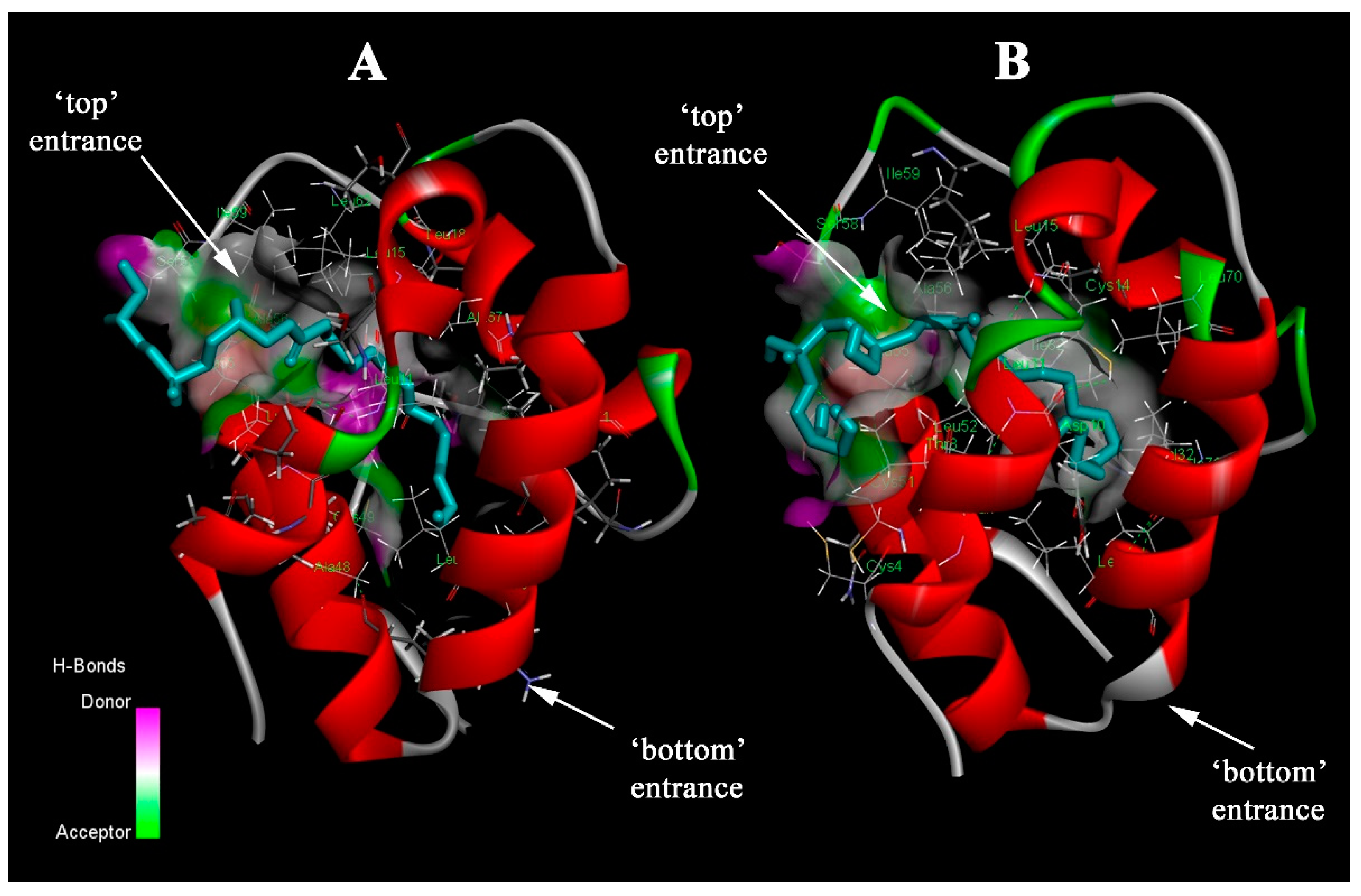
2.1. Molecular Docking
2.2. Competitive Binding Assay
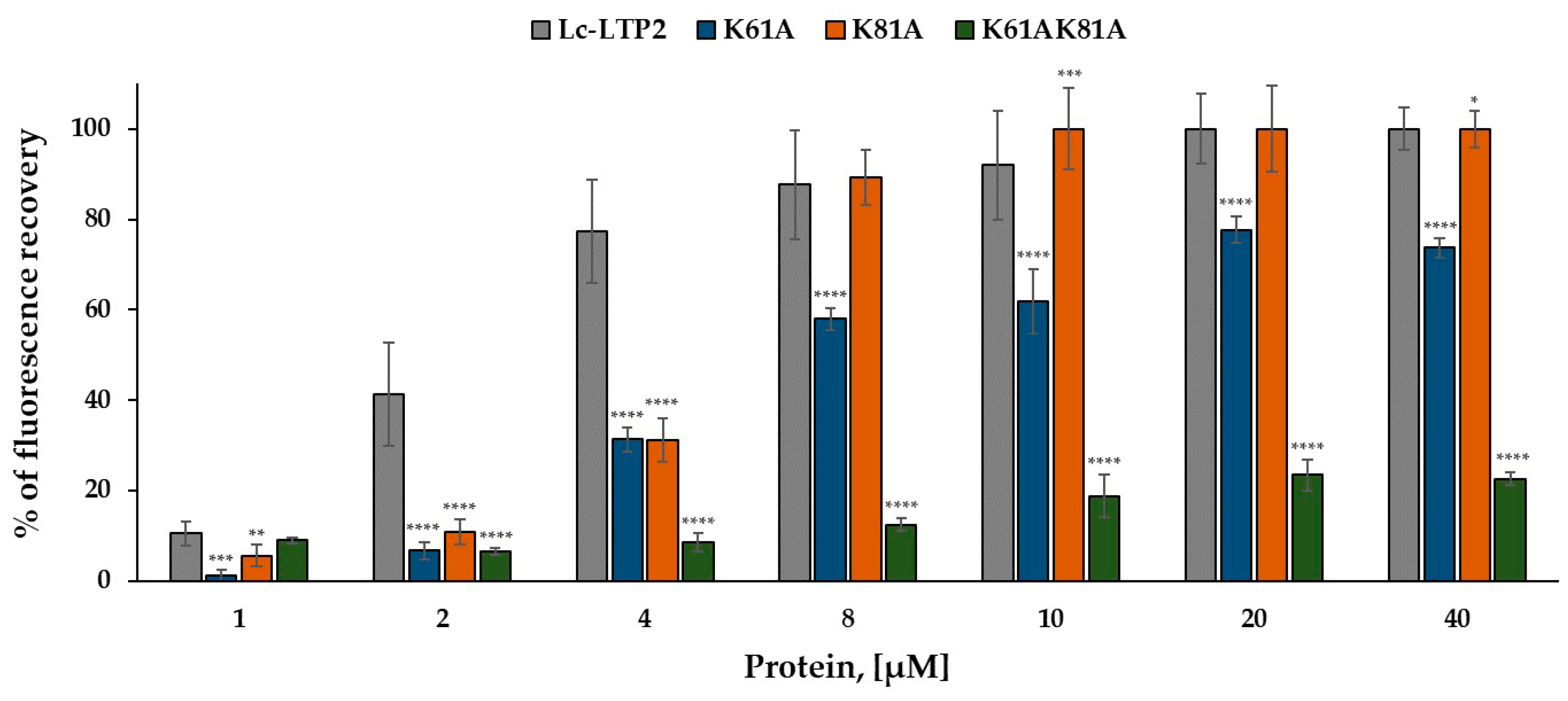
2.3. Liposome Leakage Assay
2.4. Transfer of Lipids
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cloning of Plasmids, Protein Expression, and Purification
4.3. Computational Molecular Docking
4.4. Ligand Binding
4.5. Calcein Release Assay
4.6. Lipid Transfer Assay
4.7. Dynamic Light Scattering
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| LTP | Lipid transfer protein |
| LMPC | 1-myristoyl-2-hydroxy-sn-glycero-3-phosphocholine |
| LMPG | 1-myristoyl-2-hydroxy-sn-glycero-3-phospho-(1’-rac-glycerol) |
| LPPC | 1-palmitoyl-2-hydroxy-sn-glycero-3-phosphocholine |
| LPPG | 1-palmitoyl-2-hydroxy-sn-glycero-3-phospho-(1’-rac-glycerol) |
| POPC | 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine |
| POPG | 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac-glycerol) |
| DMPG | 1,2-Dimyristoyl-sn-glycero-3-phosphoglycerol |
| FA | Fatty acid |
| LAU | Lauric acid |
| MYR | Myristic acid |
| STE | Stearic acid |
| OLE | Oleic acid |
| TNS | 2-p-toluidinonaphthalene-6-sulphonate |
| SUV | Small unilamellar vesicle |
References
- Amador, V.C.; dos Santos-Silva, C.A.; Vilela, L.M.B.; Oliveira-Lima, M.; Rêgo, M.d.S.; Roldan-Filho, R.S.; de Oliveira-Silva, R.L.; Lemos, A.B.; de Oliveira, W.D.; Ferreira-Neto, J.R.C.; et al. Lipid Transfer Proteins (LTPs)—Structure, Diversity and Roles beyond Antimicrobial Activity. Antibiotics 2021, 10, 1281. [Google Scholar] [CrossRef] [PubMed]
- Zottich, U.; Da Cunha, M.; Carvalho, A.O.; Dias, G.B.; Silva, N.C.; Santos, I.S.; Nacimento, V.V.D.; Miguel, E.C.; Machado, O.L.; Gomes, V.M. Purification, biochemical characterization and antifungal activity of a new lipid transfer protein (LTP) from Coffea canephora seeds with α-amylase inhibitor properties. Biochim. Biophys. Acta-Gen. Subj. 2011, 1810, 375–383. [Google Scholar] [CrossRef]
- Sun, J.-Y.; Gaudet, D.A.; Lu, Z.-X.; Frick, M.; Puchalski, B.; Laroche, A. Characterization and Antifungal Properties of Wheat Nonspecific Lipid Transfer Proteins. Mol. Plant-Microbe Interact. 2008, 21, 346–360. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, S.S.; Elseehy, M.M.; Aljuaid, B.S.; El-Shehawi, A.M. Transcriptome Analysis of Jojoba (Simmondsia chinensis) during Seed Development and Liquid Wax Ester Biosynthesis. Plants 2020, 9, 588. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Ma, K.; Ji, G.; Pan, L.; Zhou, Q. Lipid transfer proteins involved in plant–pathogen interactions and their molecular mechanisms. Mol. Plant Pathol. 2022, 23, 1815–1829. [Google Scholar] [CrossRef]
- Qiao, P.; Bourgault, R.; Mohammadi, M.; Gore, M.A.; Molina, I.; Scanlon, M.J. A maize LIPID TRANSFER PROTEIN may bridge the gap between PHYTOCHROME-mediated light signaling and cuticle biosynthesis. Plant Signal. Behav. 2020, 15, 1790824. [Google Scholar] [CrossRef]
- Melnikova, D.N.; Finkina, E.I.; Bogdanov, I.V.; Tagaev, A.A.; Ovchinnikova, T.V. Features and Possible Applications of Plant Lipid-Binding and Transfer Proteins. Membranes 2023, 13, 2. [Google Scholar] [CrossRef]
- Melnikova, D.N.; Finkina, E.I.; Bogdanov, I.V.; Ovchinnikova, T.V. Plant Pathogenesis-Related Proteins Binding Lipids and Other Hydrophobic Ligands. Russ. J. Bioorg. Chem. 2018, 44, 586–594. [Google Scholar] [CrossRef]
- Madni, Z.K.; Kumar, A.; Kumar, U.; Jaiswal, D.; Salunke, D.M. Dynamics of lipid displacement inside the hydrophobic cavity of a nonspecific lipid transfer protein from Solanum melongena. J. Biomol. Struct. Dyn. 2022, 41, 5839–5849. [Google Scholar] [CrossRef]
- Cheng, H.-C.; Cheng, P.-T.; Peng, P.; Lyu, P.-C.; Sun, Y.-J. Lipid binding in rice nonspecific lipid transfer protein-1 complexes from Oryza sativa. Protein Sci. 2004, 13, 2304–2315. [Google Scholar] [CrossRef]
- Salminen, T.A.; Blomqvist, K.; Edqvist, J. Lipid transfer proteins: Classification, nomenclature, structure, and function. Planta 2016, 244, 971–997. [Google Scholar] [CrossRef] [PubMed]
- Charvolin, D.; Douliez, J.-P.; Marion, D.; Cohen-Addad, C.; Pebay-Peyroula, E. The crystal structure of a wheat nonspecific lipid transfer protein (ns-LTP1) complexed with two molecules of phospholipid at 2.1 A° resolution. Eur. J. Biochem. 1999, 264, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Yeats, T.H.; Rose, J.K. The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs). Protein Sci. 2008, 17, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Tassin-Moindrot, S.; Caille, A.; Douliez, J.-P.; Marion, D.; Vovelle, F. The wide binding properties of a wheat nonspecific lipid transfer protein. Solution structure of a complex with prostaglandin B2. J. Biol. Inorg. Chem. 2000, 267, 1117–1124. [Google Scholar]
- Melnikova, D.; Bogdanov, I.; Ignatova, A.; Ovchinnikova, T.; Finkina, E. New insights into ligand binding by plant lipid transfer proteins: A case study of the lentil Lc-LTP2. Biochem. Biophys. Res. Commun. 2020, 528, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Melnikova, D.; Bogdanov, I.; Ovchinnikova, T.; Finkina, E. Interaction between the Lentil Lipid Transfer Protein Lc-LTP2 and Its Novel Signal Ligand PI(4,5)P2. Membranes 2020, 10, 357. [Google Scholar] [CrossRef] [PubMed]
- Shenkarev, Z.O.; Melnikova, D.N.; Finkina, E.I.; Sukhanov, S.V.; Boldyrev, I.A.; Gizatullina, A.K.; Mineev, K.S.; Arseniev, A.S.; Ovchinnikova, T.V. Ligand Binding Properties of the Lentil Lipid Transfer Protein: Molecular Insight into the Possible Mechanism of Lipid Uptake. Biochemistry 2017, 56, 1785–1796. [Google Scholar] [CrossRef]
- Grelaa, E.R.; Günterb, K.D. Fatty acid composition and tocopherol content of some legume seeds. Anim. Sci. Technol. 1995, 52, 325–331. [Google Scholar] [CrossRef]
- Ryan, E.; Galvin, K.; O’Connor, T.P.; Maguire, A.R.; O’Brien, N.M. Phytosterol, squalene, tocopherol content and fatty acid profile of selected seeds, grains, and legumes. Plant Foods Hum. Nutr. 2007, 62, 85–91. [Google Scholar] [CrossRef]
- Kendall, D.A.; Macdonald, R.C. A Fluorescence Assay to Monitor Vesicle Fusion and Lysis. J. Biol. Chem. 1982, 257, 3892–3895. [Google Scholar] [CrossRef]
- Connor, J.; Yatvin, M.B.; Huang, L. Ph-Sensitive Liposomes—Acid-Induced Liposome Fusion. Proc. Natl. Acad. Sci. USA 1984, 81, 1715–1718. [Google Scholar] [CrossRef] [PubMed]
- Bogdanov, I.V.; Shenkarev, Z.O.; Finkina, E.I.; Melnikova, D.N.; Rumynskiy, E.I.; Arseniev, A.S.; Ovchinnikova, T.V. A novel lipid transfer protein from the pea Pisum sativum: Isolation, recombinant expression, solution structure, antifungal activity, lipid binding, and allergenic properties. BMC Plant Biol. 2016, 16, 1186. [Google Scholar] [CrossRef] [PubMed]
- Buhot, N.; Gomès, E.; Milat, M.-L.; Ponchet, M.; Marion, D.; Lequeu, J.; Delrot, S.; Coutos-Thévenot, P.; Blein, J.-P. Modulation of the biological activity of a tobacco LTP1 by lipid complexation. Mol. Biol. Cell 2004, 15, 5047–5052. [Google Scholar] [CrossRef] [PubMed]
- Dubiela, P.; Del Conte, R.; Cantini, F.; Borowski, T.; Aina, R.; Radauer, C.; Bublin, M.; Hoffmann-Sommergruber, K.; Alessandri, S. Impact of lipid binding on the tertiary structure and allergenic potential of Jug r 3, the non-specific lipid transfer protein from walnut. Sci. Rep. 2019, 9, 2007. [Google Scholar] [CrossRef]
- Gizatullina, A.K.; Finkina, E.I.; Mineev, K.S.; Melnikova, D.N.; Bogdanov, I.V.; Telezhinskaya, I.N.; Balandin, S.V.; Shenkarev, Z.O.; Arseniev, A.S.; Ovchinnikova, T.V. Recombinant production and solution structure of lipid transfer protein from lentil Lens culinaris. Biochem. Biophys. Res. Commun. 2013, 439, 427–432. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, 363–367. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina, Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–561. [Google Scholar] [CrossRef]
- Dassault Systèmes BIOVIA, Discovery Studio Visualizer, v20.1.0.19295, Dassault Systèmes: San Diego, CA, USA, 2020.
- Dutta, S.; Watson, B.G.; Mattoo, S.; Rochet, J.-C. Calcein Release Assay to Measure Membrane Permeabilization by Recombinant Alpha-Synuclein. Bio-Protocol 2020, 10, e3690. [Google Scholar] [CrossRef]
- Alekseeva, A.S.; Tretiakova, D.S.; Melnikova, D.N.; Molotkovsky, U.G.; Boldyrev, I.A. Novel fluorescent membrane probe 2,3;5,6-bis(cyclohexyl)-BODIPY-labeled phosphatidylcholine. Russ. J. Bioorg. Chem. 2016, 42, 305–309. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melnikova, D.N.; Bogdanov, I.V.; Potapov, A.E.; Alekseeva, A.S.; Finkina, E.I.; Ovchinnikova, T.V. Molecular Insight into Ligand Binding and Transport by the Lentil Lipid Transfer Protein Lc-LTP2: The Role of Basic Amino Acid Residues at Opposite Entrances to the Hydrophobic Cavity. Biomolecules 2023, 13, 1699. https://doi.org/10.3390/biom13121699
Melnikova DN, Bogdanov IV, Potapov AE, Alekseeva AS, Finkina EI, Ovchinnikova TV. Molecular Insight into Ligand Binding and Transport by the Lentil Lipid Transfer Protein Lc-LTP2: The Role of Basic Amino Acid Residues at Opposite Entrances to the Hydrophobic Cavity. Biomolecules. 2023; 13(12):1699. https://doi.org/10.3390/biom13121699
Chicago/Turabian StyleMelnikova, Daria N., Ivan V. Bogdanov, Andrey E. Potapov, Anna S. Alekseeva, Ekaterina I. Finkina, and Tatiana V. Ovchinnikova. 2023. "Molecular Insight into Ligand Binding and Transport by the Lentil Lipid Transfer Protein Lc-LTP2: The Role of Basic Amino Acid Residues at Opposite Entrances to the Hydrophobic Cavity" Biomolecules 13, no. 12: 1699. https://doi.org/10.3390/biom13121699
APA StyleMelnikova, D. N., Bogdanov, I. V., Potapov, A. E., Alekseeva, A. S., Finkina, E. I., & Ovchinnikova, T. V. (2023). Molecular Insight into Ligand Binding and Transport by the Lentil Lipid Transfer Protein Lc-LTP2: The Role of Basic Amino Acid Residues at Opposite Entrances to the Hydrophobic Cavity. Biomolecules, 13(12), 1699. https://doi.org/10.3390/biom13121699








