CISD1 Is a Breast Cancer Prognostic Biomarker Associated with Diabetes Mellitus
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Histological Verification of CISD1 Expression
2.3. Correlation Analysis of CISD1 Expression and Clinical Features
2.4. Correlation Analysis of CISD1 Expression and Immune Infiltration
2.5. Protein-Protein, Chemical-Protein Interaction Analysis
2.6. Gene Ontology (GO) Enrichment Analysis
2.7. Transcription Regulating Prediction
2.8. Statistical Analysis
3. Results
3.1. High Expression Level of CISD1 in BRCA Tumor Correlates with Poor Survival Probability
3.2. Correlation between CISD1 Expression and Clinical Features
3.3. The Correlation of CISD1 and Immune Cell Infiltration
3.4. Co-Expression Genes, Protein-Protein Interaction, and Compound-Binding Analysis
3.5. Enrichment Analysis of Shared Genes between the Co-Expressed Genes with CISD1 and Diabetes Mellitus
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Deprey, K.; Batistatou, N.; Kritzer, J.A. A critical analysis of methods used to investigate the cellular uptake and subcellular localization of RNA therapeutics. Nucleic Acids. Res. 2020, 48, 7623–7639. [Google Scholar] [CrossRef] [PubMed]
- Pack, L.R.; Daigh, L.H.; Chung, M.; Meyer, T. Clinical CDK4/6 inhibitors induce selective and immediate dissociation of p21 from cyclin D-CDK4 to inhibit CDK2. Nat. Commun. 2021, 12, 3356. [Google Scholar] [CrossRef] [PubMed]
- Lohard, S.; Bourgeois, N.; Maillet, L.; Gautier, F.; Fétiveau, A.; Lasla, H.; Nguyen, F.; Vuillier, C.; Dumont, A.; Moreau-Aubry, A.; et al. STING-dependent paracriny shapes apoptotic priming of breast tumors in response to anti-mitotic treatment. Nat. Commun. 2020, 11, 259. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, A.E.; Goodwin, P.J. Diabetes, metformin and breast cancer: A tangled web. Ann. Oncol. 2021, 32, 285–286. [Google Scholar] [CrossRef]
- Goldberg, I.J. Clinical review 124: Diabetic dyslipidemia: Causes and consequences. J. Clin. Endocrinol. Metab. 2001, 86, 965–971. [Google Scholar] [CrossRef]
- Cerf, M.E. Beta cell dysfunction and insulin resistance. Front. Endocrinol. 2013, 4, 37. [Google Scholar] [CrossRef]
- Bodmer, M.; Meier, C.; Krähenbühl, S.; Jick, S.S.; Meier, C.R. Long-Term Metformin Use Is Associated with Decreased Risk of Breast Cancer. Diabetes Care 2010, 33, 1304–1308. [Google Scholar] [CrossRef]
- Chiarelli, F.; Di Marzio, D. Peroxisome proliferator-activated receptor-gamma agonists and diabetes: Current evidence and future perspectives. Vasc. Health Risk Manag. 2008, 4, 297–304. [Google Scholar]
- Monami, M.; Lamanna, C.; Marchionni, N.; Mannucci, E. Rosiglitazone and risk of cancer: A meta-analysis of randomized clinical trials. Diabetes Care 2008, 31, 1455–1460. [Google Scholar] [CrossRef]
- Koro, C.; Barrett, S.; Qizilbash, N. Cancer risks in thiazolidinedione users compared to other anti-diabetic agents. Pharmacoepidemiol. Drug Saf. 2007, 16, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Lin, L.; Cheng, D.; Xu, Y.; Xu, M.; Chen, Y.; Wang, W.; Bi, Y.; Li, D.; Lu, J. Thiazolidinedione therapy and breast cancer risk in diabetic women: A systematic review and meta-analysis. Diabetes Metab. Res. Rev. 2018, 34, e2961. [Google Scholar] [CrossRef] [PubMed]
- Turchi, R.; Faraonio, R.; Lettieri-Barbato, D.; Aquilano, K. An Overview of the Ferroptosis Hallmarks in Friedreich′s Ataxia. Biomolecules 2020, 10, 1489. [Google Scholar] [CrossRef]
- Li, J.; Cao, F.; Yin, H.L.; Huang, Z.J.; Lin, Z.T.; Mao, N.; Sun, B.; Wang, G. Ferroptosis: Past, present and future. Cell Death Dis. 2020, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Darash-Yahana, M.; Sohn, Y.S.; Bai, F.; Song, L.; Cabantchik, I.Z.; Jennings, P.A.; Onuchic, J.N.; Nechushtai, R. NEET Proteins: A New Link Between Iron Metabolism, Reactive Oxygen Species, and Cancer. Antioxid. Redox. Signal. 2019, 30, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Wiley, S.E.; Paddock, M.L.; Abresch, E.C.; Gross, L.; van der Geer, P.; Nechushtai, R.; Murphy, A.N.; Jennings, P.A.; Dixon, J.E. The outer mitochondrial membrane protein mitoNEET contains a novel redox-active 2Fe-2S cluster. J. Biol. Chem. 2007, 282, 23745–23749. [Google Scholar] [CrossRef]
- Sohn, Y.S.; Tamir, S.; Song, L.; Michaeli, D.; Matouk, I.; Conlan, A.R.; Harir, Y.; Holt, S.H.; Shulaev, V.; Paddock, M.L.; et al. NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth. Proc. Natl. Acad. Sci. USA 2013, 110, 14676–14681. [Google Scholar] [CrossRef]
- Corona, J.C.; Duchen, M.R. PPARgamma and PGC-1alpha as therapeutic targets in Parkinson′s. Neurochem. Res. 2015, 40, 308–316. [Google Scholar] [CrossRef]
- Landry, A.P.; Cheng, Z.; Ding, H. Reduction of mitochondrial protein mitoNEET [2Fe-2S] clusters by human glutathione reductase. Free Radic. Biol. Med. 2015, 81, 119–127. [Google Scholar] [CrossRef]
- Takahashi, T.; Yamamoto, M.; Amikura, K.; Kato, K.; Serizawa, T.; Serizawa, K.; Akazawa, D.; Aoki, T.; Kawai, K.; Ogasawara, E.; et al. A novel MitoNEET ligand, TT01001, improves diabetes and ameliorates mitochondrial function in db/db mice. J. Pharmacol. Exp. Ther. 2015, 352, 338–345. [Google Scholar] [CrossRef]
- Hsiung, K.C.; Liu, K.Y.; Tsai, T.F.; Yoshina, S.; Mitani, S.; Chin-Ming Tan, B.; Lo, S.J. Defects in CISD-1, a mitochondrial iron-sulfur protein, lower glucose level and ATP production in Caenorhabditis elegans. Biomed. J. 2020, 43, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Yevshin, I.; Sharipov, R.; Kolmykov, S.; Kondrakhin, Y.; Kolpakov, F. GTRD: A database on gene transcription regulation-2019 update. Nucleic Acids Res. 2019, 47, D100–D105. [Google Scholar] [CrossRef] [PubMed]
- Geldenhuys, W.J.; Piktel, D.; Moore, J.C.; Rellick, S.L.; Meadows, E.; Pinti, M.V.; Hollander, J.M.; Ammer, A.G.; Martin, K.H.; Gibson, L.F. Loss of the redox mitochondrial protein mitoNEET leads to mitochondrial dysfunction in B-cell acute lymphoblastic leukemia. Free Radic. Biol. Med. 2021, 175, 226–235. [Google Scholar] [CrossRef]
- Furihata, T.; Takada, S.; Kakutani, N.; Maekawa, S.; Tsuda, M.; Matsumoto, J.; Mizushima, W.; Fukushima, A.; Yokota, T.; Enzan, N.; et al. Cardiac-specific loss of mitoNEET expression is linked with age-related heart failure. Commun. Biol. 2021, 4, 138. [Google Scholar] [CrossRef]
- Geldenhuys, W.J.; Benkovic, S.A.; Lin, L.; Yonutas, H.M.; Crish, S.D.; Sullivan, P.G.; Darvesh, A.S.; Brown, C.M.; Richardson, J.R. MitoNEET (CISD1) Knockout Mice Show Signs of Striatal Mitochondrial Dysfunction and a Parkinson′s Disease Phenotype. ACS Chem. Neurosci. 2017, 8, 2759–2765. [Google Scholar] [CrossRef]
- Hua, J.; Gao, Z.; Zhong, S.; Wei, B.; Zhu, J.; Ying, R. CISD1 protects against atherosclerosis by suppressing lipid accumulation and inflammation via mediating Drp1. Biochem. Biophys. Res. Commun. 2021, 577, 80–88. [Google Scholar] [CrossRef]
- Zhang, Z.B.; Xiong, L.L.; Xue, L.L.; Deng, Y.P.; Du, R.L.; Hu, Q.; Xu, Y.; Yang, S.J.; Wang, T.H. MiR-127-3p targeting CISD1 regulates autophagy in hypoxic-ischemic cortex. Cell Death Dis. 2021, 12, 279. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Li, X.; Zhang, X.; Kang, R.; Tang, D. CISD1 inhibits ferroptosis by protection against mitochondrial lipid peroxidation. Biochem. Biophys. Res. Commun. 2016, 478, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Geldenhuys, W.J.; Long, T.E.; Saralkar, P.; Iwasaki, T.; Nunez, R.A.A.; Nair, R.R.; Konkle, M.E.; Menze, M.A.; Pinti, M.V.; Hollander, J.M.; et al. Crystal structure of the mitochondrial protein mitoNEET bound to a benze-sulfonide ligand. Commun. Chem. 2019, 2, 77. [Google Scholar] [CrossRef] [PubMed]
- Eketunde, A.O. Diabetes as a Risk Factor for Breast Cancer. Cureus 2020, 12, e8010. [Google Scholar] [CrossRef]
- Keller, M.P.; Choi, Y.; Wang, P.; Davis, D.B.; Rabaglia, M.E.; Oler, A.T.; Stapleton, D.S.; Argmann, C.; Schueler, K.L.; Edwards, S.; et al. A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res. 2008, 18, 706–716. [Google Scholar] [CrossRef]
- Zeng, C.; Mulas, F.; Sui, Y.; Guan, T.; Miller, N.; Tan, Y.; Liu, F.; Jin, W.; Carrano, A.C.; Huising, M.O.; et al. Pseudotemporal Ordering of Single Cells Reveals Metabolic Control of Postnatal β Cell Proliferation. Cell Metab. 2017, 25, 1160–1175. [Google Scholar] [CrossRef]
- Rahman, I.; Athar, M.T.; Islam, M. Type 2 Diabetes, Obesity, and Cancer Share Some Common and Critical Pathways. Front. Oncol. 2020, 10, 600824. [Google Scholar] [CrossRef]
- Marjault, H.B.; Karmi, O.; Zuo, K.; Michaeli, D.; Eisenberg-Domovich, Y.; Rossetti, G.; de Chassey, B.; Vonderscher, J.; Cabantchik, I.; Carloni, P.; et al. An anti-diabetic drug targets NEET (CISD) proteins through destabilization of their [2Fe-2S] clusters. Commun. Biol. 2022, 5, 437. [Google Scholar] [CrossRef]
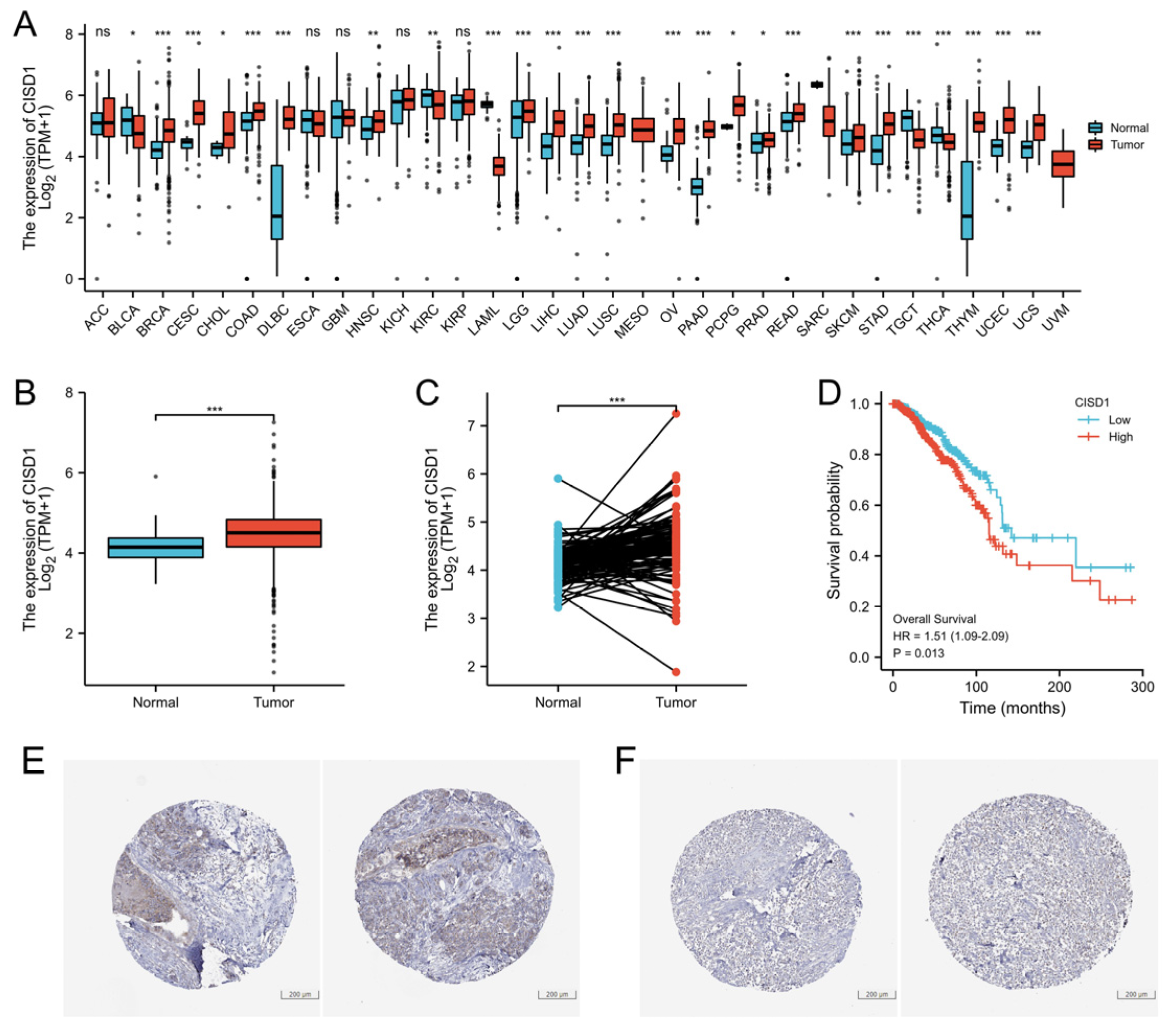
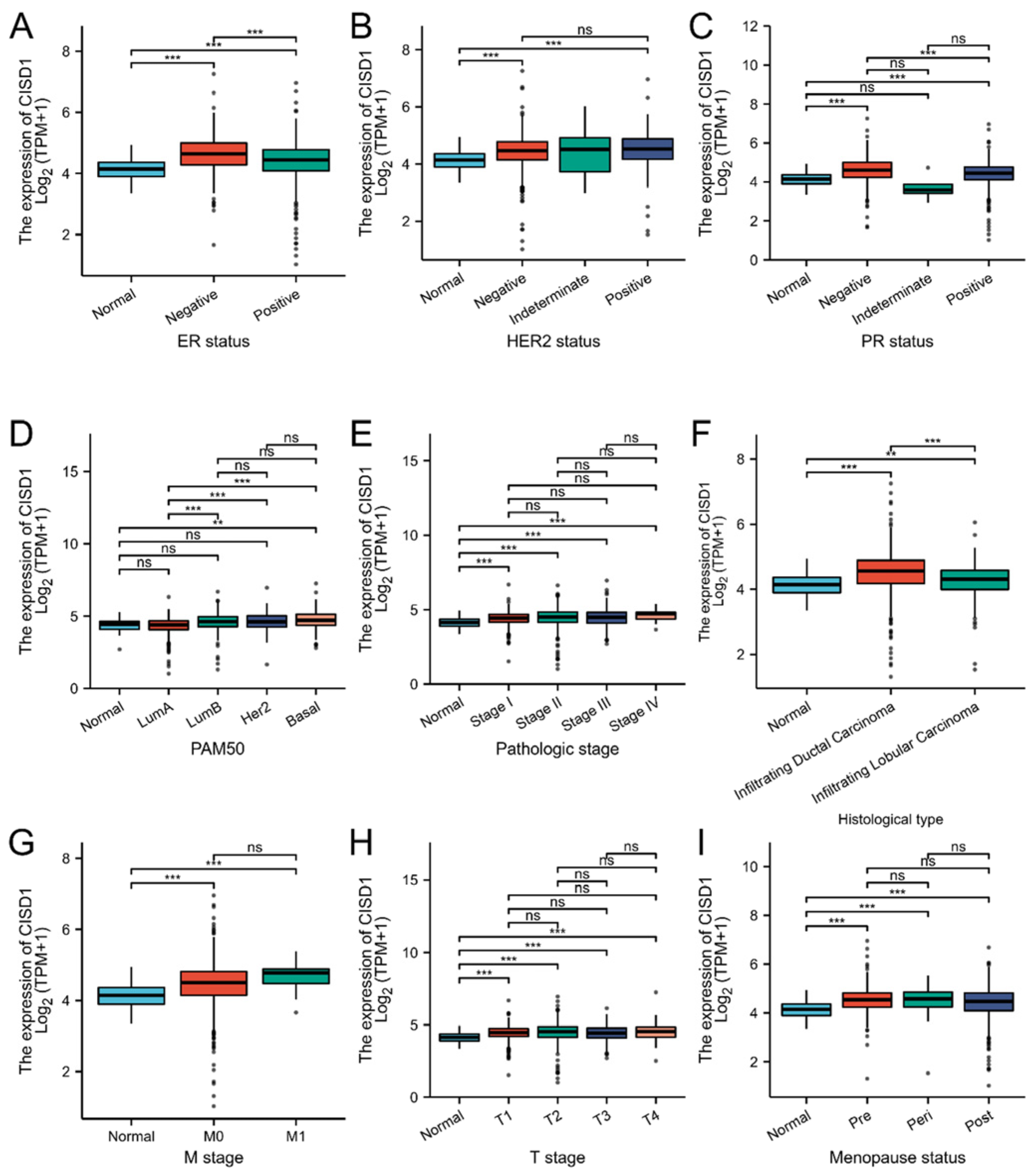
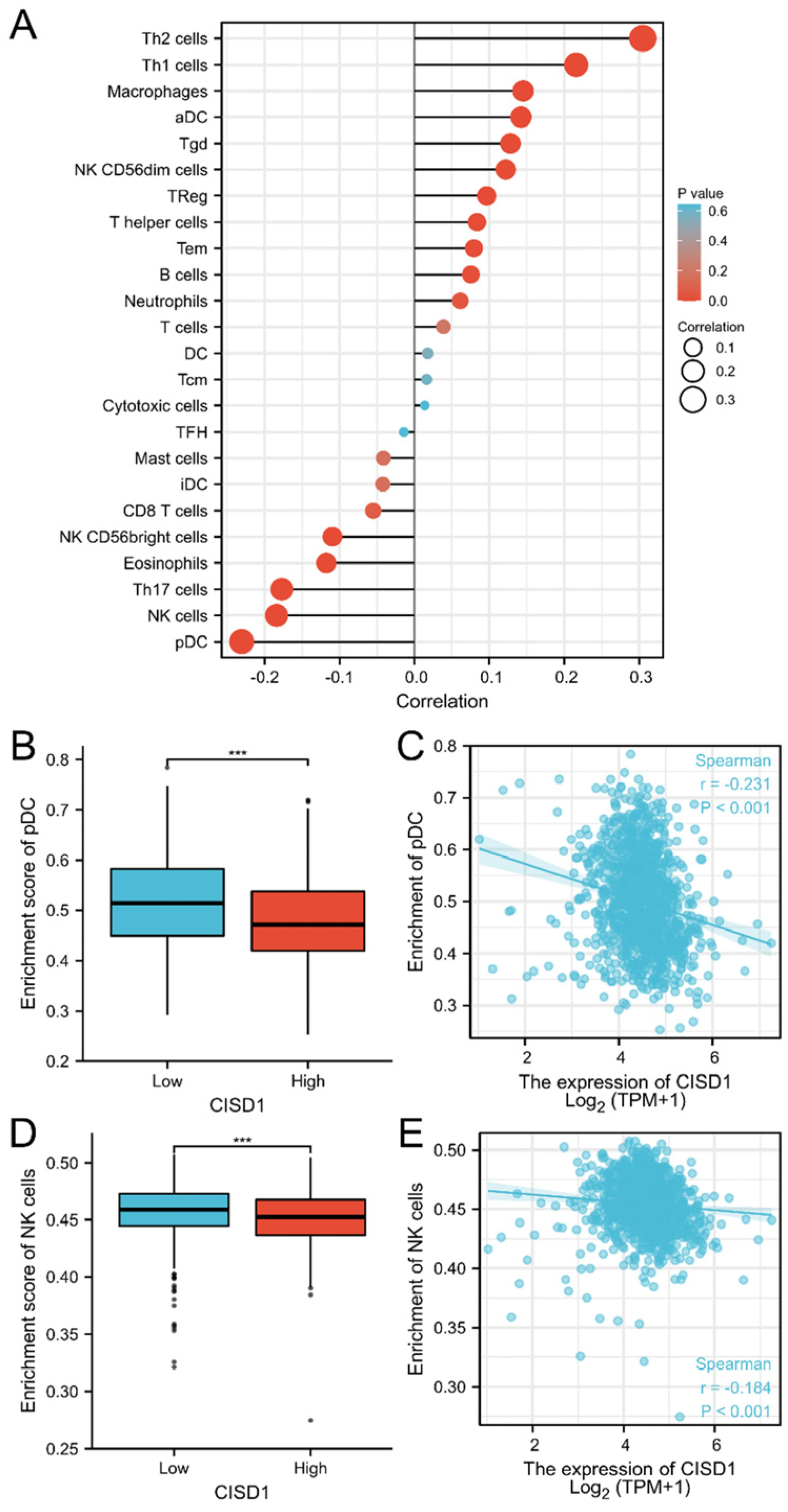

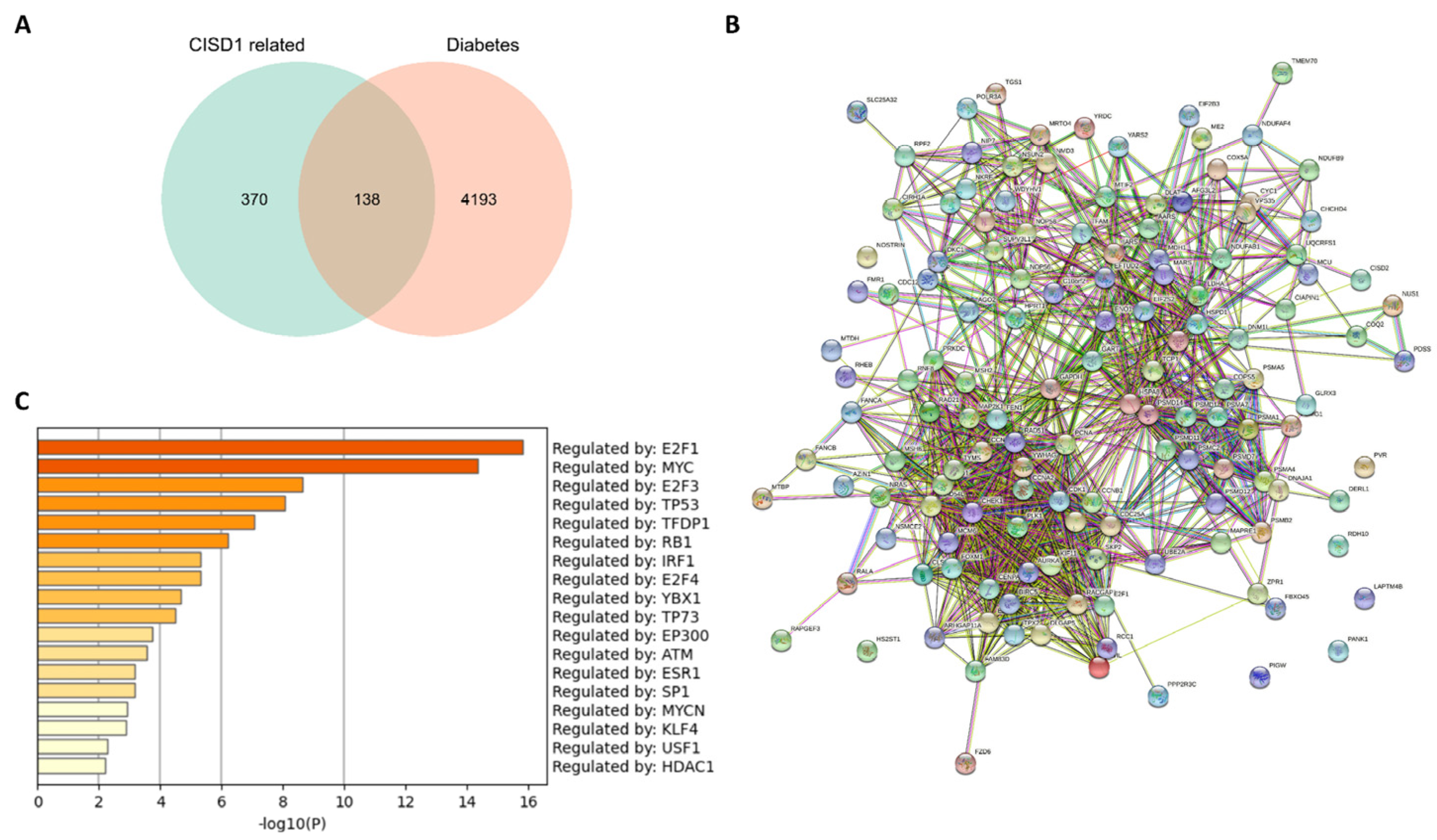

| Characteristic | Low Expression of CISD1 | High Expression of CISD1 | p |
|---|---|---|---|
| n | 541 | 542 | |
| T stage, n (%) | 0.075 | ||
| T1 | 148 (13.7%) | 129 (11.9%) | |
| T2 | 296 (27.4%) | 333 (30.8%) | |
| T3 | 80 (7.4%) | 59 (5.5%) | |
| T4 | 16 (1.5%) | 19 (1.8%) | |
| N stage, n (%) | 0.012 | ||
| N0 | 266 (25%) | 248 (23.3%) | |
| N1 | 175 (16.4%) | 183 (17.2%) | |
| N2 | 45 (4.2%) | 71 (6.7%) | |
| N3 | 47 (4.4%) | 29 (2.7%) | |
| M stage, n (%) | 0.047 | ||
| M0 | 451 (48.9%) | 451 (48.9%) | |
| M1 | 5 (0.5%) | 15 (1.6%) | |
| Pathologic stage, n (%) | 0.099 | ||
| Stage I | 101 (9.5%) | 80 (7.5%) | |
| Stage II | 304 (28.7%) | 315 (29.7%) | |
| Stage III | 124 (11.7%) | 118 (11.1%) | |
| Stage IV | 5 (0.5%) | 13 (1.2%) | |
| Race, n (%) | <0.001 | ||
| Asian | 23 (2.3%) | 37 (3.7%) | |
| Black or African American | 70 (7%) | 111 (11.2%) | |
| White | 405 (40.7%) | 348 (35%) | |
| Age, n (%) | 0.022 | ||
| ≤60 | 281 (25.9%) | 320 (29.5%) | |
| >60 | 260 (24%) | 222 (20.5%) | |
| Histological type, n (%) | <0.001 | ||
| Infiltrating Ductal Carcinoma | 346 (35.4%) | 426 (43.6%) | |
| Infiltrating Lobular Carcinoma | 139 (14.2%) | 66 (6.8%) | |
| PR status, n (%) | <0.001 | ||
| Negative | 145 (14%) | 197 (19.1%) | |
| Indeterminate | 3 (0.3%) | 1 (0.1%) | |
| Positive | 371 (35.9%) | 317 (30.7%) | |
| ER status, n (%) | <0.001 | ||
| Negative | 90 (8.7%) | 150 (14.5%) | |
| Indeterminate | 0 (0%) | 2 (0.2%) | |
| Positive | 430 (41.5%) | 363 (35.1%) | |
| HER2 status, n (%) | 0.647 | ||
| Negative | 290 (39.9%) | 268 (36.9%) | |
| Indeterminate | 6 (0.8%) | 6 (0.8%) | |
| Positive | 75 (10.3%) | 82 (11.3%) | |
| PAM50, n (%) | <0.001 | ||
| Normal | 23 (2.1%) | 17 (1.6%) | |
| LumA | 338 (31.2%) | 224 (20.7%) | |
| LumB | 84 (7.8%) | 120 (11.1%) | |
| Her2 | 33 (3%) | 49 (4.5%) | |
| Basal | 63 (5.8%) | 132 (12.2%) | |
| Menopause status, n (%) | 0.280 | ||
| Pre | 105 (10.8%) | 124 (12.8%) | |
| Peri | 19 (2%) | 21 (2.2%) | |
| Post | 364 (37.4%) | 339 (34.9%) | |
| Age, median (IQR) | 60 (50, 68) | 57 (48, 66) | 0.013 |
| Cell Type | Correlation (Pearson) | p (Pearson) | Correlation (Spearman) | p (Spearman) |
|---|---|---|---|---|
| aDC | 0.169 | <0.001 | 0.142 | <0.001 |
| B cells | 0.079 | 0.009 | 0.075 | 0.012 |
| CD8 T cells | 0.066 | 0.027 | −0.055 | 0.067 |
| Cytotoxic cells | 0.062 | 0.039 | 0.014 | 0.646 |
| DC | 0.078 | 0.010 | 0.018 | 0.552 |
| Eosinophils | −0.088 | 0.003 | −0.118 | <0.001 |
| iDC | 0.023 | 0.445 | −0.042 | 0.161 |
| Macrophages | 0.175 | <0.001 | 0.145 | <0.001 |
| Mast cells | −0.003 | 0.910 | −0.041 | 0.168 |
| Neutrophils | 0.118 | <0.001 | 0.061 | 0.041 |
| NK CD56bright cells | −0.065 | 0.031 | −0.109 | <0.001 |
| NK CD56dim cells | 0.154 | <0.001 | 0.122 | <0.001 |
| NK cells | −0.087 | 0.004 | −0.184 | <0.001 |
| pDC | −0.201 | <0.001 | −0.231 | <0.001 |
| T cells | 0.073 | 0.015 | 0.039 | 0.197 |
| T helper cells | 0.056 | 0.064 | 0.084 | 0.005 |
| Tcm | −0.041 | 0.168 | 0.016 | 0.585 |
| Tem | 0.065 | 0.030 | 0.079 | 0.008 |
| TFH | 0.037 | 0.215 | −0.014 | 0.639 |
| Tgd | 0.134 | <0.001 | 0.128 | <0.001 |
| Th1 cells | 0.240 | <0.001 | 0.216 | <0.001 |
| Th17 cells | −0.141 | <0.001 | −0.177 | <0.001 |
| Th2 cells | 0.282 | <0.001 | 0.305 | <0.001 |
| TReg | 0.111 | <0.001 | 0.097 | 0.001 |
| GO | Description | Count | % | Log10(P) | Log10(q) |
|---|---|---|---|---|---|
| M30019 | HSD17B8 TARGET GENES | 22 | 16.00 | −12.00 | −9.20 |
| M19064 | E2F Q6 | 11 | 8.00 | −7.90 | −5.10 |
| M10115 | E2F Q4 | 11 | 8.00 | −7.90 | −5.10 |
| M29943 | DLX6 TARGET GENES | 16 | 12.00 | −7.80 | −5.00 |
| M12497 | E2F Q3 | 10 | 7.20 | −7.00 | −4.40 |
| M5768 | E2F1DP1RB 01 | 10 | 7.20 | −6.90 | −4.30 |
| M9279 | E2F1 Q6 | 10 | 7.20 | −6.80 | −4.20 |
| M12402 | E2F1DP1 01 | 10 | 7.20 | −6.80 | −4.20 |
| M12555 | E2F1DP2 01 | 10 | 7.20 | −6.80 | −4.20 |
| M17867 | E2F 02 | 10 | 7.20 | −6.80 | −4.20 |
| M19298 | E2F4DP2 01 | 10 | 7.20 | −6.80 | −4.20 |
| M10526 | E2F4DP1 01 | 10 | 7.20 | −6.70 | −4.20 |
| M1485 | E2F1 Q3 | 10 | 7.20 | −6.70 | −4.10 |
| M8998 | E2F1 Q4 01 | 9 | 6.50 | −5.90 | −3.40 |
| M17117 | E2F Q3 01 | 9 | 6.50 | −5.80 | −3.40 |
| M3037 | E2F1 Q6 01 | 9 | 6.50 | −5.70 | −3.30 |
| M29995 | HES4 TARGET GENES | 13 | 9.40 | −5.40 | −3.00 |
| M40770 | ATXN7L3 TARGET GENES | 9 | 6.50 | −5.20 | −2.80 |
| M1905 | SGCGSSAAA E2F1DP2 01 | 7 | 5.10 | −4.90 | −2.60 |
| M102 | E2F Q4 01 | 8 | 5.80 | −4.80 | −2.60 |
| MCODE | GO | Description | Log10(P) |
|---|---|---|---|
| MCODE_1 | R−HSA−69613 | p53−Independent G1/S DNA damage checkpoint | −32.6 |
| MCODE_1 | R−HSA−69610 | p53−Independent DNA Damage Response | −32.6 |
| MCODE_1 | R−HSA−69601 | Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | −32.6 |
| MCODE_2 | M176 | PID FOXM1 PATHWAY | −14.5 |
| MCODE_2 | GO:0044772 | mitotic cell cycle phase transition | −12.9 |
| MCODE_2 | GO:0044770 | cell cycle phase transition | −12.7 |
| MCODE_3 | GO:0033365 | protein localization to organelle | −5.8 |
| MCODE_3 | GO:0048285 | organelle fission | −5.5 |
| MCODE_3 | GO:0000819 | sister chromatid segregation | −4.9 |
| MCODE_4 | hsa03008 | Ribosome biogenesis in eukaryotes | −9.9 |
| MCODE_4 | GO:0022613 | ribonucleoprotein complex biogenesis | −8.5 |
| MCODE_4 | R−HSA−6790901 | rRNA modification in the nucleus and cytosol | −8.5 |
| MCODE_5 | WP4016 | DNA IR−damage and cellular response via ATR | −10.5 |
| MCODE_5 | WP4946 | DNA repair pathways, full network | −7.3 |
| MCODE_5 | M258 | PID BARD1 PATHWAY | −7 |
| MCODE_6 | R−HSA−5358565 | Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) | −12 |
| MCODE_6 | R−HSA−5358508 | Mismatch Repair | −11.9 |
| MCODE_6 | CORUM:286 | PCNA−MSH2−MSH6 complex | −11.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, F.; Dong, Y.; Zhong, F.; Guo, H.; Dong, P. CISD1 Is a Breast Cancer Prognostic Biomarker Associated with Diabetes Mellitus. Biomolecules 2023, 13, 37. https://doi.org/10.3390/biom13010037
Liu F, Dong Y, Zhong F, Guo H, Dong P. CISD1 Is a Breast Cancer Prognostic Biomarker Associated with Diabetes Mellitus. Biomolecules. 2023; 13(1):37. https://doi.org/10.3390/biom13010037
Chicago/Turabian StyleLiu, Fangfang, Yifeng Dong, Fuyu Zhong, Haodan Guo, and Pengzhi Dong. 2023. "CISD1 Is a Breast Cancer Prognostic Biomarker Associated with Diabetes Mellitus" Biomolecules 13, no. 1: 37. https://doi.org/10.3390/biom13010037
APA StyleLiu, F., Dong, Y., Zhong, F., Guo, H., & Dong, P. (2023). CISD1 Is a Breast Cancer Prognostic Biomarker Associated with Diabetes Mellitus. Biomolecules, 13(1), 37. https://doi.org/10.3390/biom13010037





