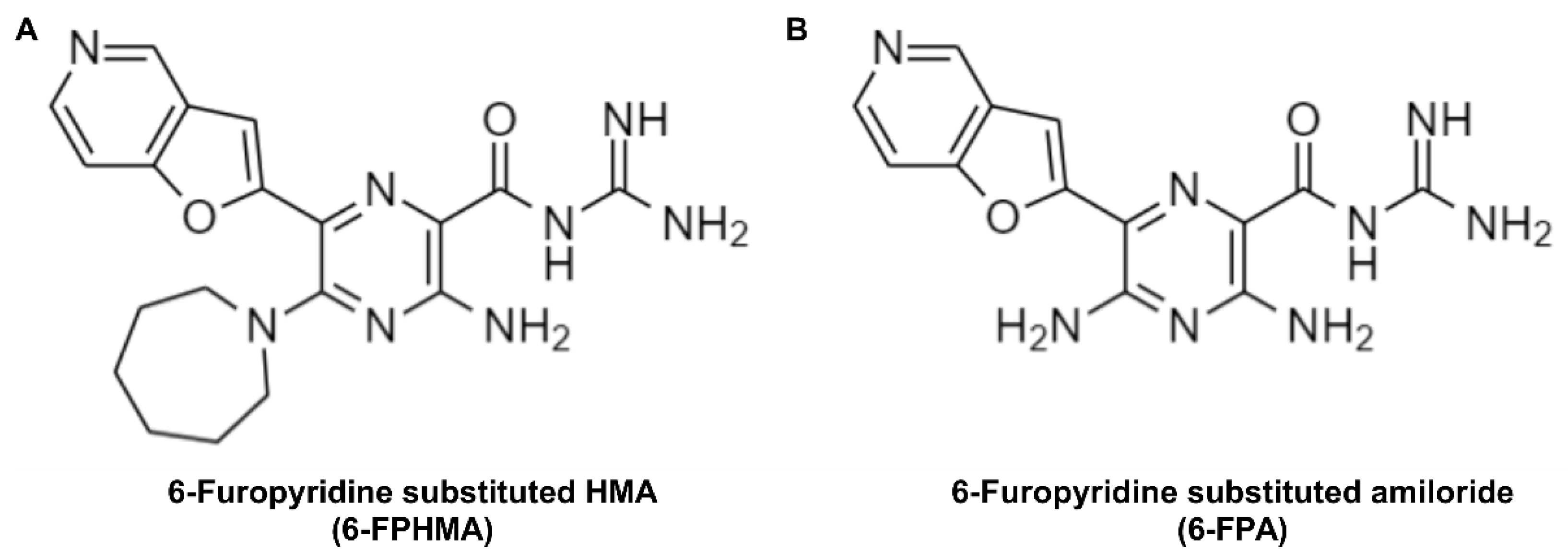
6-Furopyridine Hexamethylene Amiloride Is a Non-Selective P2X7 Receptor Antagonist
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Lines
2.3. Expression Constructs
2.4. Transfection
2.5. Plate-Based Flow Cytometric Dye Uptake for Compound Screening
2.6. Flow Cytometric Uptake Assays for Characterization
2.7. Fura-2AM Ca2+ Response Assay
2.8. Whole Blood IL-1β Release Assay
2.9. CD23 Shedding Assay
2.10. Statistical Analysis
3. Results
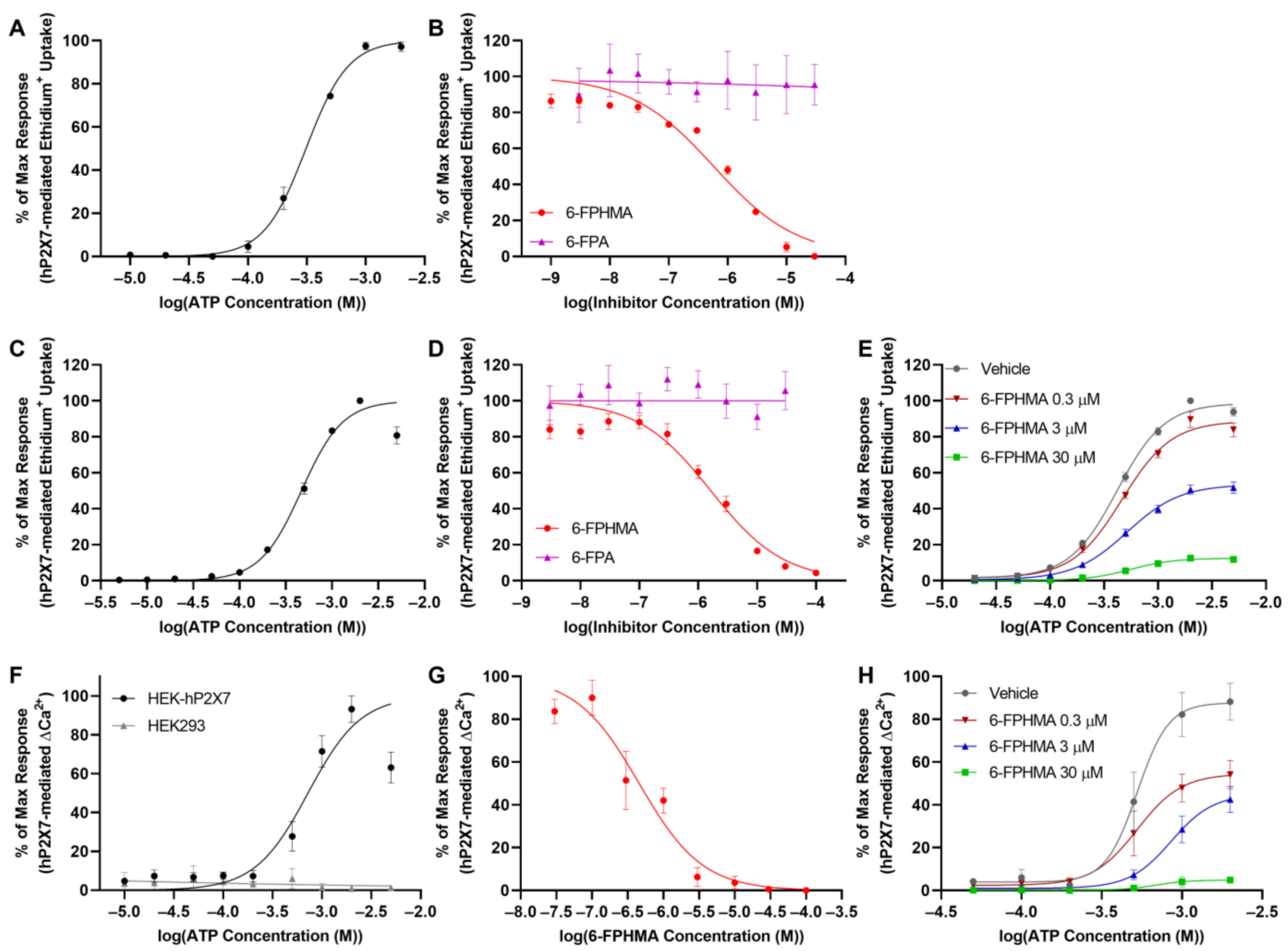
3.1. 6-FPHMA Is a Novel P2X7 Antagonist
3.2. 6-FPHMA, but Not 6-FPA, Inhibits hP2X7-Mediated Dye Uptake
3.3. 6-FPHMA Inhibits hP2X7-Mediated Ca2+ Responses
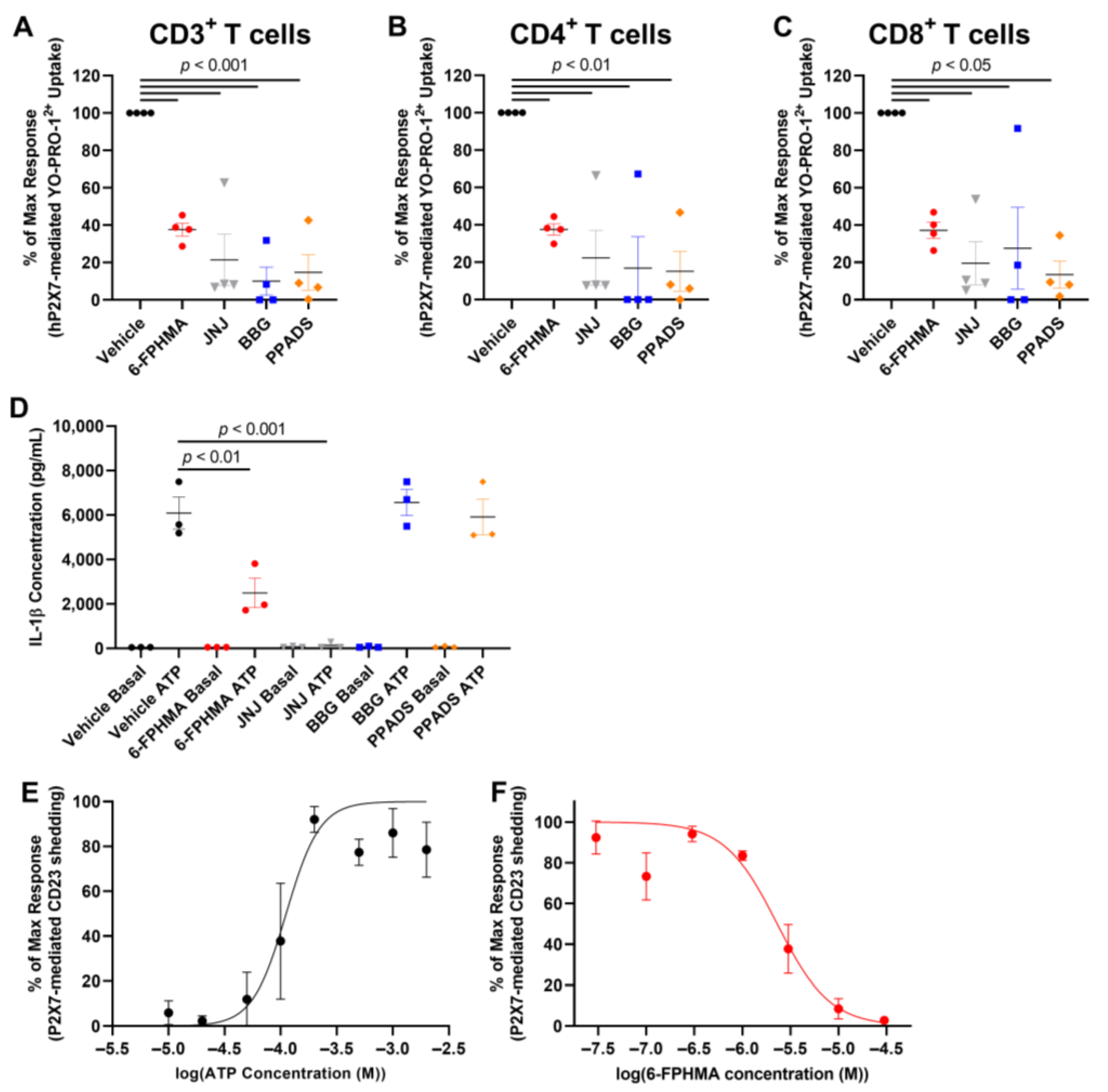
3.4. 6-FPHMA Inhibits hP2X7-Mediated Dye Uptake in Human T Cells
3.5. 6-FPHMA Inhibits hP2X7-Mediated IL-1β Release and CD23 Shedding from Human Leukocytes
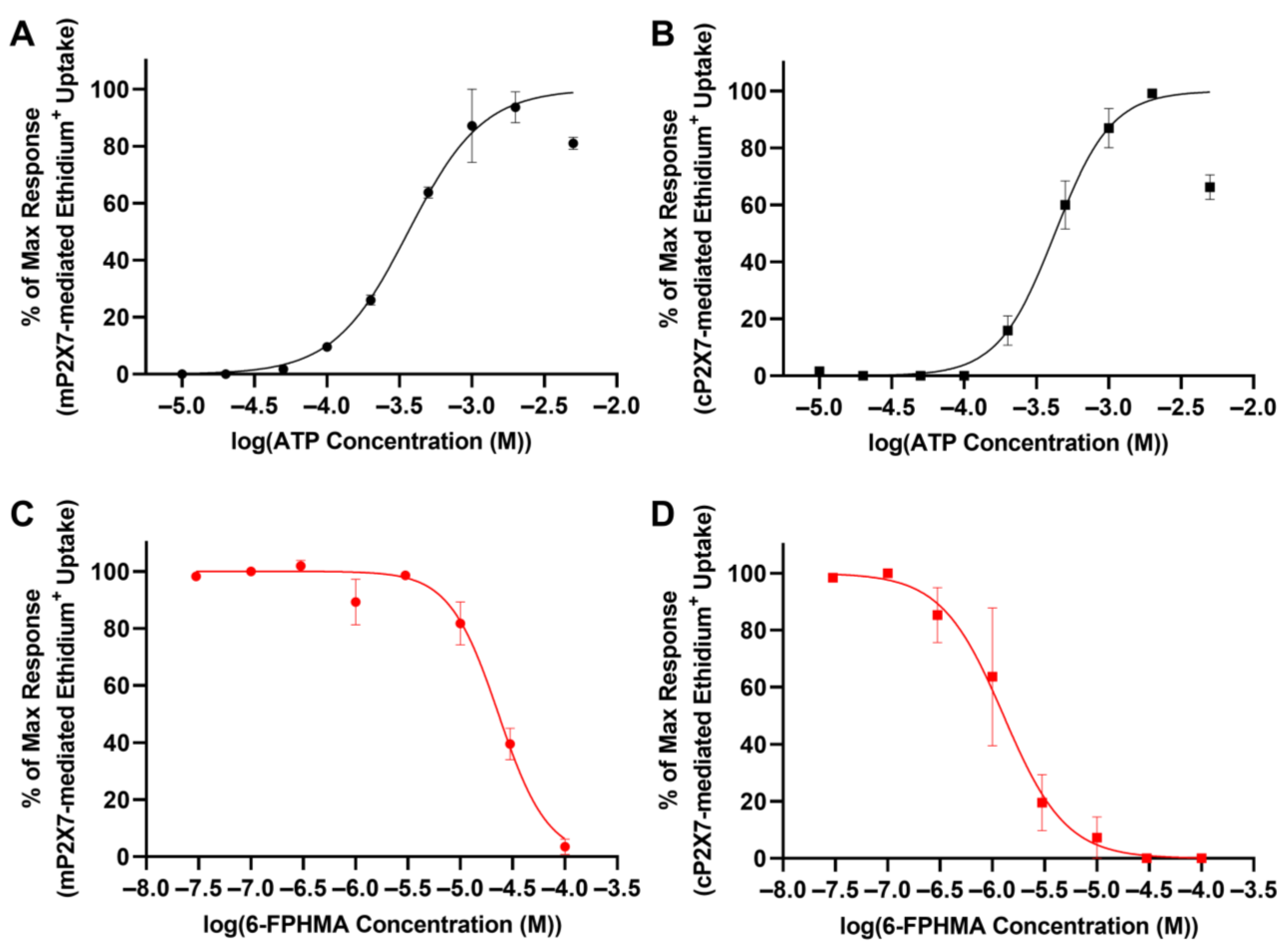
3.6. 6-FPHMA Inhibits mP2X7- and cP2X7-Mediated Dye Uptake
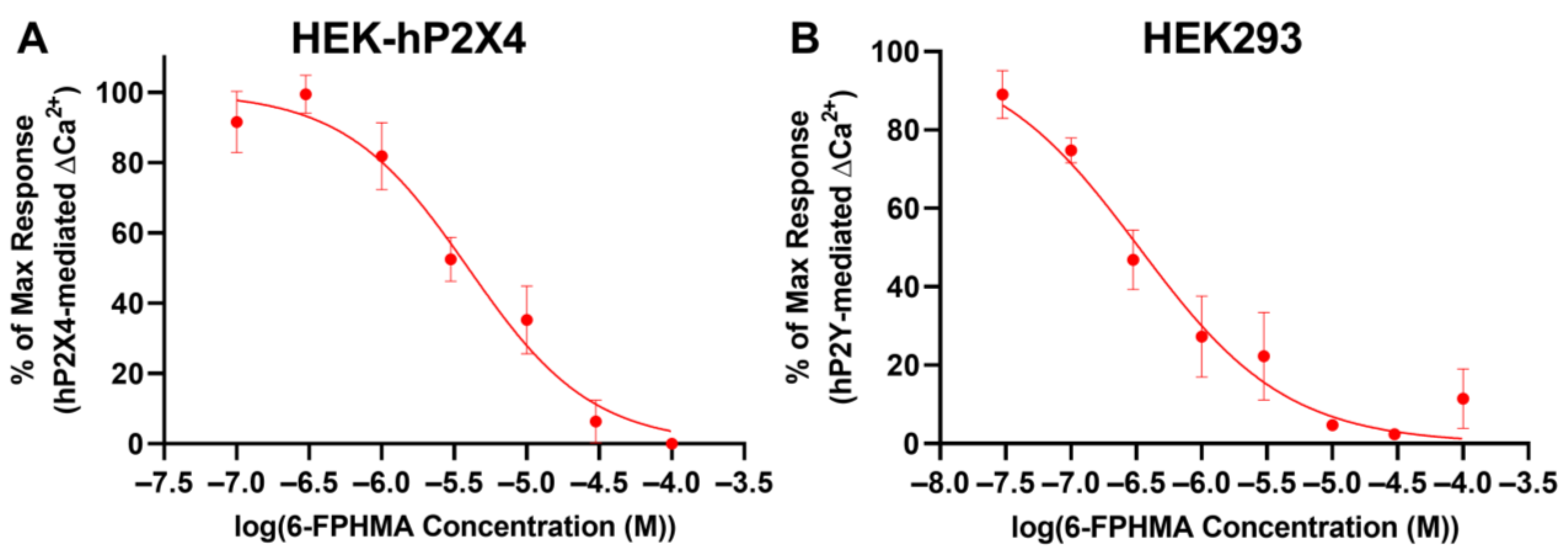
3.7. 6-FPHMA Inhibits hP2X4- and hP2Y-Mediated Ca2+ Responses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burnstock, G. Purine and purinergic receptors. Brain Neurosci. Adv. 2018, 2, 2398212818817494. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, A.L.; Sarti, A.C.; Di Virgilio, F. Extracellular nucleotides and nucleosides as signalling molecules. Immunol. Lett. 2019, 205, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Di Virgilio, F.; Dal Ben, D.; Sarti, A.C.; Giuliani, A.L.; Falzoni, S. The P2X7 Receptor in Infection and Inflammation. Immunity 2017, 47, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Sluyter, R. The P2X7 Receptor. Adv. Exp. Med. Biol. 2017, 1051, 17–53. [Google Scholar] [CrossRef]
- Giuliani, A.L.; Sarti, A.C.; Falzoni, S.; Di Virgilio, F. The P2X7 Receptor-Interleukin-1 Liaison. Front. Pharmacol. 2017, 8, 123. [Google Scholar] [CrossRef] [PubMed]
- Pupovac, A.; Sluyter, R. Roles of extracellular nucleotides and P2 receptors in ectodomain shedding. Cell Mol. Life Sci. 2016, 73, 4159–4173. [Google Scholar] [CrossRef]
- De Marchi, E.; Orioli, E.; Dal Ben, D.; Adinolfi, E. P2X7 Receptor as a Therapeutic Target. Adv. Protein Chem. Struct. Biol. 2016, 104, 39–79. [Google Scholar] [CrossRef]
- Sun, Q.; Sever, P. Amiloride: A review. J. Renin Angiotensin Aldosterone Syst. 2020, 21, 1470320320975893. [Google Scholar] [CrossRef]
- Kleyman, T.R.; Cragoe, E.J., Jr. Amiloride and its analogs as tools in the study of ion transport. J. Membr. Biol. 1988, 105, 1–21. [Google Scholar] [CrossRef]
- Wiley, J.S.; Jamieson, G.P.; Mayger, W.; Cragoe, E.J., Jr.; Jopson, M. Extracellular ATP stimulates an amiloride-sensitive sodium influx in human lymphocytes. Arch. Biochem. Biophys. 1990, 280, 263–268. [Google Scholar] [CrossRef]
- Wiley, J.S.; Gargett, C.E.; Zhang, W.; Snook, M.B.; Jamieson, G.P. Partial agonists and antagonists reveal a second permeability state of human lymphocyte P2Z/P2X7 channel. Am. J. Physiol. 1998, 275, C1224–C1231. [Google Scholar] [CrossRef] [PubMed]
- Wiley, J.S.; Chen, R.; Jamieson, G.P. The ATP4- receptor-operated channel (P2Z class) of human lymphocytes allows Ba2+ and ethidium+ uptake: Inhibition of fluxes by suramin. Arch. Biochem. Biophys. 1993, 305, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Chessell, I.P.; Michel, A.D.; Humphrey, P.P. Effects of antagonists at the human recombinant P2X7 receptor. Br. J. Pharmacol. 1998, 124, 1314–1320. [Google Scholar] [CrossRef]
- Nuttle, L.C.; Dubyak, G.R. Differential activation of cation channels and non-selective pores by macrophage P2z purinergic receptors expressed in Xenopus oocytes. J. Biol. Chem. 1994, 269, 13988–13996. [Google Scholar] [CrossRef]
- Hibell, A.D.; Thompson, K.M.; Xing, M.; Humphrey, P.P.; Michel, A.D. Complexities of measuring antagonist potency at P2X7 receptor orthologs. J. Pharmacol. Exp. Ther. 2001, 296, 947–957. [Google Scholar] [PubMed]
- Buckley, B.J.; Aboelela, A.; Minaei, E.; Jiang, L.X.; Xu, Z.; Ali, U.; Fildes, K.; Cheung, C.; Cook, S.M.; Johnson, D.C.; et al. 6-Substituted Hexamethylene Amiloride (HMA) Derivatives as Potent and Selective Inhibitors of the Human Urokinase Plasminogen Activator for Use in Cancer. J. Med. Chem. 2018, 61, 8299–8320. [Google Scholar] [CrossRef]
- Buckley, B.J.; Kumar, A.; Aboelela, A.; Bujaroski, R.S.; Li, X.; Majed, H.; Fliegel, L.; Ranson, M.; Kelso, M.J. Screening of 5- and 6-Substituted Amiloride Libraries Identifies Dual-uPA/NHE1 Active and Single Target-Selective Inhibitors. Int. J. Mol. Sci. 2021, 22, 2999. [Google Scholar] [CrossRef]
- Buckley, B.J.; Majed, H.; Aboelela, A.; Minaei, E.; Jiang, L.; Fildes, K.; Cheung, C.; Johnson, D.; Bachovchin, D.; Cook, G.M.; et al. 6-Substituted amiloride derivatives as inhibitors of the urokinase-type plasminogen activator for use in metastatic disease. Bioorg. Med. Chem. Lett. 2019, 29, 126753. [Google Scholar] [CrossRef]
- Bhaskaracharya, A.; Dao-Ung, P.; Jalilian, I.; Spildrejorde, M.; Skarratt, K.K.; Fuller, S.J.; Sluyter, R.; Stokes, L. Probenecid blocks human P2X7 receptor-induced dye uptake via a pannexin-1 independent mechanism. PLoS ONE 2014, 9, e93058. [Google Scholar] [CrossRef]
- Spildrejorde, M.; Bartlett, R.; Stokes, L.; Jalilian, I.; Peranec, M.; Sluyter, V.; Curtis, B.L.; Skarratt, K.K.; Skora, A.; Bakhsh, T.; et al. R270C polymorphism leads to loss of function of the canine P2X7 receptor. Physiol. Genom. 2014, 46, 512–522. [Google Scholar] [CrossRef]
- Sophocleous, R.A.; Berg, T.; Finol-Urdaneta, R.K.; Sluyter, V.; Keshiya, S.; Bell, L.; Curtis, S.J.; Curtis, B.L.; Seavers, A.; Bartlett, R.; et al. Pharmacological and genetic characterisation of the canine P2X4 receptor. Br. J. Pharmacol. 2020, 177, 2812–2829. [Google Scholar] [CrossRef] [PubMed]
- Ly, D.; Dongol, A.; Cuthbertson, P.; Guy, T.V.; Geraghty, N.J.; Sophocleous, R.A.; Sin, L.; Turner, B.J.; Watson, D.; Yerbury, J.J.; et al. The P2X7 receptor antagonist JNJ-47965567 administered thrice weekly from disease onset does not alter progression of amyotrophic lateral sclerosis in SOD1G93A mice. Purinergic Signal. 2020, 16, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Cuthbertson, P.; Geraghty, N.J.; Adhikary, S.R.; Casolin, S.; Watson, D.; Sluyter, R. P2X7 receptor antagonism increases regulatory T cells and reduces clinical and histological graft-versus-host disease in a humanised mouse model. Clin. Sci. 2021, 135, 495–513. [Google Scholar] [CrossRef]
- Turner, R.J.; Geraghty, N.J.; Williams, J.G.; Ly, D.; Brungs, D.; Carolan, M.G.; Guy, T.V.; Watson, D.; de Leon, J.F.; Sluyter, R. Comparison of peripheral blood mononuclear cell isolation techniques and the impact of cryopreservation on human lymphocytes expressing CD39 and CD73. Purinergic Signal. 2020, 16, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Paredes, R.M.; Etzler, J.C.; Watts, L.T.; Zheng, W.; Lechleiter, J.D. Chemical calcium indicators. Methods 2008, 46, 143–151. [Google Scholar] [CrossRef]
- Spildrejorde, M.; Curtis, S.J.; Curtis, B.L.; Sluyter, R. Extracellular adenosine 5’-triphosphate and lipopolysaccharide induce interleukin-1β release in canine blood. Vet Immunol. Immunopathol. 2014, 157, 105–110. [Google Scholar] [CrossRef]
- Farrell, A.W.; Gadeock, S.; Pupovac, A.; Wang, B.; Jalilian, I.; Ranson, M.; Sluyter, R. P2X7 receptor activation induces cell death and CD23 shedding in human RPMI 8226 multiple myeloma cells. Biochim Biophys Acta. 2010, 1800, 1173–1182. [Google Scholar] [CrossRef]
- Agrawal, A.; Kruse, L.S.; Vangsted, A.J.; Gartland, A.; Jørgensen, N.R. Human P2X7 Receptor Causes Cycle Arrest in RPMI-8226 Myeloma Cells to Alter the Interaction with Osteoblasts and Osteoclasts. Cells 2020, 9, 2341. [Google Scholar] [CrossRef]
- Gilbert, S.M.; Oliphant, C.J.; Hassan, S.; Peille, A.L.; Bronsert, P.; Falzoni, S.; di Virgilio, F.; McNulty, S.; Lara, R. ATP in the tumour microenvironment drives expression of nfP2X7, a key mediator of cancer cell survival. Oncogene 2019, 38, 194–208. [Google Scholar] [CrossRef]
- Elhage, A.; Turner, R.J.; Cuthbertson, P.; Watson, D.; Sluyter, R. Preparation of the Murine Anti-Human P2X7 Receptor Monoclonal Antibody (Clone L4). Methods Mol. Biol. 2022, 2510, 77–98. [Google Scholar]
- Fischer, W.; Franke, H.; Gröger-Arndt, H.; Illes, P. Evidence for the existence of P2Y1,2,4 receptor subtypes in HEK-293 cells: Reactivation of P2Y1 receptors after repetitive agonist application. Naunyn Schmiedebergs Arch. Pharmacol. 2005, 371, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Adhikary, S.R.; Geraghty, N.J.; Cuthbertson, P.; Sluyter, R.; Watson, D. Altered donor P2X7 activity in human leukocytes correlates with P2RX7 genotype but does not affect the development of graft-versus-host disease in humanised mice. Purinergic Signal. 2019, 15, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Dubyak, G.R. P2X7 receptor regulation of non-classical secretion from immune effector cells. Cell Microbiol. 2012, 14, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- Pupovac, A.; Stokes, L.; Sluyter, R. CAY10593 inhibits the human P2X7 receptor independently of phospholipase D1 stimulation. Purinergic Signal. 2013, 9, 609–619. [Google Scholar] [CrossRef][Green Version]
- Donnelly-Roberts, D.L.; Namovic, M.T.; Han, P.; Jarvis, M.F. Mammalian P2X7 receptor pharmacology: Comparison of recombinant mouse, rat and human P2X7 receptors. Br. J. Pharmacol. 2009, 157, 1203–1214. [Google Scholar] [CrossRef]
- Roman, S.; Cusdin, F.S.; Fonfria, E.; Goodwin, J.A.; Reeves, J.; Lappin, S.C.; Chambers, L.; Walter, D.S.; Clay, W.C.; Michel, A.D. Cloning and pharmacological characterization of the dog P2X7 receptor. Br. J. Pharmacol. 2009, 158, 1513–1526. [Google Scholar] [CrossRef]
- Stevenson, R.O.; Taylor, R.M.; Wiley, J.S.; Sluyter, R. The P2X7 receptor mediates the uptake of organic cations in canine erythrocytes and mononuclear leukocytes: Comparison to equivalent human cell types. Purinergic Signal. 2009, 5, 385–394. [Google Scholar] [CrossRef]
- Bartlett, R.; Stokes, L.; Curtis, S.J.; Curtis, B.L.; Sluyter, R. Probenecid directly impairs activation of the canine P2X7 receptor. Nucleosides Nucleotides Nucleic Acids 2017, 36, 736–744. [Google Scholar] [CrossRef]
- Karasawa, A.; Kawate, T. Structural basis for subtype-specific inhibition of the P2X7 receptor. Elife 2016, 5, e22153. [Google Scholar] [CrossRef]
- Allsopp, R.C.; Dayl, S.; Bin Dayel, A.; Schmid, R.; Evans, R.J. Mapping the Allosteric Action of Antagonists A740003 and A438079 Reveals a Role for the Left Flipper in Ligand Sensitivity at P2X7 Receptors. Mol. Pharmacol. 2018, 93, 553–562. [Google Scholar] [CrossRef]
- Bin Dayel, A.; Evans, R.J.; Schmid, R. Mapping the Site of Action of Human P2X7 Receptor Antagonists AZ11645373, Brilliant Blue G, KN-62, Calmidazolium, and ZINC58368839 to the Intersubunit Allosteric Pocket. Mol. Pharmacol. 2019, 96, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Ase, A.R.; Therrien, É.; Séguéla, P. An Allosteric Inhibitory Site Conserved in the Ectodomain of P2X Receptor Channels. Front. Cell Neurosci. 2019, 13, 121. [Google Scholar] [CrossRef] [PubMed]
- Schwiering, M.; Husmann, M.; Hellmann, N. P2X-Receptor Antagonists Inhibit the Interaction of S. aureus Hemolysin A with Membranes. Toxins 2017, 9, 332. [Google Scholar] [CrossRef] [PubMed]
- Bragança, B.; Correia-de-Sá, P. Resolving the Ionotropic P2X4 Receptor Mystery Points Towards a New Therapeutic Target for Cardiovascular Diseases. Int. J. Mol. Sci. 2020, 21, 5005. [Google Scholar] [CrossRef] [PubMed]
- Shokoples, B.G.; Paradis, P.; Schiffrin, E.L. P2X7 Receptors: An Untapped Target for the Management of Cardiovascular Disease. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Sophocleous, R.A.; Ooi, L.; Sluyter, R. The P2X4 Receptor: Cellular and Molecular Characteristics of a Promising Neuroinflammatory Target. Int. J. Mol. Sci. 2022, 23, 5739. [Google Scholar] [CrossRef]
- Calzaferri, F.; Ruiz-Ruiz, C.; de Diego, A.M.G.; de Pascual, R.; Méndez-López, I.; Cano-Abad, M.F.; Maneu, V.; de los Ríos, C.; Gandía, L.; García, A.G. The purinergic P2X7 receptor as a potential drug target to combat neuroinflammation in neurodegenerative diseases. Med. Res. Rev. 2020, 40, 2427–2465. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuthbertson, P.; Elhage, A.; Al-Rifai, D.; Sophocleous, R.A.; Turner, R.J.; Aboelela, A.; Majed, H.; Bujaroski, R.S.; Jalilian, I.; Kelso, M.J.; et al. 6-Furopyridine Hexamethylene Amiloride Is a Non-Selective P2X7 Receptor Antagonist. Biomolecules 2022, 12, 1309. https://doi.org/10.3390/biom12091309
Cuthbertson P, Elhage A, Al-Rifai D, Sophocleous RA, Turner RJ, Aboelela A, Majed H, Bujaroski RS, Jalilian I, Kelso MJ, et al. 6-Furopyridine Hexamethylene Amiloride Is a Non-Selective P2X7 Receptor Antagonist. Biomolecules. 2022; 12(9):1309. https://doi.org/10.3390/biom12091309
Chicago/Turabian StyleCuthbertson, Peter, Amal Elhage, Dena Al-Rifai, Reece A. Sophocleous, Ross J. Turner, Ashraf Aboelela, Hiwa Majed, Richard S. Bujaroski, Iman Jalilian, Michael J. Kelso, and et al. 2022. "6-Furopyridine Hexamethylene Amiloride Is a Non-Selective P2X7 Receptor Antagonist" Biomolecules 12, no. 9: 1309. https://doi.org/10.3390/biom12091309
APA StyleCuthbertson, P., Elhage, A., Al-Rifai, D., Sophocleous, R. A., Turner, R. J., Aboelela, A., Majed, H., Bujaroski, R. S., Jalilian, I., Kelso, M. J., Watson, D., Buckley, B. J., & Sluyter, R. (2022). 6-Furopyridine Hexamethylene Amiloride Is a Non-Selective P2X7 Receptor Antagonist. Biomolecules, 12(9), 1309. https://doi.org/10.3390/biom12091309





