REST Is Not Resting: REST/NRSF in Health and Disease
Abstract
1. Introduction
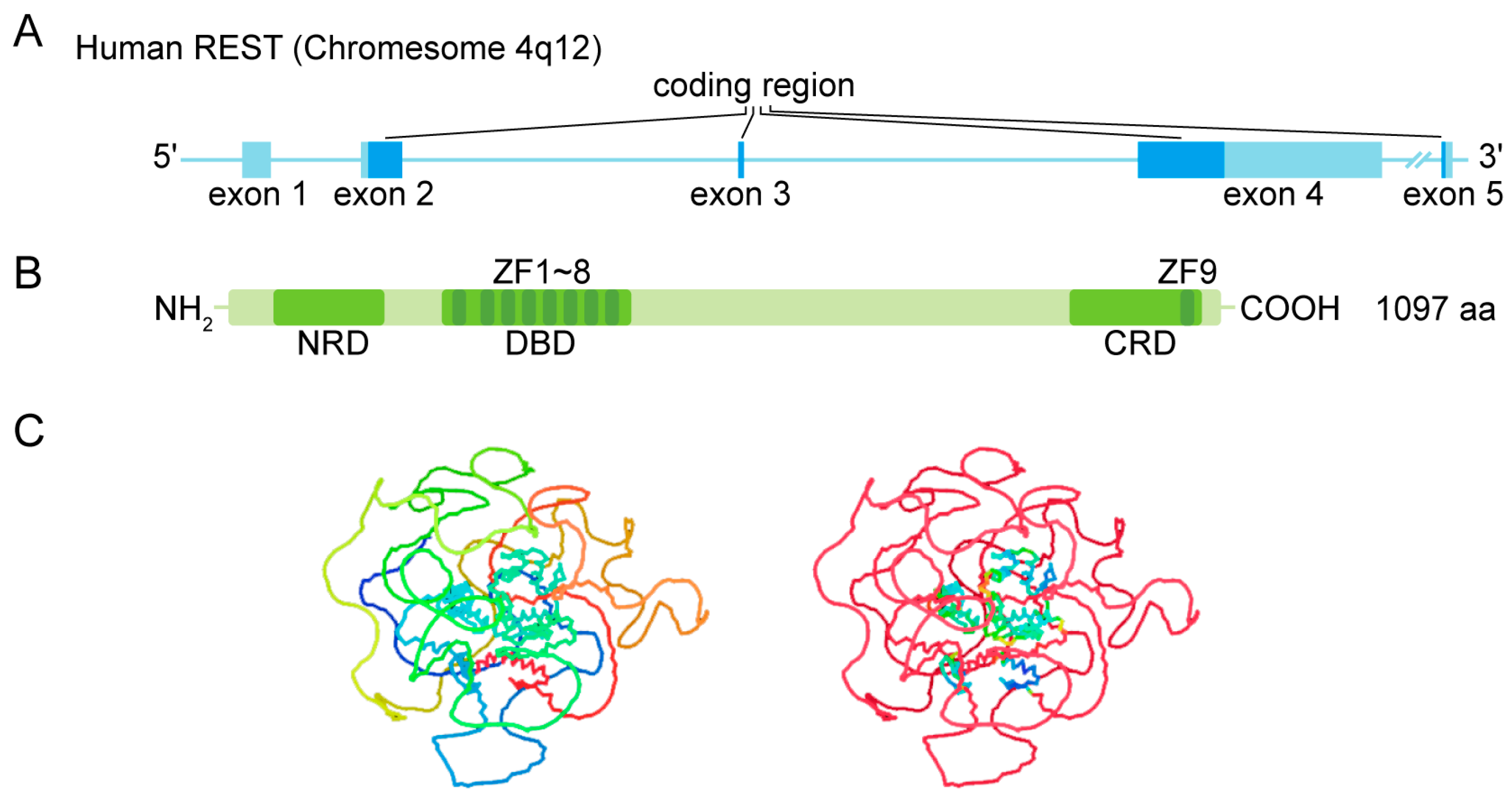
1.1. Gene and Protein Structure
1.2. Nuclear Location
1.3. DNA Binding
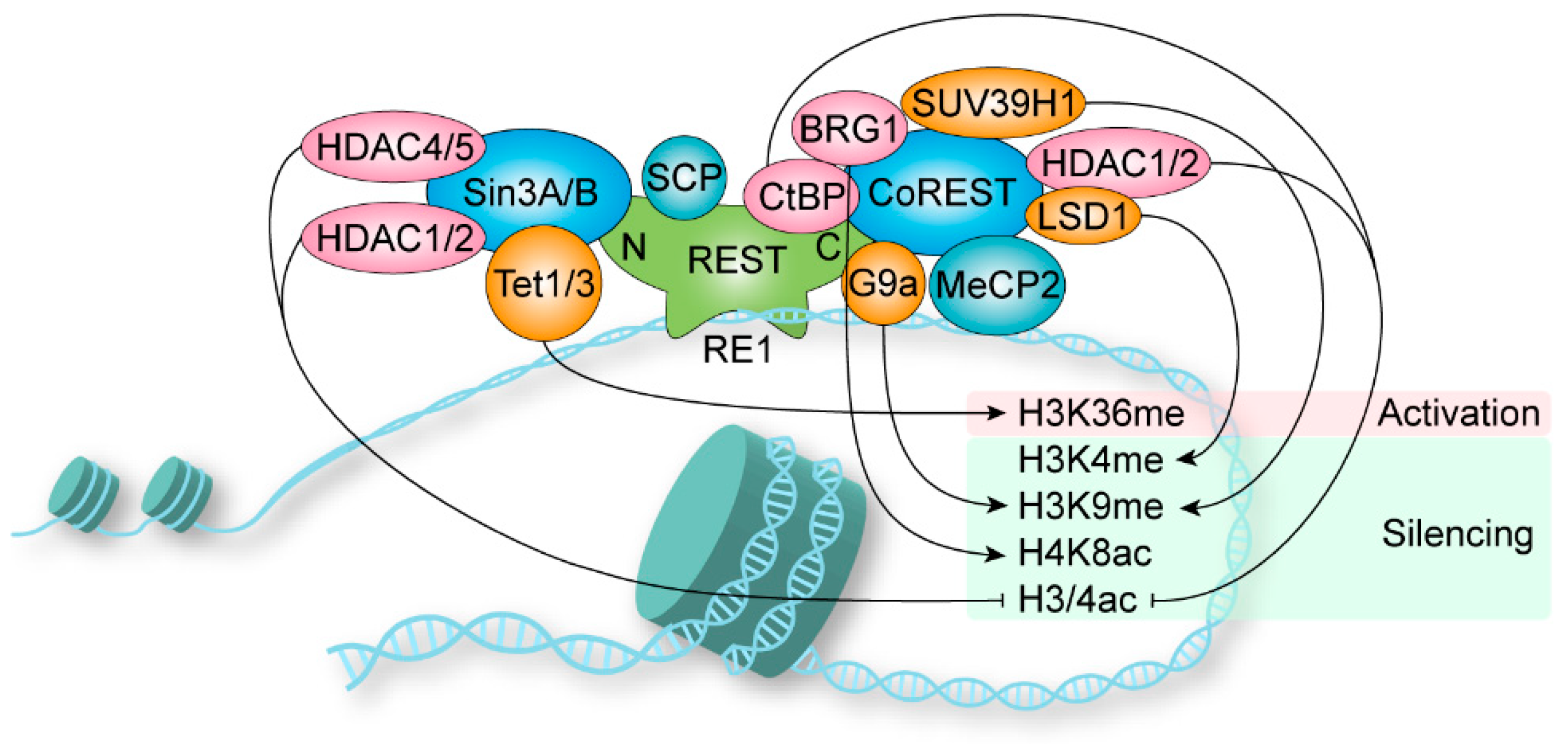
1.4. Coregulator Recruitment
2. REST in Nervous System
2.1. Embryogenesis and Neurogenesis
2.2. Neuronal Differentiation
2.3. Neuronal Survival
2.4. Neuronal Transmission and Synaptic Plasticity
2.5. Pain
2.6. Neuroendocrine
2.7. Intelligence and Memory
2.8. Aging and Alzheimer’s Disease
2.9. Parkinson’s Disease
2.10. Huntington’s Disease
2.11. Epilepsy
2.12. Ischemia
2.13. Psychiatric Disorders
3. REST in Other Systems
3.1. Heart
3.2. Pancreas
3.3. Skin
3.4. Eye
3.5. Vascular
4. REST in Cancer
4.1. Nervous System Cancer
4.2. Non-Nervous System Cancer
5. Targeting REST
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chong, J.A.; Tapia-Ramirez, J.; Kim, S.; Toledo-Aral, J.J.; Zheng, Y.; Boutros, M.C.; Altshuller, Y.M.; Frohman, M.A.; Kraner, S.D.; Mandel, G. REST: A mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 1995, 80, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Schoenherr, C.J.; Anderson, D.J. The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science 1995, 267, 1360–1363. [Google Scholar] [CrossRef]
- Chen, G.L.; Miller, G.M. Alternative REST Splicing Underappreciated. Eneuro 2018, 5, 1–6. [Google Scholar] [CrossRef]
- Shimojo, M.; Lee, J.H.; Hersh, L.B. Role of zinc finger domains of the transcription factor neuron-restrictive silencer factor/repressor element-1 silencing transcription factor in DNA binding and nuclear localization. J. Biol. Chem. 2001, 276, 13121–13126. [Google Scholar] [CrossRef] [PubMed]
- Shimojo, M. Characterization of the nuclear targeting signal of REST/NRSF. Neurosci. Lett. 2006, 398, 161–166. [Google Scholar] [CrossRef]
- Shimojo, M.; Hersh, L.B. Characterization of the REST/NRSF-interacting LIM domain protein (RILP): Localization and interaction with REST/NRSF. J. Neurochem. 2006, 96, 1130–1138. [Google Scholar] [CrossRef]
- Lee, J.H.; Shimojo, M.; Chai, Y.G.; Hersh, L.B. Studies on the interaction of REST4 with the cholinergic repressor element-1/neuron restrictive silencer element. Brain Res. Mol. Brain Res. 2000, 80, 88–98. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, W.; Shen, J.; Tong, X.; Yang, Z.; Shen, Z.; Lan, W.; Wu, H.; Cao, C. Cysteine 397 plays important roles in the folding of the neuron-restricted silencer factor/RE1-silencing transcription factor. Biochem. Biophys. Res. Commun. 2011, 414, 309–314. [Google Scholar] [CrossRef]
- Kraner, S.D.; Chong, J.A.; Tsay, H.J.; Mandel, G. Silencing the type II sodium channel gene: A model for neural-specific gene regulation. Neuron 1992, 9, 37–44. [Google Scholar] [CrossRef]
- Mori, N.; Schoenherr, C.; Vandenbergh, D.J.; Anderson, D.J. A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron 1992, 9, 45–54. [Google Scholar] [CrossRef]
- Li, L.; Suzuki, T.; Mori, N.; Greengard, P. Identification of a functional silencer element involved in neuron-specific expression of the synapsin I gene. Proc. Natl. Acad. Sci. USA 1993, 90, 1460–1464. [Google Scholar] [CrossRef]
- Schoenherr, C.J.; Paquette, A.J.; Anderson, D.J. Identification of potential target genes for the neuron-restrictive silencer factor. Proc. Natl. Acad. Sci. USA 1996, 93, 9881–9886. [Google Scholar] [CrossRef]
- Bruce, A.W.; Donaldson, I.J.; Wood, I.C.; Yerbury, S.A.; Sadowski, M.I.; Chapman, M.; Gottgens, B.; Buckley, N.J. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. USA 2004, 101, 10458–10463. [Google Scholar] [CrossRef]
- Sun, Y.M.; Greenway, D.J.; Johnson, R.; Street, M.; Belyaev, N.D.; Deuchars, J.; Bee, T.; Wilde, S.; Buckley, N.J. Distinct profiles of REST interactions with its target genes at different stages of neuronal development. Mol. Biol. Cell 2005, 16, 5630–5638. [Google Scholar] [CrossRef]
- Johnson, D.S.; Mortazavi, A.; Myers, R.M.; Wold, B. Genome-wide mapping of in vivo protein-DNA interactions. Science 2007, 316, 1497–1502. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, M.; Niu, G.; Cheng, Y.; Fei, J. Genome-wide identification of target genes repressed by the zinc finger transcription factor REST/NRSF in the HEK 293 cell line. Acta Biochim. Biophys. Sin. 2009, 41, 1008–1017. [Google Scholar] [CrossRef]
- Sedaghat, Y.; Bui, H.H.; Mazur, C.; Monia, B.P. Identification of REST-regulated genes and pathways using a REST-targeted antisense approach. Nucleic Acid Ther. 2013, 23, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.; Teh, C.H.; Kunarso, G.; Wong, K.Y.; Srinivasan, G.; Cooper, M.L.; Volta, M.; Chan, S.S.; Lipovich, L.; Pollard, S.M.; et al. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008, 6, e256. [Google Scholar] [CrossRef]
- Mortazavi, A.; Leeper Thompson, E.C.; Garcia, S.T.; Myers, R.M.; Wold, B. Comparative genomics modeling of the NRSF/REST repressor network: From single conserved sites to genome-wide repertoire. Genome Res. 2006, 16, 1208–1221. [Google Scholar] [CrossRef]
- Ding, Y.; Lorenz, W.A.; Chuang, J.H. CodingMotif: Exact determination of overrepresented nucleotide motifs in coding sequences. BMC Bioinform. 2012, 13, 32. [Google Scholar] [CrossRef] [PubMed]
- Jothi, R.; Cuddapah, S.; Barski, A.; Cui, K.; Zhao, K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008, 36, 5221–5231. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Jiang, H.; Ma, W.; Johnson, D.S.; Myers, R.M.; Wong, W.H. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008, 26, 1293–1300. [Google Scholar] [CrossRef]
- Valouev, A.; Johnson, D.S.; Sundquist, A.; Medina, C.; Anton, E.; Batzoglou, S.; Myers, R.M.; Sidow, A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods 2008, 5, 829–834. [Google Scholar] [CrossRef]
- Nix, D.A.; Courdy, S.J.; Boucher, K.M. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinform. 2008, 9, 523. [Google Scholar] [CrossRef]
- Boeva, V.; Surdez, D.; Guillon, N.; Tirode, F.; Fejes, A.P.; Delattre, O.; Barillot, E. De novo motif identification improves the accuracy of predicting transcription factor binding sites in ChIP-Seq data analysis. Nucleic Acids Res. 2010, 38, e126. [Google Scholar] [CrossRef]
- McLean, C.Y.; Bristor, D.; Hiller, M.; Clarke, S.L.; Schaar, B.T.; Lowe, C.B.; Wenger, A.M.; Bejerano, G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010, 28, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.K.; Jayatillake, R.V.; Spouge, J.L. NEXT-peak: A normal-exponential two-peak model for peak-calling in ChIP-seq data. BMC Genom. 2013, 14, 349. [Google Scholar] [CrossRef]
- Yu, H.B.; Johnson, R.; Kunarso, G.; Stanton, L.W. Coassembly of REST and its cofactors at sites of gene repression in embryonic stem cells. Genome Res. 2011, 21, 1284–1293. [Google Scholar] [CrossRef]
- Grimes, J.A.; Nielsen, S.J.; Battaglioli, E.; Miska, E.A.; Speh, J.C.; Berry, D.L.; Atouf, F.; Holdener, B.C.; Mandel, G.; Kouzarides, T. The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J. Biol. Chem. 2000, 275, 9461–9467. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Myers, S.J.; Dingledine, R. Transcriptional repression by REST: Recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 1999, 2, 867–872. [Google Scholar] [CrossRef]
- Roopra, A.; Sharling, L.; Wood, I.C.; Briggs, T.; Bachfischer, U.; Paquette, A.J.; Buckley, N.J. Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Mol. Cell Biol. 2000, 20, 2147–2157. [Google Scholar] [CrossRef]
- Adams, G.E.; Chandru, A.; Cowley, S.M. Co-repressor, co-activator and general transcription factor: The many faces of the Sin3 histone deacetylase (HDAC) complex. Biochem. J. 2018, 475, 3921–3932. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Kuwahara, K.; Harada, M.; Takahashi, N.; Yasuno, S.; Adachi, Y.; Kawakami, R.; Nakanishi, M.; Tanimoto, K.; Usami, S.; et al. Class II HDACs mediate CaMK-dependent signaling to NRSF in ventricular myocytes. J. Mol. Cell. Cardiol. 2006, 41, 1010–1022. [Google Scholar] [CrossRef]
- Naruse, Y.; Aoki, T.; Kojima, T.; Mori, N. Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc. Natl. Acad. Sci. USA 1999, 96, 13691–13696. [Google Scholar] [CrossRef]
- Ooi, L.; Wood, I.C. Chromatin crosstalk in development and disease: Lessons from REST. Nat. Rev. Genet. 2007, 8, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Nomura, M.; Uda-Tochio, H.; Murai, K.; Mori, N.; Nishimura, Y. The neural repressor NRSF/REST binds the PAH1 domain of the Sin3 corepressor by using its distinct short hydrophobic helix. J. Mol. Biol. 2005, 354, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Higo, J.; Nishimura, Y.; Nakamura, H. A free-energy landscape for coupled folding and binding of an intrinsically disordered protein in explicit solvent from detailed all-atom computations. J. Am. Chem. Soc. 2011, 133, 10448–10458. [Google Scholar] [CrossRef]
- Chandru, A.; Bate, N.; Vuister, G.W.; Cowley, S.M. Sin3A recruits Tet1 to the PAH1 domain via a highly conserved Sin3-Interaction Domain. Sci. Rep. 2018, 8, 14689. [Google Scholar] [CrossRef] [PubMed]
- Perera, A.; Eisen, D.; Wagner, M.; Laube, S.K.; Kunzel, A.F.; Koch, S.; Steinbacher, J.; Schulze, E.; Splith, V.; Mittermeier, N.; et al. TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Rep. 2015, 11, 283–294. [Google Scholar] [CrossRef]
- Yeo, M.; Lee, S.K.; Lee, B.; Ruiz, E.C.; Pfaff, S.L.; Gill, G.N. Small CTD phosphatases function in silencing neuronal gene expression. Science 2005, 307, 596–600. [Google Scholar] [CrossRef]
- Andres, M.E.; Burger, C.; Peral-Rubio, M.J.; Battaglioli, E.; Anderson, M.E.; Grimes, J.; Dallman, J.; Ballas, N.; Mandel, G. CoREST: A functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. USA 1999, 96, 9873–9878. [Google Scholar] [CrossRef] [PubMed]
- Lunyak, V.V.; Burgess, R.; Prefontaine, G.G.; Nelson, C.; Sze, S.H.; Chenoweth, J.; Schwartz, P.; Pevzner, P.A.; Glass, C.; Mandel, G.; et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science 2002, 298, 1747–1752. [Google Scholar] [CrossRef] [PubMed]
- Ballas, N.; Mandel, G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr. Opin. Neurobiol. 2005, 15, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Roopra, A.; Qazi, R.; Schoenike, B.; Daley, T.J.; Morrison, J.F. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell 2004, 14, 727–738. [Google Scholar] [CrossRef]
- Ding, N.; Tomomori-Sato, C.; Sato, S.; Conaway, R.C.; Conaway, J.W.; Boyer, T.G. MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression. J. Biol. Chem. 2009, 284, 2648–2656. [Google Scholar] [CrossRef]
- Ding, N.; Zhou, H.; Esteve, P.O.; Chin, H.G.; Kim, S.; Xu, X.; Joseph, S.M.; Friez, M.J.; Schwartz, C.E.; Pradhan, S.; et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol. Cell 2008, 31, 347–359. [Google Scholar] [CrossRef]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef]
- Ooi, L.; Belyaev, N.D.; Miyake, K.; Wood, I.C.; Buckley, N.J. BRG1 chromatin remodeling activity is required for efficient chromatin binding by repressor element 1-silencing transcription factor (REST) and facilitates REST-mediated repression. J. Biol. Chem. 2006, 281, 38974–38980. [Google Scholar] [CrossRef]
- Shi, Y.; Sawada, J.; Sui, G.; Affarel, B.; Whetstine, J.R.; Lan, F.; Ogawa, H.; Luke, M.P.; Nakatani, Y.; Shi, Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 2003, 422, 735–738. [Google Scholar] [CrossRef]
- Garriga-Canut, M.; Schoenike, B.; Qazi, R.; Bergendahl, K.; Daley, T.J.; Pfender, R.M.; Morrison, J.F.; Ockuly, J.; Stafstrom, C.; Sutula, T.; et al. 2-Deoxy-D-glucose reduces epilepsy progression by NRSF-CtBP-dependent metabolic regulation of chromatin structure. Nat. Neurosci. 2006, 9, 1382–1387. [Google Scholar] [CrossRef]
- Ballas, N.; Grunseich, C.; Lu, D.D.; Speh, J.C.; Mandel, G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 2005, 121, 645–657. [Google Scholar] [CrossRef]
- Chen, Z.F.; Paquette, A.J.; Anderson, D.J. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 1998, 20, 136–142. [Google Scholar] [CrossRef]
- Wang, Y.C.; Liu, P.; Yue, L.Y.; Huang, F.; Xu, Y.X.; Zhu, C.Q. NRSF deficiency leads to abnormal postnatal development of dentate gyrus and impairment of progenitors in subgranular zone of hippocampus. Hippocampus 2021, 31, 935–9563. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Lee, Y.K.; Lim, Y.C.; Zheng, Z.; Lin, X.M.; Ng, D.P.; Holbrook, J.D.; Law, H.Y.; Kwek, K.Y.; Yeo, G.S.; et al. Global DNA hypermethylation in down syndrome placenta. PLoS Genet. 2013, 9, e1003515. [Google Scholar] [CrossRef]
- El Hajj, N.; Dittrich, M.; Bock, J.; Kraus, T.F.; Nanda, I.; Muller, T.; Seidmann, L.; Tralau, T.; Galetzka, D.; Schneider, E.; et al. Epigenetic dysregulation in the developing down syndrome cortex. Epigenetics 2016, 11, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Canzonetta, C.; Mulligan, C.; Deutsch, S.; Ruf, S.; O’Doherty, A.; Lyle, R.; Borel, C.; Lin-Marq, N.; Delom, F.; Groet, J.; et al. DYRK1A-dosage imbalance perturbs NRSF/REST levels, deregulating pluripotency and embryonic stem cell fate in Down syndrome. Am. J. Hum. Genet. 2008, 83, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Lepagnol-Bestel, A.M.; Zvara, A.; Maussion, G.; Quignon, F.; Ngimbous, B.; Ramoz, N.; Imbeaud, S.; Loe-Mie, Y.; Benihoud, K.; Agier, N.; et al. DYRK1A interacts with the REST/NRSF-SWI/SNF chromatin remodelling complex to deregulate gene clusters involved in the neuronal phenotypic traits of Down syndrome. Hum. Mol. Genet. 2009, 18, 1405–1414. [Google Scholar] [CrossRef]
- Nishihara, S.; Tsuda, L.; Ogura, T. The canonical Wnt pathway directly regulates NRSF/REST expression in chick spinal cord. Biochem. Biophys. Res. Commun. 2003, 311, 55–63. [Google Scholar] [CrossRef]
- Li, H.; Malbon, C.C.; Wang, H.Y. Gene profiling of Frizzled-1 and Frizzled-2 signaling: Expression of G-protein-coupled receptor chimeras in mouse F9 teratocarcinoma embryonal cells. Mol. Pharmacol. 2004, 65, 45–55. [Google Scholar] [CrossRef]
- Gates, K.P.; Mentzer, L.; Karlstrom, R.O.; Sirotkin, H.I. The transcriptional repressor REST/NRSF modulates hedgehog signaling. Dev. Biol. 2010, 340, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ren, J.; Wang, Z.; Yao, J.; Fei, J. NRSF/REST is required for gastrulation and neurogenesis during zebrafish development. Acta Biochim. Biophys. Sin. 2012, 44, 385–393. [Google Scholar] [CrossRef][Green Version]
- Olguin, P.; Oteiza, P.; Gamboa, E.; Gomez-Skarmeta, J.L.; Kukuljan, M. RE-1 silencer of transcription/neural restrictive silencer factor modulates ectodermal patterning during Xenopus development. J. Neurosci. 2006, 26, 2820–2829. [Google Scholar] [CrossRef]
- Arora, D.K.; Cosgrave, A.S.; Howard, M.R.; Bubb, V.; Quinn, J.P.; Thippeswamy, T. Evidence of postnatal neurogenesis in dorsal root ganglion: Role of nitric oxide and neuronal restrictive silencer transcription factor. J. Mol. Neurosci. 2007, 32, 97–107. [Google Scholar] [CrossRef]
- Kok, F.O.; Taibi, A.; Wanner, S.J.; Xie, X.; Moravec, C.E.; Love, C.E.; Prince, V.E.; Mumm, J.S.; Sirotkin, H.I. Zebrafish rest regulates developmental gene expression but not neurogenesis. Development 2012, 139, 3838–3848. [Google Scholar] [CrossRef][Green Version]
- Moravec, C.E.; Li, E.; Maaswinkel, H.; Kritzer, M.F.; Weng, W.; Sirotkin, H.I. Rest mutant zebrafish swim erratically and display atypical spatial preferences. Behav. Brain Res. 2015, 284, 238–248. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aoki, H.; Hara, A.; Era, T.; Kunisada, T.; Yamada, Y. Genetic ablation of Rest leads to in vitro-specific derepression of neuronal genes during neurogenesis. Development 2012, 139, 667–677. [Google Scholar] [CrossRef]
- Cheng, Y.; Yin, Y.; Zhang, A.; Bernstein, A.M.; Kawaguchi, R.; Gao, K.; Potter, K.; Gilbert, H.Y.; Ao, Y.; Ou, J.; et al. Transcription factor network analysis identifies REST/NRSF as an intrinsic regulator of CNS regeneration in mice. Nat. Commun. 2022, 13, 4418. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Kagalwala, M.N.; Parker-Thornburg, J.; Adams, H.; Majumder, S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature 2008, 453, 223–227. [Google Scholar] [CrossRef]
- Gupta, S.K.; Gressens, P.; Mani, S. NRSF downregulation induces neuronal differentiation in mouse embryonic stem cells. Differentiation 2009, 77, 19–28. [Google Scholar] [CrossRef]
- Thakore-Shah, K.; Koleilat, T.; Jan, M.; John, A.; Pyle, A.D. REST/NRSF Knockdown Alters Survival, Lineage Differentiation and Signaling in Human Embryonic Stem Cells. PLoS ONE 2015, 10, e0145280. [Google Scholar] [CrossRef]
- Tyler, C.R.; Labrecque, M.T.; Solomon, E.R.; Guo, X.; Allan, A.M. Prenatal arsenic exposure alters REST/NRSF and microRNA regulators of embryonic neural stem cell fate in a sex-dependent manner. Neurotoxicol. Teratol. 2017, 59, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Satoh, J.; Kawana, N.; Yamamoto, Y. ChIP-Seq Data Mining: Remarkable Differences in NRSF/REST Target Genes between Human ESC and ESC-Derived Neurons. Bioinform. Biol. Insights 2013, 7, 357–368. [Google Scholar] [CrossRef]
- Gao, Z.; Ure, K.; Ding, P.; Nashaat, M.; Yuan, L.; Ma, J.; Hammer, R.E.; Hsieh, J. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 2011, 31, 9772–9786. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Kameoka, S.; Lentz, S.; Majumder, S. Activation of REST/NRSF target genes in neural stem cells is sufficient to cause neuronal differentiation. Mol. Cell. Biol. 2004, 24, 8018–8025. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Zhuang, Z.; Liu, A.; Chai, Y.; Hoffman, P.W. The role of the RE1 element in activation of the NR1 promoter during neuronal differentiation. J. Neurochem. 2003, 86, 992–1005. [Google Scholar] [CrossRef]
- Kim, S.M.; Yang, J.W.; Park, M.J.; Lee, J.K.; Kim, S.U.; Lee, Y.S.; Lee, M.A. Regulation of human tyrosine hydroxylase gene by neuron-restrictive silencer factor. Biochem. Biophys. Res. Commun. 2006, 346, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Abrajano, J.J.; Qureshi, I.A.; Gokhan, S.; Zheng, D.; Bergman, A.; Mehler, M.F. Differential deployment of REST and CoREST promotes glial subtype specification and oligodendrocyte lineage maturation. PLoS ONE 2009, 4, e7665. [Google Scholar] [CrossRef]
- Kohyama, J.; Sanosaka, T.; Tokunaga, A.; Takatsuka, E.; Tsujimura, K.; Okano, H.; Nakashima, K. BMP-induced REST regulates the establishment and maintenance of astrocytic identity. J. Cell Biol. 2010, 189, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, Y.; Lv, Y.; Zhang, S.; Chen, L.; Bai, C.; Nan, X.; Yue, W.; Pei, X. NRSF silencing induces neuronal differentiation of human mesenchymal stem cells. Exp. Cell Res. 2008, 314, 2257–2265. [Google Scholar] [CrossRef]
- Kim, H.J.; Denli, A.M.; Wright, R.; Baul, T.D.; Clemenson, G.D.; Morcos, A.S.; Zhao, C.; Schafer, S.T.; Gage, F.H.; Kagalwala, M.N. REST Regulates Non-Cell-Autonomous Neuronal Differentiation and Maturation of Neural Progenitor Cells via Secretogranin II. J. Neurosci. 2015, 35, 14872–14884. [Google Scholar] [CrossRef]
- Zhao, X.; Glass, Z.; Chen, J.; Yang, L.; Kaplan, D.L.; Xu, Q. mRNA Delivery Using Bioreducible Lipidoid Nanoparticles Facilitates Neural Differentiation of Human Mesenchymal Stem Cells. Adv. Healthc. Mater. 2021, 10, e2000938. [Google Scholar] [CrossRef] [PubMed]
- Aoki, H.; Hara, A.; Oomori, Y.; Shimizu, Y.; Yamada, Y.; Kunisada, T. Neonatal lethality of neural crest cell-specific Rest knockout mice is associated with gastrointestinal distension caused by aberrations of myenteric plexus. Genes Cells 2014, 19, 723–742. [Google Scholar] [CrossRef] [PubMed]
- Tomasoni, R.; Negrini, S.; Fiordaliso, S.; Klajn, A.; Tkatch, T.; Mondino, A.; Meldolesi, J.; D’Alessandro, R. A signaling loop of REST, TSC2 and beta-catenin governs proliferation and function of PC12 neural cells. J. Cell Sci. 2011, 124, 3174–3186. [Google Scholar] [CrossRef]
- Toshiyuki, N.; Ichiro, M. Molecular mechanisms regulating cell type specific expression of BMP/RA Inducible Neural-specific Protein-1 that suppresses cell cycle progression: Roles of NRSF/REST and DNA methylation. Brain Res. Mol. Brain Res. 2004, 125, 47–59. [Google Scholar] [CrossRef]
- Yoo, A.S.; Staahl, B.T.; Chen, L.; Crabtree, G.R. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 2009, 460, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Tabuchi, A.; Yamada, T.; Sasagawa, S.; Naruse, Y.; Mori, N.; Tsuda, M. REST4-mediated modulation of REST/NRSF-silencing function during BDNF gene promoter activation. Biochem. Biophys. Res. Commun. 2002, 290, 415–420. [Google Scholar] [CrossRef]
- Qiang, M.; Rani, C.S.; Ticku, M.K. Neuron-restrictive silencer factor regulates the N-methyl-D-aspartate receptor 2B subunit gene in basal and ethanol-induced gene expression in fetal cortical neurons. Mol. Pharmacol. 2005, 67, 2115–2125. [Google Scholar] [CrossRef] [PubMed]
- Tateno, M.; Ukai, W.; Hashimoto, E.; Ikeda, H.; Saito, T. Implication of increased NRSF/REST binding activity in the mechanism of ethanol inhibition of neuronal differentiation. J. Neural Transm. 2006, 113, 283–293. [Google Scholar] [CrossRef]
- Tateno, M.; Saito, T. Biological studies on alcohol-induced neuronal damage. Psychiatry Investig. 2008, 5, 21–27. [Google Scholar] [CrossRef]
- Cai, L.; Bian, M.; Liu, M.; Sheng, Z.; Suo, H.; Wang, Z.; Huang, F.; Fei, J. Ethanol-induced neurodegeneration in NRSF/REST neuronal conditional knockout mice. Neuroscience 2011, 181, 196–205. [Google Scholar] [CrossRef]
- Formisano, L.; Guida, N.; Laudati, G.; Boscia, F.; Esposito, A.; Secondo, A.; Di Renzo, G.; Canzoniero, L.M. Extracellular signal-related kinase 2/specificity protein 1/specificity protein 3/repressor element-1 silencing transcription factor pathway is involved in Aroclor 1254-induced toxicity in SH-SY5Y neuronal cells. J. Neurosci. Res. 2015, 93, 167–177. [Google Scholar] [CrossRef]
- Rivera-Cervantes, M.C.; Castaneda-Arellano, R.; Castro-Torres, R.D.; Gudino-Cabrera, G.; Feria y Velasco, A.I.; Camins, A.; Beas-Zarate, C. P38 MAPK inhibition protects against glutamate neurotoxicity and modifies NMDA and AMPA receptor subunit expression. J. Mol. Neurosci. 2015, 55, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Pajarillo, E.; Rizor, A.; Son, D.S.; Aschner, M.; Lee, E. The transcription factor REST up-regulates tyrosine hydroxylase and antiapoptotic genes and protects dopaminergic neurons against manganese toxicity. J. Biol. Chem. 2020, 295, 3040–3054. [Google Scholar] [CrossRef]
- Kallunki, P.; Edelman, G.M.; Jones, F.S. Tissue-specific expression of the L1 cell adhesion molecule is modulated by the neural restrictive silencer element. J. Cell Biol. 1997, 138, 1343–1354. [Google Scholar] [CrossRef] [PubMed]
- Schulte, C.; Racchetti, G.; D’Alessandro, R.; Meldolesi, J. A new form of neurite outgrowth sustained by the exocytosis of enlargeosomes expressed under the control of REST. Traffic 2010, 11, 1304–1314. [Google Scholar] [CrossRef] [PubMed]
- Paquette, A.J.; Perez, S.E.; Anderson, D.J. Constitutive expression of the neuron-restrictive silencer factor (NRSF)/REST in differentiating neurons disrupts neuronal gene expression and causes axon pathfinding errors in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 12318–12323. [Google Scholar] [CrossRef]
- Schoch, S.; Cibelli, G.; Thiel, G. Neuron-specific gene expression of synapsin I. Major role of a negative regulatory mechanism. J. Biol. Chem. 1996, 271, 3317–3323. [Google Scholar] [CrossRef]
- Chin, L.S.; Weigel, C.; Li, L. Transcriptional regulation of gene expression of sec6, a component of mammalian exocyst complex at the synapse. Brain Res. Mol. Brain Res. 2000, 79, 127–137. [Google Scholar] [CrossRef]
- Pozzi, D.; Menna, E.; Canzi, A.; Desiato, G.; Mantovani, C.; Matteoli, M. The Communication Between the Immune and Nervous Systems: The Role of IL-1β in Synaptopathies. Front. Mol. Neurosci. 2018, 11, 111. [Google Scholar] [CrossRef]
- Buffolo, F.; Petrosino, V.; Albini, M.; Moschetta, M.; Carlini, F.; Floss, T.; Kerlero de Rosbo, N.; Cesca, F.; Rocchi, A.; Uccelli, A.; et al. Neuroinflammation induces synaptic scaling through IL-1β-mediated activation of the transcriptional repressor REST/NRSF. Cell Death Dis. 2021, 12, 180. [Google Scholar] [CrossRef]
- Bersten, D.C.; Wright, J.A.; McCarthy, P.J.; Whitelaw, M.L. Regulation of the neuronal transcription factor NPAS4 by REST and microRNAs. Biochim. Biophys. Acta 2014, 1839, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Prestigio, C.; Ferrante, D.; Marte, A.; Romei, A.; Lignani, G.; Onofri, F.; Valente, P.; Benfenati, F.; Baldelli, P. REST/NRSF drives homeostatic plasticity of inhibitory synapses in a target-dependent fashion. eLife 2021, 10, e69058. [Google Scholar] [CrossRef]
- Moon, S.M.; Kim, J.S.; Park, B.R.; Kim, D.K.; Kim, S.G.; Kim, H.J.; Chun, H.S.; Lee, B.K.; Kim, C.S. Transcriptional regulation of the neuropeptide VGF by the neuron-restrictive silencer factor/neuron-restrictive silencer element. NeuroReport 2015, 26, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, D.; Lignani, G.; Ferrea, E.; Contestabile, A.; Paonessa, F.; D’Alessandro, R.; Lippiello, P.; Boido, D.; Fassio, A.; Meldolesi, J.; et al. REST/NRSF-mediated intrinsic homeostasis protects neuronal networks from hyperexcitability. EMBO J. 2013, 32, 2994–3007. [Google Scholar] [CrossRef]
- Drews, V.L.; Shi, K.; de Haan, G.; Meisler, M.H. Identification of evolutionarily conserved, functional noncoding elements in the promoter region of the sodium channel gene SCN8A. Mamm. Genome 2007, 18, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Ariano, P.; Zamburlin, P.; D’Alessandro, R.; Meldolesi, J.; Lovisolo, D. Differential repression by the transcription factor REST/NRSF of the various Ca2+ signalling mechanisms in pheochromocytoma PC12 cells. Cell Calcium 2010, 47, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Antoniotti, S.; Ruffinatti, F.A.; Torriano, S.; Luganini, A.; D’Alessandro, R.; Lovisolo, D. REST levels affect the functional expression of voltage dependent calcium channels and the migratory activity in immortalized GnRH neurons. Neurosci. Lett. 2016, 629, 19–25. [Google Scholar] [CrossRef]
- Kim, D.S. No effect of expression of neuron-restrictive silencer factor (NRSF) protein on N-type Ca2+ channel alpha 1B gene promoter activity in NS20Y cells. Mol. Cells 1998, 8, 600–605. [Google Scholar]
- Centonze, E.; Marte, A.; Albini, M.; Rocchi, A.; Cesca, F.; Chiacchiaretta, M.; Floss, T.; Baldelli, P.; Ferroni, S.; Benfenati, F.; et al. Neuron-restrictive silencer factor/repressor element 1-silencing transcription factor (NRSF/REST) controls spatial K(+) buffering in primary cortical astrocytes. J. Neurochem. 2023, 165, 701–721. [Google Scholar] [CrossRef]
- Lonnerberg, P.; Schoenherr, C.J.; Anderson, D.J.; Ibanez, C.F. Cell type-specific regulation of choline acetyltransferase gene expression. Role of the neuron-restrictive silencer element and cholinergic-specific enhancer sequences. J. Biol. Chem. 1996, 271, 33358–33365. [Google Scholar] [CrossRef][Green Version]
- Nawa, Y.; Kaneko, H.; Oda, M.; Tsubonoya, M.; Hiroi, T.; Gentile, M.T.; Colucci-D’Amato, L.; Takahashi, R.; Matsui, H. Functional characterization of the neuron-restrictive silencer element in the human tryptophan hydroxylase 2 gene expression. J. Neurochem. 2017, 142, 827–840. [Google Scholar] [CrossRef]
- Wood, I.C.; Roopra, A.; Buckley, N.J. Neural specific expression of the m4 muscarinic acetylcholine receptor gene is mediated by a RE1/NRSE-type silencing element. J. Biol. Chem. 1996, 271, 14221–14225. [Google Scholar] [CrossRef]
- Mieda, M.; Haga, T.; Saffen, D.W. Expression of the rat m4 muscarinic acetylcholine receptor gene is regulated by the neuron-restrictive silencer element/repressor element 1. J. Biol. Chem. 1997, 272, 5854–5860. [Google Scholar] [CrossRef]
- Mu, W.; Burt, D.R. Transcriptional regulation of GABAA receptor gamma2 subunit gene. Brain Res. Mol. Brain Res. 1999, 67, 137–147. [Google Scholar] [CrossRef]
- Okamoto, S.; Sherman, K.; Lipton, S.A. Absence of binding activity of neuron-restrictive silencer factor is necessary, but not sufficient for transcription of NMDA receptor subunit type 1 in neuronal cells. Brain Res. Mol. Brain Res. 1999, 74, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Lemonde, S.; Rogaeva, A.; Albert, P.R. Cell type-dependent recruitment of trichostatin A-sensitive repression of the human 5-HT1A receptor gene. J. Neurochem. 2004, 88, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Brene, S.; Messer, C.; Okado, H.; Hartley, M.; Heinemann, S.F.; Nestler, E.J. Regulation of GluR2 promoter activity by neurotrophic factors via a neuron-restrictive silencer element. Eur. J. Neurosci. 2000, 12, 1525–1533. [Google Scholar] [CrossRef]
- Lee, K.S.; Chatterjee, P.; Choi, E.Y.; Sung, M.K.; Oh, J.; Won, H.; Park, S.M.; Kim, Y.J.; Yi, S.V.; Choi, J.K. Selection on the regulation of sympathetic nervous activity in humans and chimpanzees. PLoS Genet. 2018, 14, e1007311. [Google Scholar] [CrossRef]
- Kim, C.S.; Hwang, C.K.; Song, K.Y.; Choi, H.S.; Kim, D.K.; Law, P.Y.; Wei, L.N.; Loh, H.H. Novel function of neuron-restrictive silencer factor (NRSF) for posttranscriptional regulation. Biochim. Biophys. Acta 2008, 1783, 1835–1846. [Google Scholar] [CrossRef][Green Version]
- Kim, C.S.; Hwang, C.K.; Choi, H.S.; Song, K.Y.; Law, P.Y.; Wei, L.N.; Loh, H.H. Neuron-restrictive silencer factor (NRSF) functions as a repressor in neuronal cells to regulate the mu opioid receptor gene. J. Biol. Chem. 2004, 279, 46464–46473. [Google Scholar] [CrossRef]
- Kim, C.S.; Choi, H.S.; Hwang, C.K.; Song, K.Y.; Lee, B.K.; Law, P.Y.; Wei, L.N.; Loh, H.H. Evidence of the neuron-restrictive silencer factor (NRSF) interaction with Sp3 and its synergic repression to the mu opioid receptor (MOR) gene. Nucleic Acids Res. 2006, 34, 6392–6403. [Google Scholar] [CrossRef] [PubMed]
- Kong, M.; Shi, L.; Zhou, Y.; He, J.; Zhang, W.; Gu, X.; Zhang, J.; Ma, Z. Changes of Mu-opioid receptor and neuron-restrictive silencer factor in periaquductal gray in mouse models of remifentanil-induced postoperative hyperalgesia. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2014, 39, 901–906. [Google Scholar] [CrossRef]
- Shudo, Y.; Shimojo, M.; Fukunaga, M.; Ito, S. Pituitary adenylate cyclase-activating polypeptide is regulated by alternative splicing of transcriptional repressor REST/NRSF in nerve injury. Life Sci. 2015, 143, 174–181. [Google Scholar] [CrossRef]
- Wang, D.; Yu, J. Negative regulation of REST on NR2B in spinal cord contributes to the development of bone cancer pain in mice. Oncotarget 2016, 7, 85564–85572. [Google Scholar] [CrossRef][Green Version]
- Zhu, C.; Tang, J.; Ding, T.; Chen, L.; Wang, W.; Mei, X.P.; He, X.T.; Wang, W.; Zhang, L.D.; Dong, Y.L.; et al. Neuron-restrictive silencer factor-mediated downregulation of mu-opioid receptor contributes to the reduced morphine analgesia in bone cancer pain. Pain 2017, 158, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Xiao-Die, X.; Xiao-Hong, W.; Cheng-Feng, H.; Zhong-Yu, Y.; Jian-Tao, W.; Hou-Guang, Z.; Jing-Chun, G. Increased NRSF/REST in anterior cingulate cortex contributes to diabetes-related neuropathic pain. Biochem. Biophys. Res. Commun. 2020, 527, 785–790. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, S.R.; Chen, H.; Pan, H.L. RE1-silencing transcription factor controls the acute-to-chronic neuropathic pain transition and Chrm2 receptor gene expression in primary sensory neurons. J. Biol. Chem. 2018, 293, 19078–19091. [Google Scholar] [CrossRef]
- Zhang, F.; Gigout, S.; Liu, Y.; Wang, Y.; Hao, H.; Buckley, N.J.; Zhang, H.; Wood, I.C.; Gamper, N. Repressor element 1-silencing transcription factor drives the development of chronic pain states. Pain 2019, 160, 2398–2408. [Google Scholar] [CrossRef] [PubMed]
- Uchida, H.; Sasaki, K.; Ma, L.; Ueda, H. Neuron-restrictive silencer factor causes epigenetic silencing of Kv4.3 gene after peripheral nerve injury. Neuroscience 2010, 166, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Uchida, H.; Ma, L.; Ueda, H. Epigenetic gene silencing underlies C-fiber dysfunctions in neuropathic pain. J. Neurosci. 2010, 30, 4806–4814. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Q.; Yang, Y.; Lv, Y.; Chen, L.; Bai, C.; Nan, X.; Wang, Y.; Pei, X. Regulatory role of neuron-restrictive silencing factor in the specific expression of cocaine- and amphetamine-regulated transcript gene. J. Neurochem. 2008, 106, 1314–1324. [Google Scholar] [CrossRef]
- D’Alessandro, R.; Klajn, A.; Stucchi, L.; Podini, P.; Malosio, M.L.; Meldolesi, J. Expression of the neurosecretory process in PC12 cells is governed by REST. J. Neurochem. 2008, 105, 1369–1383. [Google Scholar] [CrossRef]
- Prada, I.; Marchaland, J.; Podini, P.; Magrassi, L.; D’Alessandro, R.; Bezzi, P.; Meldolesi, J. REST/NRSF governs the expression of dense-core vesicle gliosecretion in astrocytes. J. Cell Biol. 2011, 193, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Valiya Veettil, M.; Krishna, G.; Roy, A.; Ghosh, A.; Dutta, D.; Kumar, B.; Chakraborty, S.; Anju, T.R.; Sharma-Walia, N.; Chandran, B. Kaposi’s Sarcoma-Associated Herpesvirus Infection Induces the Expression of Neuroendocrine Genes in Endothelial Cells. J. Virol. 2020, 94, e01692-19. [Google Scholar] [CrossRef] [PubMed]
- Mbikay, M.; Raffin-Sanson, M.L.; Sirois, F.; Kalenga, L.; Chretien, M.; Seidah, N.G. Characterization of a repressor element in the promoter region of proprotein convertase 2 (PC2) gene. Brain Res. Mol. Brain Res. 2002, 102, 35–47. [Google Scholar] [CrossRef]
- Seth, K.A.; Majzoub, J.A. Repressor element silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) can act as an enhancer as well as a repressor of corticotropin-releasing hormone gene transcription. J. Biol. Chem. 2001, 276, 13917–13923. [Google Scholar] [CrossRef] [PubMed]
- Rockowitz, S.; Zheng, D. Significant expansion of the REST/NRSF cistrome in human versus mouse embryonic stem cells: Potential implications for neural development. Nucleic Acids Res. 2015, 43, 5730–5743. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, D.; Rockowitz, S.; Zheng, D. Divergence and rewiring of regulatory networks for neural development between human and other species. Neurogenesis 2016, 3, e1231495. [Google Scholar] [CrossRef]
- Bolton, J.L.; Schulmann, A.; Garcia-Curran, M.M.; Regev, L.; Chen, Y.; Kamei, N.; Shao, M.; Singh-Taylor, A.; Jiang, S.; Noam, Y.; et al. Unexpected Transcriptional Programs Contribute to Hippocampal Memory Deficits and Neuronal Stunting after Early-Life Adversity. Cell Rep. 2020, 33, 108511. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, X.; Li, D.; Fang, R.; Wang, Z.; Yun, D.; Wang, M.; Wang, J.; Dong, H.; Fei, Z.; et al. NRSF regulates age-dependently cognitive ability and its conditional knockout in APP/PS1 mice moderately alters ad-like pathology. Hum. Mol. Genet. 2022, 32, 2558–2575. [Google Scholar] [CrossRef]
- Yang, M.; Li, Y.; Hu, L.; Luo, D.; Zhang, Y.; Xiao, X.; Li, G.; Zhang, L.; Zhu, G. Lead exposure inhibits expression of SV2C through NRSF. Toxicology 2018, 398–399, 23–30. [Google Scholar] [CrossRef]
- Verma, P.; Gupta, R.K.; Gandhi, B.S.; Singh, P. CDRI-08 Attenuates REST/NRSF-Mediated Expression of NMDAR1 Gene in PBDE-209-Exposed Mice Brain. Evid. Based Complement. Alternat Med. 2015, 2015, 403840. [Google Scholar] [CrossRef]
- Gupta, P.; Gupta, R.K.; Gandhi, B.S.; Singh, P. Differential binding of CREB and REST/NRSF to NMDAR1 promoter is associated with the sex-selective cognitive deficit following postnatal PBDE-209 exposure in mice. Environ. Sci. Pollut. Res. Int. 2023. ahead-of-print. [Google Scholar] [CrossRef]
- Gong, P.; Zhang, F.; Lei, X.; Wu, X.; Chen, D.; Zhang, W.; Zhang, K.; Zheng, A.; Gao, X. No observable relationship between the 12 genes of nervous system and reasoning skill in a young Chinese Han population. Cell Mol. Neurobiol. 2011, 31, 519–526. [Google Scholar] [CrossRef]
- Mori, N.; Mizuno, T.; Murai, K.; Nakano, I.; Yamashita, H. Effect of age on the gene expression of neural-restrictive silencing factor NRSF/REST. Neurobiol. Aging 2002, 23, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, A.; Carminati, E.; De Fusco, A.; Kowalska, J.A.; Floss, T.; Benfenati, F. REST/NRSF deficiency impairs autophagy and leads to cellular senescence in neurons. Aging Cell 2021, 20, e13471. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Sun, R.; Yao, M.; Chen, W.; Wang, Z.; Fei, J. A C-terminal truncated mutation of spr-3 gene extends lifespan in Caenorhabditis elegans. Acta Biochim. Biophys. Sin. 2013, 45, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.J.; He, C.F.; Zhou, R.Y.; Xu, X.D.; Xiang, L.X.; Wang, J.T.; Wang, X.R.; Zhou, H.G.; Guo, J.C. High glucose and palmitic acid induces neuronal senescence by NRSF/REST elevation and the subsequent mTOR-related autophagy suppression. Mol. Brain 2022, 15, 61. [Google Scholar] [CrossRef] [PubMed]
- Obata, H.; Naito, K.; Kikui, A.; Nakamura, S.; Suzuki, K.; Kawakita, S.; Suzuki, T.; Goto, K.; Nagura, N.; Sugiyama, Y.; et al. Age-related differences for expression of the nerve-specific proteins after peripheral nerve injury. Exp. Ther. Med. 2022, 24, 682. [Google Scholar] [CrossRef]
- Gonzalez-Castaneda, R.E.; Sanchez-Gonzalez, V.J.; Flores-Soto, M.; Vazquez-Camacho, G.; Macias-Islas, M.A.; Ortiz, G.G. Neural restrictive silencer factor and choline acetyltransferase expression in cerebral tissue of Alzheimer’s Disease patients: A pilot study. Genet. Mol. Biol. 2013, 36, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Orta-Salazar, E.; Aguilar-Vazquez, A.; Martinez-Coria, H.; Luquin-De Anda, S.; Rivera-Cervantes, M.; Beas-Zarate, C.; Feria-Velasco, A.; Diaz-Cintra, S. REST/NRSF-induced changes of ChAT protein expression in the neocortex and hippocampus of the 3xTg-AD mouse model for Alzheimer’s disease. Life Sci. 2014, 116, 83–89. [Google Scholar] [CrossRef]
- Lu, T.; Aron, L.; Zullo, J.; Pan, Y.; Kim, H.; Chen, Y.; Yang, T.H.; Kim, H.M.; Drake, D.; Liu, X.S.; et al. REST and stress resistance in ageing and Alzheimer’s disease. Nature 2014, 507, 448–454. [Google Scholar] [CrossRef]
- Kawamura, M.; Sato, S.; Matsumoto, G.; Fukuda, T.; Shiba-Fukushima, K.; Noda, S.; Takanashi, M.; Mori, N.; Hattori, N. Loss of nuclear REST/NRSF in aged-dopaminergic neurons in Parkinson’s disease patients. Neurosci. Lett. 2019, 699, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Cai, L.; Liang, M.; Huang, Y.; Gao, H.; Lu, S.; Fei, J.; Huang, F. Alteration of NRSF expression exacerbating 1-methyl-4-phenyl-pyridinium ion-induced cell death of SH-SY5Y cells. Neurosci. Res. 2009, 65, 236–244. [Google Scholar] [CrossRef]
- Yu, M.; Suo, H.; Liu, M.; Cai, L.; Liu, J.; Huang, Y.; Xu, J.; Wang, Y.; Zhu, C.; Fei, J.; et al. NRSF/REST neuronal deficient mice are more vulnerable to the neurotoxin MPTP. Neurobiol. Aging 2013, 34, 916–927. [Google Scholar] [CrossRef] [PubMed]
- Suo, H.; Wang, P.; Tong, J.; Cai, L.; Liu, J.; Huang, D.; Huang, L.; Wang, Z.; Huang, Y.; Xu, J.; et al. NRSF is an essential mediator for the neuroprotection of trichostatin A in the MPTP mouse model of Parkinson’s disease. Neuropharmacology 2015, 99, 67–78. [Google Scholar] [CrossRef]
- Huang, D.; Li, Q.; Wang, Y.; Liu, Z.; Wang, Z.; Li, H.; Wang, J.; Su, J.; Ma, Y.; Yu, M.; et al. Brain-specific NRSF deficiency aggravates dopaminergic neurodegeneration and impairs neurogenesis in the MPTP mouse model of Parkinson’s disease. Aging 2019, 11, 3280–3297. [Google Scholar] [CrossRef]
- Li, H.; Liu, Z.; Wu, Y.; Chen, Y.; Wang, J.; Wang, Z.; Huang, D.; Wang, M.; Yu, M.; Fei, J.; et al. The deficiency of NRSF/REST enhances the pro-inflammatory function of astrocytes in a model of Parkinson’s disease. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165590. [Google Scholar] [CrossRef]
- Zuccato, C.; Tartari, M.; Crotti, A.; Goffredo, D.; Valenza, M.; Conti, L.; Cataudella, T.; Leavitt, B.R.; Hayden, M.R.; Timmusk, T.; et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 2003, 35, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Marullo, M.; Valenza, M.; Mariotti, C.; Di Donato, S.; Cattaneo, E.; Zuccato, C. Analysis of the repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy of non-neuronal genes in peripheral lymphocytes from patients with Huntington’s disease. Brain Pathol. 2010, 20, 96–105. [Google Scholar] [CrossRef]
- Schiffer, D.; Caldera, V.; Mellai, M.; Conforti, P.; Cattaneo, E.; Zuccato, C. Repressor element-1 silencing transcription factor (REST) is present in human control and Huntington’s disease neurones. Neuropathol. Appl. Neurobiol. 2014, 40, 899–910. [Google Scholar] [CrossRef] [PubMed]
- Zuccato, C.; Belyaev, N.; Conforti, P.; Ooi, L.; Tartari, M.; Papadimou, E.; MacDonald, M.; Fossale, E.; Zeitlin, S.; Buckley, N.; et al. Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington’s disease. J. Neurosci. 2007, 27, 6972–6983. [Google Scholar] [CrossRef]
- Shimojo, M. Huntingtin regulates RE1-silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) nuclear trafficking indirectly through a complex with REST/NRSF-interacting LIM domain protein (RILP) and dynactin p150 Glued. J. Biol. Chem. 2008, 283, 34880–34886. [Google Scholar] [CrossRef] [PubMed]
- Datta, M.; Bhattacharyya, N.P. Regulation of RE1 protein silencing transcription factor (REST) expression by HIP1 protein interactor (HIPPI). J. Biol. Chem. 2011, 286, 33759–33769. [Google Scholar] [CrossRef]
- Chen, G.L.; Ma, Q.; Goswami, D.; Shang, J.; Miller, G.M. Modulation of nuclear REST by alternative splicing: A potential therapeutic target for Huntington’s disease. J. Cell. Mol. Med. 2017, 21, 2974–2984. [Google Scholar] [CrossRef] [PubMed]
- de Souza, J.M.; Abd-Elrahman, K.S.; Ribeiro, F.M.; Ferguson, S.S.G. mGluR5 regulates REST/NRSF signaling through N-cadherin/beta-catenin complex in Huntington’s disease. Mol. Brain 2020, 13, 118. [Google Scholar] [CrossRef] [PubMed]
- Navarrete-Modesto, V.; Orozco-Suarez, S.; Alonso-Vanegas, M.; Feria-Romero, I.A.; Rocha, L. REST/NRSF transcription factor is overexpressed in hippocampus of patients with drug-resistant mesial temporal lobe epilepsy. Epilepsy Behav. 2019, 94, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.L.; Cheng, X.; Cai, L.; Tan, G.H.; Xu, L.; Feng, X.Y.; Lu, T.J.; Xiong, H.; Fei, J.; Xiong, Z.Q. Conditional deletion of NRSF in forebrain neurons accelerates epileptogenesis in the kindling model. Cereb. Cortex 2011, 21, 2158–2165. [Google Scholar] [CrossRef]
- Liu, M.; Sheng, Z.; Cai, L.; Zhao, K.; Tian, Y.; Fei, J. Neuronal conditional knockout of NRSF decreases vulnerability to seizures induced by pentylenetetrazol in mice. Acta Biochim. Biophys. Sin. 2012, 44, 476–482. [Google Scholar] [CrossRef]
- Patterson, K.P.; Barry, J.M.; Curran, M.M.; Singh-Taylor, A.; Brennan, G.; Rismanchi, N.; Page, M.; Noam, Y.; Holmes, G.L.; Baram, T.Z. Enduring Memory Impairments Provoked by Developmental Febrile Seizures Are Mediated by Functional and Structural Effects of Neuronal Restrictive Silencing Factor. J. Neurosci. 2017, 37, 3799–3812. [Google Scholar] [CrossRef]
- McClelland, S.; Flynn, C.; Dubé, C.; Richichi, C.; Zha, Q.; Ghestem, A.; Esclapez, M.; Bernard, C.; Baram, T.Z. Neuron-restrictive silencer factor-mediated hyperpolarization-activated cyclic nucleotide gated channelopathy in experimental temporal lobe epilepsy. Ann. Neurol. 2011, 70, 454–464. [Google Scholar] [CrossRef]
- Carminati, E.; Buffolo, F.; Rocchi, A.; Michetti, C.; Cesca, F.; Benfenati, F. Mild Inactivation of RE-1 Silencing Transcription Factor (REST) Reduces Susceptibility to Kainic Acid-Induced Seizures. Front. Cell. Neurosci. 2019, 13, 580. [Google Scholar] [CrossRef]
- Hu, X.L.; Cheng, X.; Fei, J.; Xiong, Z.Q. Neuron-restrictive silencer factor is not required for the antiepileptic effect of the ketogenic diet. Epilepsia 2011, 52, 1609–1616. [Google Scholar] [CrossRef]
- Bassuk, A.G.; Wallace, R.H.; Buhr, A.; Buller, A.R.; Afawi, Z.; Shimojo, M.; Miyata, S.; Chen, S.; Gonzalez-Alegre, P.; Griesbach, H.L.; et al. A homozygous mutation in human PRICKLE1 causes an autosomal-recessive progressive myoclonus epilepsy-ataxia syndrome. Am. J. Hum. Genet. 2008, 83, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.P.; Bubb, V.J.; Marshall-Jones, Z.V.; Coulson, J.M. Neuron restrictive silencer factor as a modulator of neuropeptide gene expression. Regul. Pept. 2002, 108, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Spencer, E.M.; Chandler, K.E.; Haddley, K.; Howard, M.R.; Hughes, D.; Belyaev, N.D.; Coulson, J.M.; Stewart, J.P.; Buckley, N.J.; Kipar, A.; et al. Regulation and role of REST and REST4 variants in modulation of gene expression in in vivo and in vitro in epilepsy models. Neurobiol. Dis. 2006, 24, 41–52. [Google Scholar] [CrossRef]
- Gillies, S.; Haddley, K.; Vasiliou, S.; Bubb, V.J.; Quinn, J.P. The human neurokinin B gene, TAC3, and its promoter are regulated by Neuron Restrictive Silencing Factor (NRSF) transcription factor family. Neuropeptides 2009, 43, 333–340. [Google Scholar] [CrossRef]
- Hall, A.M.; Brennan, G.P.; Nguyen, T.M.; Singh-Taylor, A.; Mun, H.S.; Sargious, M.J.; Baram, T.Z. The Role of Sirt1 in Epileptogenesis. Eneuro 2017, preprint. [Google Scholar] [CrossRef] [PubMed]
- Brennan, G.P.; Dey, D.; Chen, Y.; Patterson, K.P.; Magnetta, E.J.; Hall, A.M.; Dube, C.M.; Mei, Y.T.; Baram, T.Z. Dual and Opposing Roles of MicroRNA-124 in Epilepsy Are Mediated through Inflammatory and NRSF-Dependent Gene Networks. Cell Rep. 2016, 14, 2402–2412. [Google Scholar] [CrossRef] [PubMed]
- Calderone, A.; Jover, T.; Noh, K.M.; Tanaka, H.; Yokota, H.; Lin, Y.; Grooms, S.Y.; Regis, R.; Bennett, M.V.; Zukin, R.S. Ischemic insults derepress the gene silencer REST in neurons destined to die. J. Neurosci. 2003, 23, 2112–2121. [Google Scholar] [CrossRef]
- Formisano, L.; Noh, K.M.; Miyawaki, T.; Mashiko, T.; Bennett, M.V.; Zukin, R.S. Ischemic insults promote epigenetic reprogramming of mu opioid receptor expression in hippocampal neurons. Proc. Natl. Acad. Sci. USA 2007, 104, 4170–4175. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.M.; Geng, L.J.; Shi, X.Y.; Zhang, C.G.; Wang, S.Y.; Zhang, G.M. By up-regulating mu- and delta-opioid receptors, neuron-restrictive silencer factor knockdown promotes neurological recovery after ischemia. Oncotarget 2017, 8, 101012–101025. [Google Scholar] [CrossRef] [PubMed]
- He, C.F.; Xue, W.J.; Xu, X.D.; Wang, J.T.; Wang, X.R.; Feng, Y.; Zhou, H.G.; Guo, J.C. Knockdown of NRSF Alleviates Ischemic Brain Injury and Microvasculature Defects in Diabetic MCAO Mice. Front. Neurol. 2022, 13, 869220. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, S.; Yuan, L.; Yang, Y.; Zhang, B.; Liu, Q.; Chen, L.; Yue, W.; Li, Y.; Pei, X. Neuron-restrictive silencer factor (NRSF) represses cocaine- and amphetamine-regulated transcript (CART) transcription and antagonizes cAMP-response element-binding protein signaling through a dual NRSE mechanism. J. Biol. Chem. 2012, 287, 42574–42587. [Google Scholar] [CrossRef]
- Kaneko, N.; Hwang, J.Y.; Gertner, M.; Pontarelli, F.; Zukin, R.S. Casein kinase 1 suppresses activation of REST in insulted hippocampal neurons and halts ischemia-induced neuronal death. J. Neurosci. 2014, 34, 6030–6039. [Google Scholar] [CrossRef]
- Hwang, J.Y.; Kaneko, N.; Noh, K.M.; Pontarelli, F.; Zukin, R.S. The gene silencing transcription factor REST represses miR-132 expression in hippocampal neurons destined to die. J. Mol. Biol. 2014, 426, 3454–3466. [Google Scholar] [CrossRef]
- Yang, B.; Zang, L.E.; Cui, J.W.; Zhang, M.Y.; Ma, X.; Wei, L.L. Melatonin Plays a Protective Role by Regulating miR-26a-5p-NRSF and JAK2-STAT3 Pathway to Improve Autophagy, Inflammation and Oxidative Stress of Cerebral Ischemia-Reperfusion Injury. Drug Des. Dev. Ther. 2020, 14, 3177–3188. [Google Scholar] [CrossRef] [PubMed]
- Loe-Mie, Y.; Lepagnol-Bestel, A.M.; Maussion, G.; Doron-Faigenboim, A.; Imbeaud, S.; Delacroix, H.; Aggerbeck, L.; Pupko, T.; Gorwood, P.; Simonneau, M.; et al. SMARCA2 and other genome-wide supported schizophrenia-associated genes: Regulation by REST/NRSF, network organization and primate-specific evolution. Hum. Mol. Genet. 2010, 19, 2841–2857. [Google Scholar] [CrossRef] [PubMed]
- Gulchina, Y.; Xu, S.J.; Snyder, M.A.; Elefant, F.; Gao, W.J. Epigenetic mechanisms underlying NMDA receptor hypofunction in the prefrontal cortex of juvenile animals in the MAM model for schizophrenia. J. Neurochem. 2017, 143, 320–333. [Google Scholar] [CrossRef]
- Patel, P.D.; Bochar, D.A.; Turner, D.L.; Meng, F.; Mueller, H.M.; Pontrello, C.G. Regulation of tryptophan hydroxylase-2 gene expression by a bipartite RE-1 silencer of transcription/neuron restrictive silencing factor (REST/NRSF) binding motif. J. Biol. Chem. 2007, 282, 26717–26724. [Google Scholar] [CrossRef]
- Ishii, T.; Hashimoto, E.; Ukai, W.; Tateno, M.; Yoshinaga, T.; Saito, S.; Sohma, H.; Saito, T. Lithium-induced suppression of transcription repressor NRSF/REST: Effects on the dysfunction of neuronal differentiation by ethanol. Eur. J. Pharmacol. 2008, 593, 36–43. [Google Scholar] [CrossRef]
- Warburton, A.; Savage, A.L.; Myers, P.; Peeney, D.; Bubb, V.J.; Quinn, J.P. Molecular signatures of mood stabilisers highlight the role of the transcription factor REST/NRSF. J. Affect. Disord. 2015, 172, 63–73. [Google Scholar] [CrossRef]
- Henriksson, R.; Backman, C.M.; Harvey, B.K.; Kadyrova, H.; Bazov, I.; Shippenberg, T.S.; Bakalkin, G. PDYN, a gene implicated in brain/mental disorders, is targeted by REST in the adult human brain. Biochim. Biophys. Acta 2014, 1839, 1226–1232. [Google Scholar] [CrossRef]
- Korosi, A.; Shanabrough, M.; McClelland, S.; Liu, Z.W.; Borok, E.; Gao, X.B.; Horvath, T.L.; Baram, T.Z. Early-life experience reduces excitation to stress-responsive hypothalamic neurons and reprograms the expression of corticotropin-releasing hormone. J. Neurosci. 2010, 30, 703–713. [Google Scholar] [CrossRef]
- Hai-Ying, C.; Nagano, K.; Ezzikouri, S.; Yamaguchi, C.; Kayesh, M.E.; Rebbani, K.; Kitab, B.; Nakano, H.; Kouji, H.; Kohara, M.; et al. Establishment of an intermittent cold stress model using Tupaia belangeri and evaluation of compound C737 targeting neuron-restrictive silencer factor. Exp. Anim. 2016, 65, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Singh-Taylor, A.; Molet, J.; Jiang, S.; Korosi, A.; Bolton, J.L.; Noam, Y.; Simeone, K.; Cope, J.; Chen, Y.; Mortazavi, A.; et al. NRSF-dependent epigenetic mechanisms contribute to programming of stress-sensitive neurons by neonatal experience, promoting resilience. Mol. Psychiatry 2018, 23, 648–657. [Google Scholar] [CrossRef]
- Wen, X.H.; Guo, Q.L.; Guo, J.C. Effect of 9—PAHSA on cognitive dysfunction in diabetic mice and its possible mechanism. Biochem. Biophys. Res. Commun. 2020, 524, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, K.; Saito, Y.; Takano, M.; Arai, Y.; Yasuno, S.; Nakagawa, Y.; Takahashi, N.; Adachi, Y.; Takemura, G.; Horie, M.; et al. NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. EMBO J. 2003, 22, 6310–6321. [Google Scholar] [CrossRef]
- Kuratomi, S.; Kuratomi, A.; Kuwahara, K.; Ishii, T.M.; Nakao, K.; Saito, Y.; Takano, M. NRSF regulates the developmental and hypertrophic changes of HCN4 transcription in rat cardiac myocytes. Biochem. Biophys. Res. Commun. 2007, 353, 67–73. [Google Scholar] [CrossRef]
- Schweizer, P.A.; Yampolsky, P.; Malik, R.; Thomas, D.; Zehelein, J.; Katus, H.A.; Koenen, M. Transcription profiling of HCN-channel isotypes throughout mouse cardiac development. Basic. Res. Cardiol. 2009, 104, 621–629. [Google Scholar] [CrossRef]
- Zhang, D.; Wu, B.; Wang, P.; Wang, Y.; Lu, P.; Nechiporuk, T.; Floss, T.; Greally, J.M.; Zheng, D.; Zhou, B. Non-CpG methylation by DNMT3B facilitates REST binding and gene silencing in developing mouse hearts. Nucleic Acids Res. 2017, 45, 3102–3115. [Google Scholar] [CrossRef]
- Liu, J.; Liu, S.; Gao, H.; Han, L.; Chu, X.; Sheng, Y.; Shou, W.; Wang, Y.; Liu, Y.; Wan, J.; et al. Genome-wide studies reveal the essential and opposite roles of ARID1A in controlling human cardiogenesis and neurogenesis from pluripotent stem cells. Genome Biol. 2020, 21, 169. [Google Scholar] [CrossRef]
- D’Souza, A.; Bucchi, A.; Johnsen, A.B.; Logantha, S.J.; Monfredi, O.; Yanni, J.; Prehar, S.; Hart, G.; Cartwright, E.; Wisloff, U.; et al. Exercise training reduces resting heart rate via downregulation of the funny channel HCN4. Nat. Commun. 2014, 5, 3775. [Google Scholar] [CrossRef]
- Koyama, R.; Mannic, T.; Ito, J.; Amar, L.; Zennaro, M.C.; Rossier, M.F.; Maturana, A.D. MicroRNA-204 Is Necessary for Aldosterone-Stimulated T-Type Calcium Channel Expression in Cardiomyocytes. Int. J. Mol. Sci. 2018, 19, 2941. [Google Scholar] [CrossRef] [PubMed]
- Rossier, M.F. The Cardiac Mineralocorticoid Receptor (MR): A Therapeutic Target Against Ventricular Arrhythmias. Front. Endocrinol. 2021, 12, 694758. [Google Scholar] [CrossRef] [PubMed]
- Somekawa, S.; Imagawa, K.; Naya, N.; Takemoto, Y.; Onoue, K.; Okayama, S.; Takeda, Y.; Kawata, H.; Horii, M.; Nakajima, T.; et al. Regulation of aldosterone and cortisol production by the transcriptional repressor neuron restrictive silencer factor. Endocrinology 2009, 150, 3110–3117. [Google Scholar] [CrossRef] [PubMed]
- Inazumi, H.; Kuwahara, K.; Nakagawa, Y.; Kuwabara, Y.; Numaga-Tomita, T.; Kashihara, T.; Nakada, T.; Kurebayashi, N.; Oya, M.; Nonaka, M.; et al. NRSF-GNAO1 Pathway Contributes to the Regulation of Cardiac Ca2+ Homeostasis. Circ. Res. 2022, 130, 234–248. [Google Scholar] [CrossRef]
- Lugenbiel, P.; Govorov, K.; Syren, P.; Rahm, A.K.; Wieder, T.; Wunsch, M.; Weiberg, N.; Manolova, E.; Gramlich, D.; Rivinius, R.; et al. Epigenetic regulation of cardiac electrophysiology in atrial fibrillation: HDAC2 determines action potential duration and suppresses NRSF in cardiomyocytes. Basic Res. Cardiol. 2021, 116, 13. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, E.; Saito, Y.; Kuwahara, K.; Harada, M.; Miyamoto, Y.; Hamanaka, I.; Kajiyama, N.; Takahashi, N.; Izumi, T.; Kawakami, R.; et al. Fibronectin signaling stimulates BNP gene transcription by inhibiting neuron-restrictive silencer element-dependent repression. Cardiovasc. Res. 2002, 53, 451–459. [Google Scholar] [CrossRef]
- Kuwahara, K.; Saito, Y.; Ogawa, E.; Takahashi, N.; Nakagawa, Y.; Naruse, Y.; Harada, M.; Hamanaka, I.; Izumi, T.; Miyamoto, Y.; et al. The neuron-restrictive silencer element-neuron-restrictive silencer factor system regulates basal and endothelin 1-inducible atrial natriuretic peptide gene expression in ventricular myocytes. Mol. Cell Biol. 2001, 21, 2085–2097. [Google Scholar] [CrossRef]
- Ohba, T.; Watanabe, H.; Takahashi, Y.; Suzuki, T.; Miyoshi, I.; Nakayama, S.; Satoh, E.; Iino, K.; Sasano, H.; Mori, Y.; et al. Regulatory role of neuron-restrictive silencing factor in expression of TRPC1. Biochem. Biophys. Res. Commun. 2006, 351, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Hata, L.; Murakami, M.; Kuwahara, K.; Nakagawa, Y.; Kinoshita, H.; Usami, S.; Yasuno, S.; Fujiwara, M.; Kuwabara, Y.; Minami, T.; et al. Zinc-finger protein 90 negatively regulates neuron-restrictive silencer factor-mediated transcriptional repression of fetal cardiac genes. J. Mol. Cell. Cardiol. 2011, 50, 972–981. [Google Scholar] [CrossRef]
- Atouf, F.; Czernichow, P.; Scharfmann, R. Expression of neuronal traits in pancreatic beta cells. Implication of neuron-restrictive silencing factor/repressor element silencing transcription factor, a neuron-restrictive silencer. J. Biol. Chem. 1997, 272, 1929–1934. [Google Scholar] [CrossRef]
- Martin, D.; Kim, Y.H.; Sever, D.; Mao, C.A.; Haefliger, J.A.; Grapin-Botton, A. REST represses a subset of the pancreatic endocrine differentiation program. Dev. Biol. 2015, 405, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Kemp, D.M.; Lin, J.C.; Habener, J.F. Regulation of Pax4 paired homeodomain gene by neuron-restrictive silencer factor. J. Biol. Chem. 2003, 278, 35057–35062. [Google Scholar] [CrossRef]
- Martin, D.; Tawadros, T.; Meylan, L.; Abderrahmani, A.; Condorelli, D.F.; Waeber, G.; Haefliger, J.A. Critical role of the transcriptional repressor neuron-restrictive silencer factor in the specific control of connexin36 in insulin-producing cell lines. J. Biol. Chem. 2003, 278, 53082–53089. [Google Scholar] [CrossRef]
- Campa, D.; Capurso, G.; Pastore, M.; Talar-Wojnarowska, R.; Milanetto, A.C.; Landoni, L.; Maiello, E.; Lawlor, R.T.; Malecka-Panas, E.; Funel, N.; et al. Common germline variants within the CDKN2A/2B region affect risk of pancreatic neuroendocrine tumors. Sci. Rep. 2016, 6, 39565. [Google Scholar] [CrossRef]
- Li, B.; Wang, S.; Liu, H.; Liu, D.; Zhang, J.; Zhang, B.; Yao, H.; Lv, Y.; Wang, R.; Chen, L.; et al. Neuronal restrictive silencing factor silencing induces human amniotic fluid-derived stem cells differentiation into insulin-producing cells. Stem Cells Dev. 2011, 20, 1223–1231. [Google Scholar] [CrossRef]
- Li, H.T.; Jiang, F.X.; Shi, P.; Zhang, T.; Liu, X.Y.; Lin, X.W.; Pang, X.N. In vitro reprogramming of rat bone marrow-derived mesenchymal stem cells into insulin-producing cells by genetically manipulating negative and positive regulators. Biochem. Biophys. Res. Commun. 2012, 420, 793–798. [Google Scholar] [CrossRef]
- Kurschat, P.; Bielenberg, D.; Rossignol-Tallandier, M.; Stahl, A.; Klagsbrun, M. Neuron restrictive silencer factor NRSF/REST is a transcriptional repressor of neuropilin-1 and diminishes the ability of semaphorin 3A to inhibit keratinocyte migration. J. Biol. Chem. 2006, 281, 2721–2729. [Google Scholar] [CrossRef]
- Aoki, H.; Hara, A.; Kunisada, T. White spotting phenotype induced by targeted REST disruption during neural crest specification to a melanocyte cell lineage. Genes Cells 2015, 20, 439–449. [Google Scholar] [CrossRef]
- Lee, H.J.; Wall, B.A.; Wangari-Talbot, J.; Chen, S. Regulation of mGluR1 expression in human melanocytes and melanoma cells. Biochim. Biophys. Acta 2012, 1819, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Aoki, H.; Ogino, H.; Tomita, H.; Hara, A.; Kunisada, T. Disruption of Rest Leads to the Early Onset of Cataracts with the Aberrant Terminal Differentiation of Lens Fiber Cells. PLoS ONE 2016, 11, e0163042. [Google Scholar] [CrossRef]
- Tsuda, L.; Kaido, M.; Lim, Y.M.; Kato, K.; Aigaki, T.; Hayashi, S. An NRSF/REST-like repressor downstream of Ebi/SMRTER/Su(H) regulates eye development in Drosophila. EMBO J. 2006, 25, 3191–3202. [Google Scholar] [CrossRef]
- Cheong, A.; Bingham, A.J.; Li, J.; Kumar, B.; Sukumar, P.; Munsch, C.; Buckley, N.J.; Neylon, C.B.; Porter, K.E.; Beech, D.J.; et al. Downregulated REST transcription factor is a switch enabling critical potassium channel expression and cell proliferation. Mol. Cell 2005, 20, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Tharp, D.L.; Bowles, D.K. KCa3.1 Inhibition Decreases Size and Alters Composition of Atherosclerotic Lesions Induced by Low, Oscillatory Flow. Artery Res. 2021, 27, 93–100. [Google Scholar] [CrossRef]
- Nishimura, E.; Sasaki, K.; Maruyama, K.; Tsukada, T.; Yamaguchi, K. Decrease in neuron-restrictive silencer factor (NRSF) mRNA levels during differentiation of cultured neuroblastoma cells. Neurosci. Lett. 1996, 211, 101–104. [Google Scholar] [CrossRef]
- Lepagnol-Bestel, A.M.; Maussion, G.; Ramoz, N.; Moalic, J.M.; Gorwood, P.; Simonneau, M. Nrsf silencing induces molecular and subcellular changes linked to neuronal plasticity. NeuroReport 2007, 18, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.K.; Mitra, S.; Subhash, S.; Hertwig, F.; Kanduri, M.; Mishra, K.; Fransson, S.; Ganeshram, A.; Mondal, T.; Bandaru, S.; et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell 2014, 26, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Conti, L.; Crisafulli, L.; Caldera, V.; Tortoreto, M.; Brilli, E.; Conforti, P.; Zunino, F.; Magrassi, L.; Schiffer, D.; Cattaneo, E. REST controls self-renewal and tumorigenic competence of human glioblastoma cells. PLoS ONE 2012, 7, e38486. [Google Scholar] [CrossRef]
- Ren, H.; Gao, Z.; Wu, N.; Zeng, L.; Tang, X.; Chen, X.; Liu, Z.; Zhang, W.; Wang, L.; Li, Z. Expression of REST4 in human gliomas in vivo and influence of pioglitazone on REST in vitro. Biochem. Biophys. Res. Commun. 2015, 463, 504–509. [Google Scholar] [CrossRef]
- Dobson, T.H.W.; Tao, R.H.; Swaminathan, J.; Maegawa, S.; Shaik, S.; Bravo-Alegria, J.; Sharma, A.; Kennis, B.; Yang, Y.; Callegari, K.; et al. Transcriptional repressor REST drives lineage stage-specific chromatin compaction at Ptch1 and increases AKT activation in a mouse model of medulloblastoma. Sci. Signal 2019, 12, eaan8680. [Google Scholar] [CrossRef]
- Su, X.; Gopalakrishnan, V.; Stearns, D.; Aldape, K.; Lang, F.F.; Fuller, G.; Snyder, E.; Eberhart, C.G.; Majumder, S. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol. Cell. Biol. 2006, 26, 1666–1678. [Google Scholar] [CrossRef]
- Lawinger, P.; Venugopal, R.; Guo, Z.S.; Immaneni, A.; Sengupta, D.; Lu, W.; Rastelli, L.; Marin Dias Carneiro, A.; Levin, V.; Fuller, G.N.; et al. The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nat. Med. 2000, 6, 826–831. [Google Scholar] [CrossRef]
- Fuller, G.N.; Su, X.; Price, R.E.; Cohen, Z.R.; Lang, F.F.; Sawaya, R.; Majumder, S. Many human medulloblastoma tumors overexpress repressor element-1 silencing transcription (REST)/neuron-restrictive silencer factor, which can be functionally countered by REST-VP16. Mol. Cancer Ther. 2005, 4, 343–349. [Google Scholar] [CrossRef]
- Callegari, K.; Maegawa, S.; Bravo-Alegria, J.; Gopalakrishnan, V. Pharmacological inhibition of LSD1 activity blocks REST-dependent medulloblastoma cell migration. Cell Commun. Signal 2018, 16, 60. [Google Scholar] [CrossRef]
- Shaik, S.; Maegawa, S.; Haltom, A.R.; Wang, F.; Xiao, X.; Dobson, T.; Sharma, A.; Yang, Y.; Swaminathan, J.; Kundra, V.; et al. REST promotes ETS1-dependent vascular growth in medulloblastoma. Mol. Oncol. 2021, 15, 1486–1506. [Google Scholar] [CrossRef]
- Palm, K.; Metsis, M.; Timmusk, T. Neuron-specific splicing of zinc finger transcription factor REST/NRSF/XBR is frequent in neuroblastomas and conserved in human, mouse and rat. Brain Res. Mol. Brain Res. 1999, 72, 30–39. [Google Scholar] [CrossRef]
- Blom, T.; Tynninen, O.; Puputti, M.; Halonen, M.; Paetau, A.; Haapasalo, H.; Tanner, M.; Nupponen, N.N. Molecular genetic analysis of the REST/NRSF gene in nervous system tumors. Acta Neuropathol. 2006, 112, 483–490. [Google Scholar] [CrossRef]
- Lee, J.H.; Chai, Y.G.; Hersh, L.B. Expression patterns of mouse repressor element-1 silencing transcription factor 4 (REST4) and its possible function in neuroblastoma. J. Mol. Neurosci. 2000, 15, 205–214. [Google Scholar] [CrossRef]
- Coulson, J.M.; Edgson, J.L.; Woll, P.J.; Quinn, J.P. A splice variant of the neuron-restrictive silencer factor repressor is expressed in small cell lung cancer: A potential role in derepression of neuroendocrine genes and a useful clinical marker. Cancer Res. 2000, 60, 1840–1844. [Google Scholar] [PubMed]
- Gurrola-Diaz, C.; Lacroix, J.; Dihlmann, S.; Becker, C.M.; von Knebel Doeberitz, M. Reduced expression of the neuron restrictive silencer factor permits transcription of glycine receptor alpha1 subunit in small-cell lung cancer cells. Oncogene 2003, 22, 5636–5645. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Neumann, S.B.; Seitz, R.; Gorzella, A.; Heister, A.; Doeberitz, M.; Becker, C.M. Relaxation of glycine receptor and onconeural gene transcription control in NRSF deficient small cell lung cancer cell lines. Brain Res. Mol. Brain Res. 2004, 120, 173–181. [Google Scholar] [CrossRef]
- Chang, L.; Schwarzenbach, H.; Meyer-Staeckling, S.; Brandt, B.; Mayr, G.W.; Weitzel, J.M.; Windhorst, S. Expression Regulation of the Metastasis-Promoting Protein InsP3-Kinase-A in Tumor Cells. Mol. Cancer Res. 2011, 9, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Kreisler, A.; Strissel, P.L.; Strick, R.; Neumann, S.B.; Schumacher, U.; Becker, C.M. Regulation of the NRSF/REST gene by methylation and CREB affects the cellular phenotype of small-cell lung cancer. Oncogene 2010, 29, 5828–5838. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Oda, C.; Mishima, K.; Tsuji, I.; Obika, S.; Shimojo, M. An antisense amido-bridged nucleic acid gapmer oligonucleotide targeting SRRM4 alters REST splicing and exhibits anti-tumor effects in small cell lung cancer and prostate cancer cells. Cancer Cell Int. 2023, 23, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Xu, K.; Leng, X.; Han, Y.; Fang, Q. miRNA-9 Inhibits Proliferation and Migration of Lung Squamous Cell Carcinoma Cells by Regulating NRSF/EGFR. Technol. Cancer Res. Treat. 2020, 19, 1533033820945807. [Google Scholar] [CrossRef]
- Lim, J.S.; Ibaseta, A.; Fischer, M.M.; Cancilla, B.; O’Young, G.; Cristea, S.; Luca, V.C.; Yang, D.; Jahchan, N.S.; Hamard, C.; et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 2017, 545, 360–364. [Google Scholar] [CrossRef]
- Moss, A.C.; Jacobson, G.M.; Walker, L.E.; Blake, N.W.; Marshall, E.; Coulson, J.M. SCG3 transcript in peripheral blood is a prognostic biomarker for REST-deficient small cell lung cancer. Clin. Cancer Res. 2009, 15, 274–283. [Google Scholar] [CrossRef]
- Watanabe, H.; Mizutani, T.; Haraguchi, T.; Yamamichi, N.; Minoguchi, S.; Yamamichi-Nishina, M.; Mori, N.; Kameda, T.; Sugiyama, T.; Iba, H. SWI/SNF complex is essential for NRSF-mediated suppression of neuronal genes in human nonsmall cell lung carcinoma cell lines. Oncogene 2006, 25, 470–479. [Google Scholar] [CrossRef][Green Version]
- Nair, S.; Bora-Singhal, N.; Perumal, D.; Chellappan, S. Nicotine-mediated invasion and migration of non-small cell lung carcinoma cells by modulating STMN3 and GSPT1 genes in an ID1-dependent manner. Mol. Cancer 2014, 13, 173. [Google Scholar] [CrossRef] [PubMed]
- Westbrook, T.F.; Martin, E.S.; Schlabach, M.R.; Leng, Y.; Liang, A.C.; Feng, B.; Zhao, J.J.; Roberts, T.M.; Mandel, G.; Hannon, G.J.; et al. A genetic screen for candidate tumor suppressors identifies REST. Cell 2005, 121, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, X. HMGA2 rs968697 T > C polymorphism is associated with the risk of colorectal cancer. Nucleosides Nucleotides Nucleic Acids 2021, 40, 821–828. [Google Scholar] [CrossRef]
- Ma, M.; Zhou, Y.; Sun, R.; Shi, J.; Tan, Y.; Yang, H.; Zhang, M.; Shen, R.; Xu, L.; Wang, Z.; et al. STAT3 and AKT signaling pathways mediate oncogenic role of NRSF in hepatocellular carcinoma. Acta Biochim. Biophys. Sin. 2020, 52, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Li, S.; Zhang, H.; Zhang, X.; Guo, D.; Zhang, J. NRSF/REST levels are decreased in cholangiocellular carcinoma but not hepatocellular carcinoma compared with normal liver tissues: A tissue microarray study. Oncol. Lett. 2018, 15, 6592–6598. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Pan, G.; Zheng, G.; Wu, X.; Ren, H.; Liu, Y.; Wen, J. Expression and functions of the repressor element 1 (RE-1)-silencing transcription factor (REST) in breast cancer. J. Cell. Biochem. 2010, 110, 968–974. [Google Scholar] [CrossRef]
- Bronson, M.W.; Hillenmeyer, S.; Park, R.W.; Brodsky, A.S. Estrogen coordinates translation and transcription, revealing a role for NRSF in human breast cancer cells. Mol. Endocrinol. 2010, 24, 1120–1135. [Google Scholar] [CrossRef][Green Version]
- Sankar, S.; Gomez, N.C.; Bell, R.; Patel, M.; Davis, I.J.; Lessnick, S.L.; Luo, W. EWS and RE1-Silencing Transcription Factor Inhibit Neuronal Phenotype Development and Oncogenic Transformation in Ewing Sarcoma. Genes Cancer 2013, 4, 213–223. [Google Scholar] [CrossRef]
- Varghese, B.V.; Koohestani, F.; McWilliams, M.; Colvin, A.; Gunewardena, S.; Kinsey, W.H.; Nowak, R.A.; Nothnick, W.B.; Chennathukuzhi, V.M. Loss of the repressor REST in uterine fibroids promotes aberrant G protein-coupled receptor 10 expression and activates mammalian target of rapamycin pathway. Proc. Natl. Acad. Sci. USA 2013, 110, 2187–2192. [Google Scholar] [CrossRef]
- Cho, E.; Moon, S.M.; Park, B.R.; Kim, D.K.; Lee, B.K.; Kim, C.S. NRSF/REST regulates the mTOR signaling pathway in oral cancer cells. Oncol. Rep. 2015, 33, 1459–1464. [Google Scholar] [CrossRef]
- Tawadros, T.; Martin, D.; Abderrahmani, A.; Leisinger, H.J.; Waeber, G.; Haefliger, J.A. IB1/JIP-1 controls JNK activation and increased during prostatic LNCaP cells neuroendocrine differentiation. Cell Signal 2005, 17, 929–939. [Google Scholar] [CrossRef]
- Cortes-Sarabia, K.; Alarcon-Romero, L.D.C.; Mendoza-Catalan, M.A.; Carpio-Pedroza, J.C.; Castaneda-Saucedo, E.; Ortuno-Pineda, C. REST/NRSF Silencing Modifies Neuronal Gene Expression in siRNA-Treated HeLa Cells: A Preliminary Exploration in the Search for Neuronal Biomarkers of Cervical Cancer. Medicina 2023, 59, 537. [Google Scholar] [CrossRef]
- Zhang, D.; Li, Y.; Wang, R.; Li, Y.; Shi, P.; Kan, Z.; Pang, X. Inhibition of REST Suppresses Proliferation and Migration in Glioblastoma Cells. Int. J. Mol. Sci. 2016, 17, 664. [Google Scholar] [CrossRef]
- Deng, P.; Zuo, Y.; Feng, S.; Li, Z.; Chen, W.; Li, H.; Wang, X. Knockdown of NRSF inhibits cell proliferation of ovarian cancer via activating Hippo pathway. Life Sci. 2018, 215, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Degirmenci, V.; Xin, H.; Li, Y.; Wang, L.; Chen, J.; Hu, X.; Zhang, D. PEI-Coated Fe3O4 Nanoparticles Enable Efficient Delivery of Therapeutic siRNA Targeting REST into Glioblastoma Cells. Int. J. Mol. Sci. 2018, 19, 2230. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, D.; Zhang, T.; Zhao, F.; Lang, H.; Lin, X.; Pang, X. The differentiation of human MSCs derived from adipose and amniotic tissues into insulin-producing cells, induced by PEI@Fe3O4 nanoparticles-mediated NRSF and SHH silencing. Int. J. Mol. Med. 2018, 42, 2831–2838. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, T.; Hsieh, J.; Nakashima, K.; Taira, K.; Gage, F.H. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 2004, 116, 779–793. [Google Scholar] [CrossRef]
- Kuwabara, T.; Hsieh, J.; Nakashima, K.; Warashina, M.; Taira, K.; Gage, F.H. The NRSE smRNA specifies the fate of adult hippocampal neural stem cells. Nucleic Acids Symp. Ser. 2005, 49, 87–88. [Google Scholar] [CrossRef]
- Immaneni, A.; Lawinger, P.; Zhao, Z.; Lu, W.; Rastelli, L.; Morris, J.H.; Majumder, S. REST-VP16 activates multiple neuronal differentiation genes in human NT2 cells. Nucleic Acids Res. 2000, 28, 3403–3410. [Google Scholar] [CrossRef][Green Version]
- Watanabe, Y.; Kameoka, S.; Gopalakrishnan, V.; Aldape, K.D.; Pan, Z.Z.; Lang, F.F.; Majumder, S. Conversion of myoblasts to physiologically active neuronal phenotype. Genes Dev. 2004, 18, 889–900. [Google Scholar] [CrossRef]
- Ueda, H.; Kurita, J.I.; Neyama, H.; Hirao, Y.; Kouji, H.; Mishina, T.; Kasai, M.; Nakano, H.; Yoshimori, A.; Nishimura, Y. A mimetic of the mSin3-binding helix of NRSF/REST ameliorates abnormal pain behavior in chronic pain models. Bioorg. Med. Chem. Lett. 2017, 27, 4705–4709. [Google Scholar] [CrossRef]
- Kawase, H.; Ago, Y.; Naito, M.; Higuchi, M.; Hara, Y.; Hasebe, S.; Tsukada, S.; Kasai, A.; Nakazawa, T.; Mishina, T.; et al. mS-11, a mimetic of the mSin3-binding helix in NRSF, ameliorates social interaction deficits in a prenatal valproic acid-induced autism mouse model. Pharmacol. Biochem. Behav. 2019, 176, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Higo, J.; Takashima, H.; Fukunishi, Y.; Yoshimori, A. Generalized-ensemble method study: A helix-mimetic compound inhibits protein-protein interaction by long-range and short-range intermolecular interactions. J. Comput. Chem. 2021, 42, 956–969. [Google Scholar] [CrossRef]
- Conforti, P.; Zuccato, C.; Gaudenzi, G.; Ieraci, A.; Camnasio, S.; Buckley, N.J.; Mutti, C.; Cotelli, F.; Contini, A.; Cattaneo, E. Binding of the repressor complex REST-mSIN3b by small molecules restores neuronal gene transcription in Huntington’s disease models. J. Neurochem. 2013, 127, 22–35. [Google Scholar] [CrossRef]
- Kurita, J.I.; Hirao, Y.; Nakano, H.; Fukunishi, Y.; Nishimura, Y. Sertraline, chlorprothixene, and chlorpromazine characteristically interact with the REST-binding site of the corepressor mSin3, showing medulloblastoma cell growth inhibitory activities. Sci. Rep. 2018, 8, 13763. [Google Scholar] [CrossRef]
- Leone, S.; Mutti, C.; Kazantsev, A.; Sturlese, M.; Moro, S.; Cattaneo, E.; Rigamonti, D.; Contini, A. SAR and QSAR study on 2-aminothiazole derivatives, modulators of transcriptional repression in Huntington’s disease. Bioorg. Med. Chem. 2008, 16, 5695–5703. [Google Scholar] [CrossRef] [PubMed]
- Charbord, J.; Poydenot, P.; Bonnefond, C.; Feyeux, M.; Casagrande, F.; Brinon, B.; Francelle, L.; Auregan, G.; Guillermier, M.; Cailleret, M.; et al. High throughput screening for inhibitors of REST in neural derivatives of human embryonic stem cells reveals a chemical compound that promotes expression of neuronal genes. Stem Cells 2013, 31, 1816–1828. [Google Scholar] [CrossRef] [PubMed]
- Paonessa, F.; Criscuolo, S.; Sacchetti, S.; Amoroso, D.; Scarongella, H.; Pecoraro Bisogni, F.; Carminati, E.; Pruzzo, G.; Maragliano, L.; Cesca, F.; et al. Regulation of neural gene transcription by optogenetic inhibition of the RE1-silencing transcription factor. Proc. Natl. Acad. Sci. USA 2016, 113, E91–E100. [Google Scholar] [CrossRef]
- Criscuolo, S.; Gatti Iou, M.; Merolla, A.; Maragliano, L.; Cesca, F.; Benfenati, F. Engineering REST-Specific Synthetic PUF Proteins to Control Neuronal Gene Expression: A Combined Experimental and Computational Study. ACS Synth. Biol. 2020, 9, 2039–2054. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, L.; Liu, Y.; Wu, Y.; Huang, Y.; Zhang, D. REST Is Not Resting: REST/NRSF in Health and Disease. Biomolecules 2023, 13, 1477. https://doi.org/10.3390/biom13101477
Jin L, Liu Y, Wu Y, Huang Y, Zhang D. REST Is Not Resting: REST/NRSF in Health and Disease. Biomolecules. 2023; 13(10):1477. https://doi.org/10.3390/biom13101477
Chicago/Turabian StyleJin, Lili, Ying Liu, Yifan Wu, Yi Huang, and Dianbao Zhang. 2023. "REST Is Not Resting: REST/NRSF in Health and Disease" Biomolecules 13, no. 10: 1477. https://doi.org/10.3390/biom13101477
APA StyleJin, L., Liu, Y., Wu, Y., Huang, Y., & Zhang, D. (2023). REST Is Not Resting: REST/NRSF in Health and Disease. Biomolecules, 13(10), 1477. https://doi.org/10.3390/biom13101477






